Introduction
Colon cancer is the third most common type of
cancer, and the leading cause of cancer-related mortality worldwide
(1). There is increasing evidence to
suggest that the progression of colon cancer is a multiple-step
process, involving genetic and epigenetic abnormalities (2). By comparison, epigenetic alterations
are more frequently observed, and carry greater effects compared
with gene mutations. For example, the silencing micro RNA-137 can
target and decrease the expression level of >500 protein coding
genes (3).
Long non-coding RNAs (lncRNAs) are a type of
non-coding transcript of >200 nucleotides in length. lncRNAs
have been associated with a number of physiological and
pathological processes in various human diseases, regulating gene
expression via epigenetic, transcriptional and post-transcriptional
modification (4). Recent studies
revealed that lncRNA was an important regulator in colon cancer
pathogenesis, modulating numerous processes including cell
proliferation, differentiation, tumor migration, invasion and
angiogenesis. It has been reported that lncRNA SUMO1P3 stimulated
the proliferation, metastasis and angiogenesis of colon cancer
cells, suggesting it's potential as a novel prognostic indicator
and therapeutic target for the disease (5).
lncRNAs can target different steps in the
transcription process, regulating the activation or regression of
gene transcription by modulating different transcriptional
components, including RNA polymerase (RNAP) II and the DNA duplex
(6). These lncRNAs may be part of
the regulatory network that, together with transcription factors,
regulate the level of gene expression. Reportedly, certain lncRNAs
acted as regulators of general transcription factors which were
required for the activity of RNAP II (7). These transcription factors, in addition
to components of the initiation complex, assemble at promoter
regions and participate in transcription elongation and the
regulation of cis elements (8); it was reported that an lncRNA
transcribed from the dihydrofolate reductase (DHFR) gene was able
to form a RNA-DNA triplex in the promoter region of DHFR,
inhibiting its transcription by blocking the binding of
transcription initiation factor IIB (9). Furthermore, the gene coding lncRNA
ANRIL is able to run antisense, silencing P15 with which it
overlaps.
However, survival-related lncRNAs, particularly in
colon cancer remained to be identified. In order to select
therapeutically relevant lncRNAs, a linear regression model was
used to identify survival-related lncRNAs in colon cancer and
further detect MIR210HG-related genes. Gene regulatory network and
pathway analyses revealed that MIR210HG may exert its effects in
colon cancer through the regulation of cancer cell metabolism and
adhesion. Furthermore, bioinformatics predictions indicated that
MIR210HG may affect colon cancer by regulating transcription and
post-transcriptional processing.
Materials and methods
Screening of survival-related lncRNAs
in colon cancer
The RNA-seq data of colon cancer-associated lncRNAs,
which consisted of 471 tumor samples and 41 normal samples, were
downloaded from The Cancer Genome Atlas (TCGA) data portal
(https://cancergenome.nih.gov/) (10). The data were divided into high and
low expression groups according to a cut-off of 75 and 25%,
respectively. Homo_sapiens.GRCh38.84 was used for annotation. The
RNA-seq data of other gastrointestinal cancers, including
esophageal cancer (162 tumor and 11 normal samples), rectal cancer
(167 tumor and 10 normal samples) and stomach cancer (375 tumor and
32 normal samples), were also downloaded, and assigned into high
and low expression groups using the same classification criteria as
in colon cancer. The association between lncRNA expression and
patient survival was determined using the Kaplan-Meier method, and
evaluated using the log-rank test. The R package survival
(https://github.com/therneau/survival)
was used to perform these statistical analyses. P<0.05 was set
as the significance threshold.
Genes significantly associated with
MIR210HG expression in colon cancer
Based on the above screening, the most significant
survival-related lncRNA in colon cancer was identified as MIR210HG;
this lncRNA was therefore selected for use in further analysis.
Associations between the expression of MIR210HG and other genes
were calculated using a linear regression model. It was
hypothesized that genes whose expression levels were linearly
associated with MIR201HG were likely to be located nearby, upstream
or downstream. This method was widely used to screen target-related
genes. RNA expression was processed using log2(counts
+1) prior to analysis. Subsequently, multiple selection criteria
(P-value and adj.r2) were applied to determine the most
appropriate candidates. P<0.05 and adj.r2 >0.16
were set as the significance threshold. R was used to perform these
statistical analyses.
Functional and pathway enrichment
analysis
Gene ontology (GO) enrichment analysis (http://www.geneontology.org/) was used to detect
biological processes, molecular functions and cellular components.
Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis (http://www.kegg.jp/), which is used for pathway
investigations, was performed using the Database for Annotation,
Visualization, and Integrated Discovery (DAVID; https://david.ncifcrf.gov/, version 6.8) (11–13).
P<0.05 was set as the significance threshold.
Investigation of the correlation
between MIRE210HG expression and lymphatic metastasis
All clinical data were obtained from TCGA. A
normality test, the Kolmogorov-Smirnov test, was performed and a
Mann Whitney U test was subsequently used to assess the association
between MIRE210HG expression and metastasis in colon cancer.
P<0.05 was considered to indicate a statistically significant
difference. SPSS Statistics 22.0 (IBM Corp.) was used to perform
statistical analysis.
Protein-protein interaction (PPI)
network construction and identification of hub genes
All MIR210HG-related genes were imported into the
Search Tool for the Retrieval of Interacting Genes/Proteins
(STRING) to construct a PPI network (14). A combined score ≥0.4 was set as the
significance threshold. The network was visualized using Cytoscape
(version 3.5.1, http://www.cytoscape.org/) (15). The top 10 hub genes were filtered
using cytoHubba, an app in Cytoscape, using the Maximal Clique
Centrality (MCC) method (16).
Module analysis of the PPI
network
To investigate the most significant modules,
Clustering with Overlapping Neighborhood Expansion (ClusterONE), an
app in Cytoscape, was used to screen modules within the PPI network
with a minimum size of 6, and the minimum density set to auto
(17). P<0.05 was considered to
indicate a statistically significant difference. GO and KEGG
analyses of genes in the screened modules were performed using
DAVID.
Transcription and post-transcriptional
modulation prediction
To further investigate the mechanism of MIR210HG in
colon cancer, known MIR210HG-protein interactions were summarized
using starBase v2.0 (http://starbase.sysu.edu.cn/index.php) (18). Novel lncRNA-protein interactions
between MIR210HG and its associated transcription factors were also
predicted using RPIseq (http://pridb.gdcb.iastate.edu/RPISeq/index.html)
(19). Candidates with a Random
Forest classifier value >0.5 and Support Vector Machine
classifier value >0.5 were considered to be positive
results.
Interaction probabilities generated using RPISeq
range between 0 and 1. In performance evaluation experiments,
predictions with probabilities >0.5 were considered ‘positive’,
indicating that the corresponding RNA and protein were likely to
interact. Neighboring mRNAs located within 1 Mbps of the MIR210HG
loci were investigated to predict possible cis target genes
of MIR210HG. Candidate gene information was downloaded from USCS
(GRCh38) (20).
Results
MIR210HG expression correlates with
survival in patients with colon cancer
lncRNAs have been reported to contribute to patient
survival in multiple cancers, including colon cancer (20), breast cancer (21) and larynx squamous cell carcinoma
(22), and therefore, in the present
study, survival-related lncRNAs in colon cancer were investigated.
RNA-seq and clinical data from colon cancer cohorts (obtained from
TCGA) were analyzed to assess the association between various
lncRNAs and patient survival. A total of 226 survival-related
lncRNAs were selected using P<0.05 as the cut-off threshold (The
top 10 survival-related lncRNAs are presented in Table I). A number of lncRNAs were reported
to contribute to tumor promotion, including MIR210HG
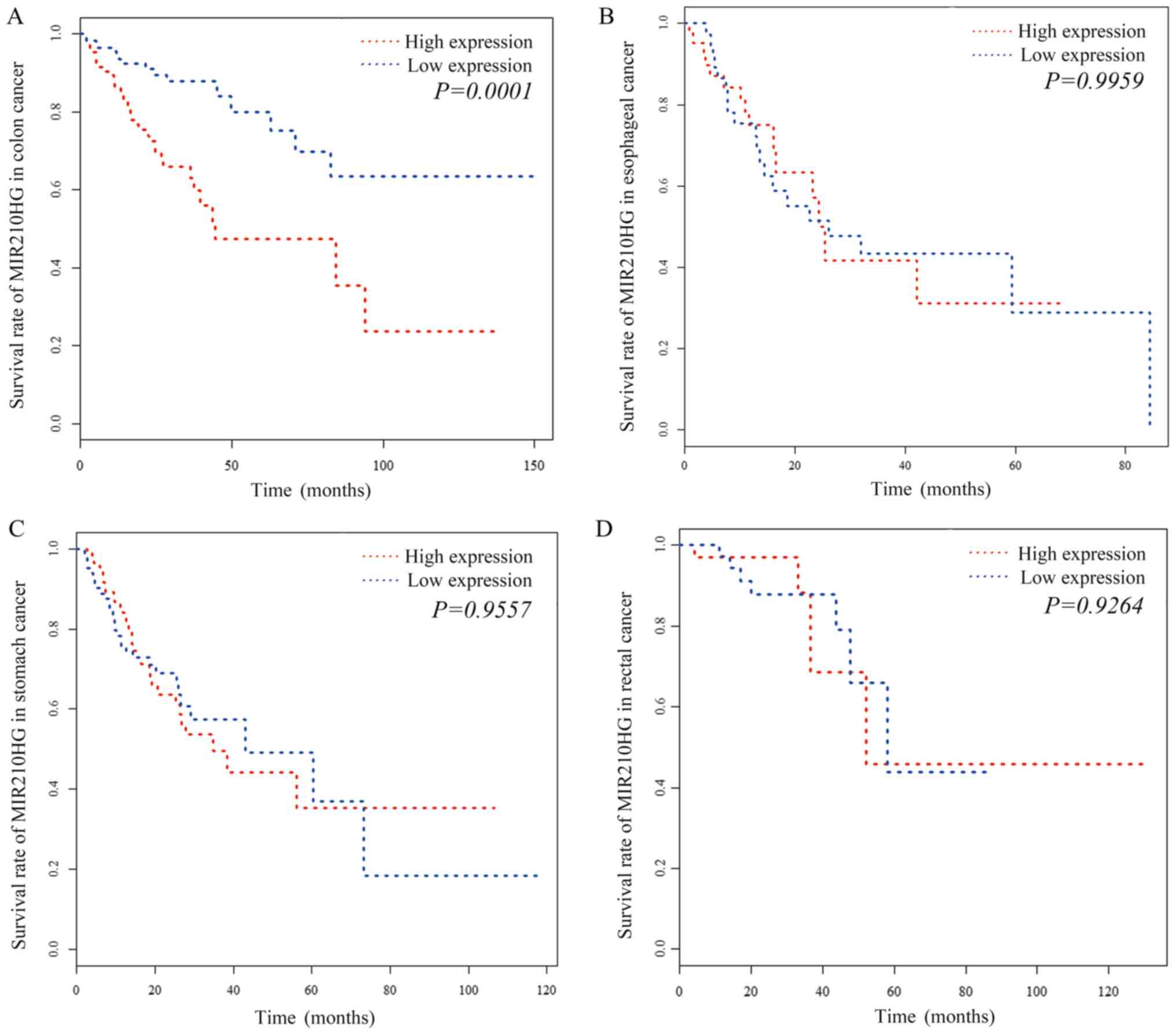
(P=1.19×10−3; log-rank test), which is a diagnostic
biomarker in glioma (23). A high
expression level of MIR210HG was significantly associated with
shorter overall survival in patients with colon cancer, suggesting
a potential prognostic role for MIR210HG in the disease (Fig. 1A). However, MIR210HG showed a weak
association with survival from esophageal, rectal and stomach
cancers, indicating that this particular effect of MIR210HG
overexpression is specific to colon cancer (Fig. 1).
 | Table I.Top 10 survival-related lncRNAs in
colon cancer. |
Table I.
Top 10 survival-related lncRNAs in
colon cancer.
| lncRNA | Expression level
[log2 (counts +1)] | P-value |
|---|
| RP11-66B24.2 | 4.35 (1.58,
8.63) |
1.08×10−4 |
| MIR210HG | 208.82 (111.73,
352.98) |
1.19×10−4 |
| RP11-367H1.1 | 5.29 (2.42,
9.54) |
1.20×10−4 |
| CTC-573N18.1 | 0.68 (0.08,
2.9) |
1.30×10−4 |
| LA16c-380A1.1 | 1.36 (0.14,
3.41) |
1.31×10−4 |
| RP11-108K3.1 | 7.79 (4.3,
13.05) |
1.35×10−4 |
| RP11-93I21.3 | 0.52 (0.08,
2.15) |
1.49×10−4 |
| LINC00174 | 179.71 (122.2,
269.39) |
2.11×10−4 |
| RP11-108K3.2 | 5.38 (1.97,
9.91) |
2.96×10−4 |
| AP006621.5 | 68.84 (44.66,
95.88) |
3.80×10−4 |
Investigation of MIR210HG-related
genes in colon cancer
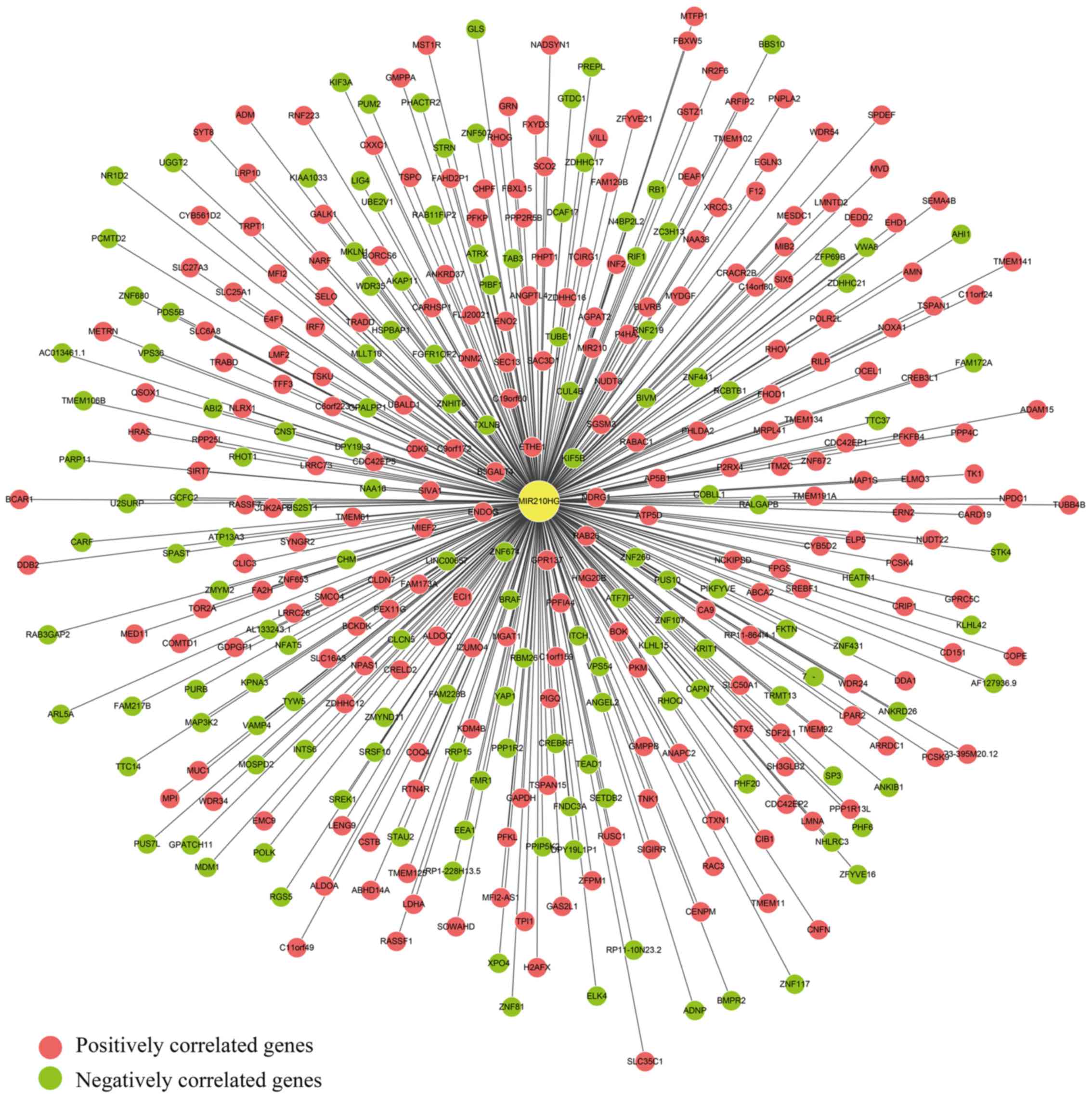
TCGA RNA-seq expression data for MIR210HG was
downloaded, and the correlation between MIR210HG expression and
other genes in colon cancer was calculated using a linear
regression model. A total of 373 genes with P<0.05 and
adj.r2 >0.16 were filtered out for further analysis
(Fig. 2). The top 10 genes
significantly associated with MIR210HG expression in colon cancer
are presented in Table II. A total
of 231 and 142 genes were positively, and negatively associated
with MIR210HG expression in colon cancer, respectively.
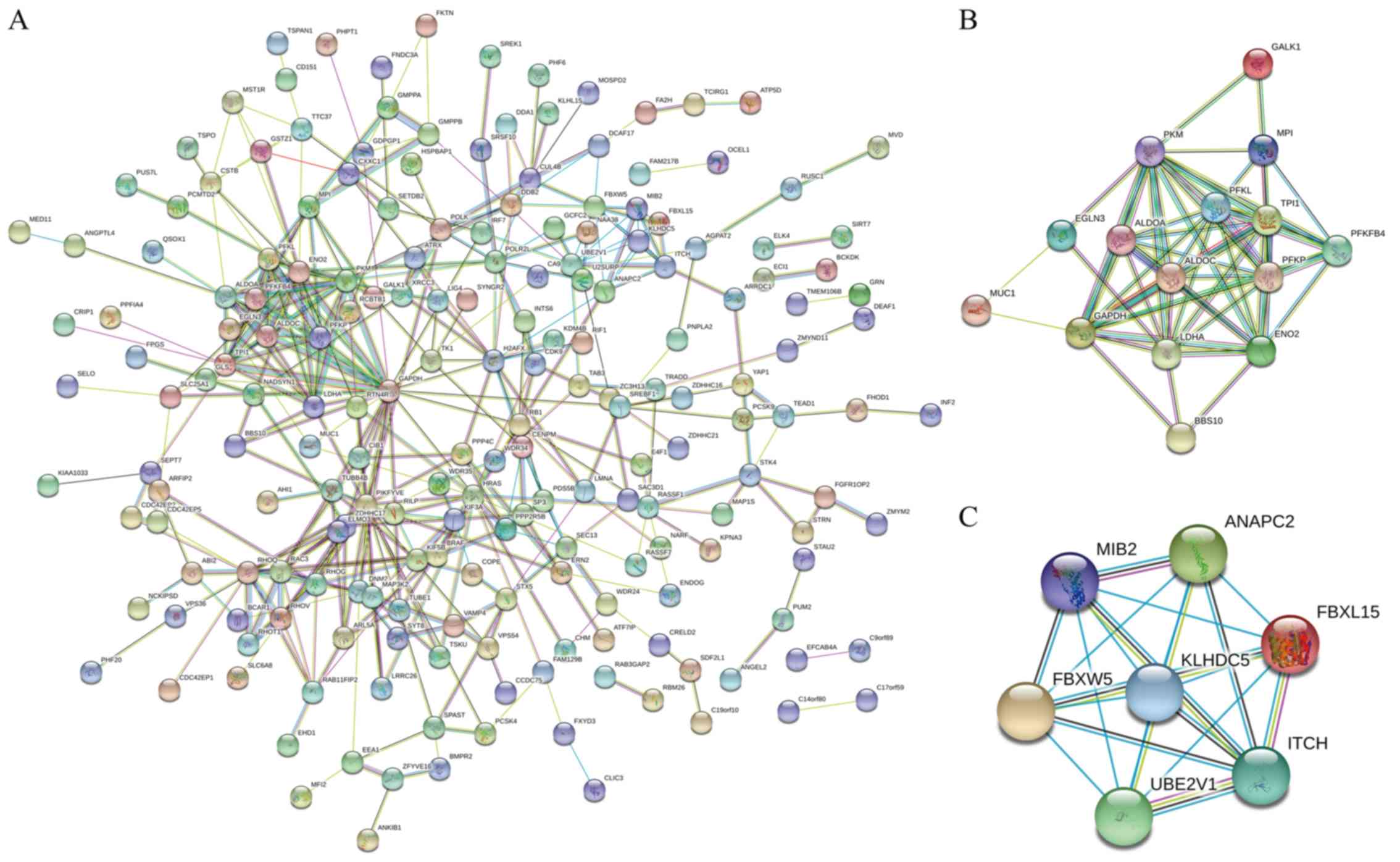
Additionally, the known PPIs among these MIR210HG-related
candidates were detected using STRING (Fig. 3). A total of 359 nodes were noted in
the network, and their interactions are presented in Fig. 3A. To identify the key nodes that most
affected this network, MCC analyses were conducted to rank the
strongest hub genes. Hub genes act as cores within the regulatory
network, and the results revealed complex and strong links with
various MIR210HG-related candidates. It was speculated that the
expression of these hub genes may be directly regulated by MIR210HG
or its downstream targets, and the results of linear regression
analysis showed a significant correlation between the expression of
the 10 hub genes and MIR210HG (Table
III). Modules were regarded as a group of proteins that
participate in the same biological process, or composition of the
same complex (24). Modularity
analysis was conducted, and 2 modules were selected from the
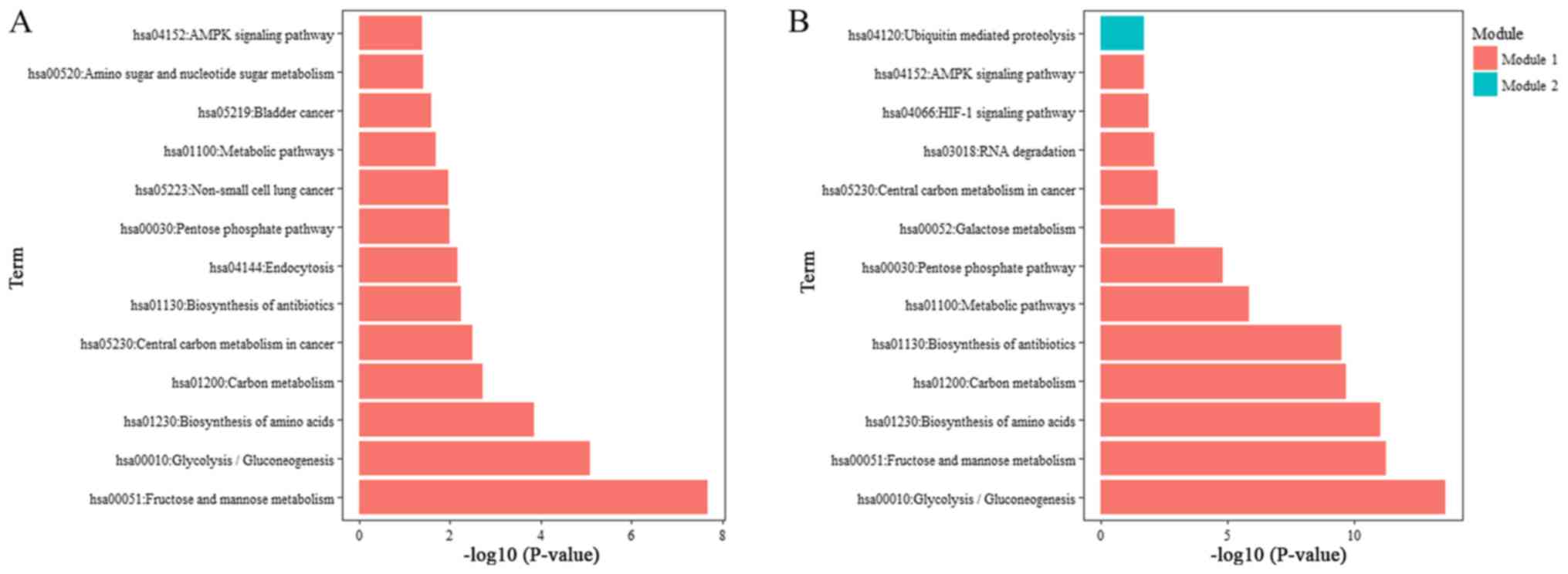
constructed PPI network, as illustrated in Fig. 3B and C. GO and KEGG analysis
indicated that module 1 may be associated with glycolysis and cell
adhesion (e.g. triosephosphate isomerase 1, GAPDH, enolase 2, mucin
1, cell surface associated, egl-9 family hypoxia inducible factor
3, Bardet-Biedl syndrome 10, and phosphofructokinase, liver type),
while module 2 may be associated with ubiquitination (e.g. kelch
like family member 42, anaphase promoting complex subunit 2, F-box
and WD repeat domain containing 5, and F-box and leucine rich
repeat protein 15) (Figs. 4B and C;
5B).
 | Table II.Top 10 genes significantly associated
with MIR210HG expression in colon cancer. |
Table II.
Top 10 genes significantly associated
with MIR210HG expression in colon cancer.
| Gene | P-value |
Adj.r2 |
|---|
| NDRG1 |
7.96×10−51 | 0.38 |
| ALDOA |
1.43×10−51 | 0.38 |
| EGLN3 |
2.49×10−51 | 0.36 |
| ALDOC |
1.09×10−51 | 0.35 |
| SLC16A3 |
5.78×10−51 | 0.34 |
| PFKFB4 |
3.23×10−51 | 0.33 |
| MIR210 |
1.02×10−51 | 0.30 |
| SLC6A8 |
1.85×10−51 | 0.30 |
| NARF |
3.34×10−51 | 0.30 |
| ANGPTL4 |
7.93×10−51 | 0.30 |
 | Table III.Top 10 hub genes in protein-protein
interaction networks. |
Table III.
Top 10 hub genes in protein-protein
interaction networks.
| No. | Hub genes | P-value | Adj.r.squared |
|---|
| 1 | PKM |
6.14×10−26 | 0.21 |
| 2 | TPI1 |
1.01×10−20 | 0.17 |
| 3 | PFKL |
1.18×10−25 | 0.21 |
| 4 | PFKP |
9.47×10−27 | 0.22 |
| 5 | GAPDH |
1.65×10−21 | 0.17 |
| 6 | ENO2 |
7.55×10−27 | 0.22 |
| 7 | LDHA |
1.88×10−29 | 0.24 |
| 8 | ALDOA |
1.43×10−50 | 0.38 |
| 9 | ALDOC |
1.09×10−46 | 0.35 |
| 10 | PFKFB4 |
3.23×10−42 | 0.33 |
MIR210HG may impact colon cancer by
targeting energy metabolism and cell adhesion
In order to determine the possible mechanism of
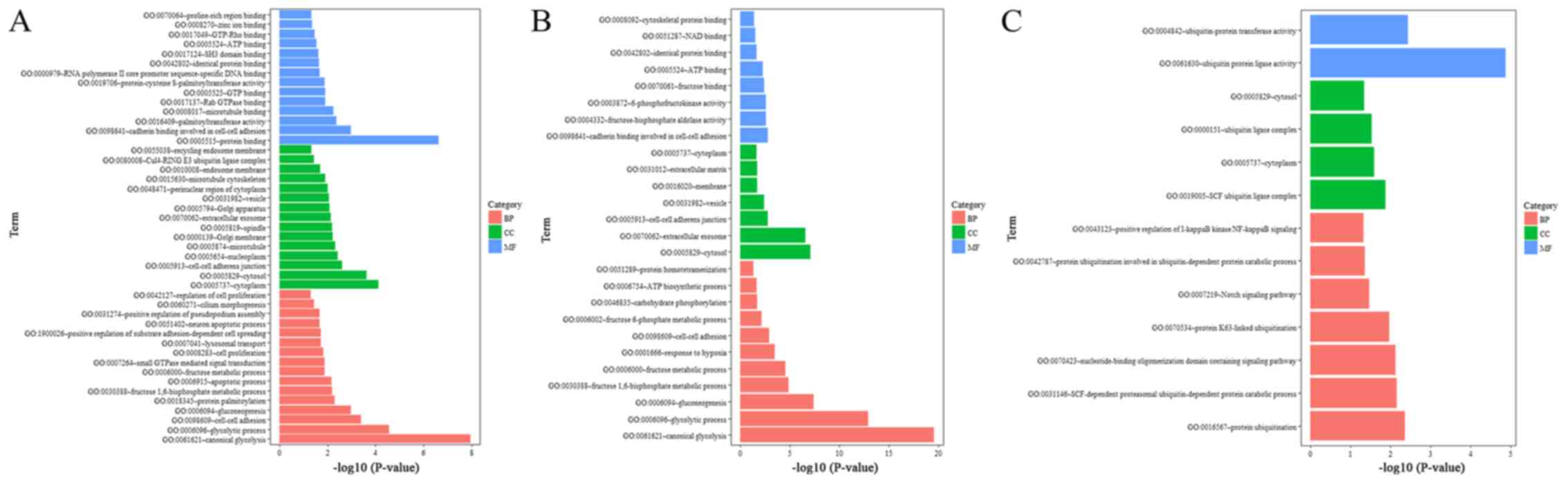
MIR210HG in colon cancer, GO and KEGG pathway analyses were
conducted. The GO results revealed that MIR210HG expression was
strongly associated with energy metabolism in colon cancer, as
evidenced by the enrichment of genes involved in canonical
glycolysis (P=1.15×10−8), glycolytic process
(P=2.78×10−5) and gluconeogenesis (P=0.001) (Fig. 4A). Additionally, the KEGG results
revealed that MIR210HG was significantly associated with the
biosynthesis of glycolysis or gluconeogenesis
(P=8.33×10−6) and central carbon metabolism in cancer
(P=0.003) (Fig. 5A). Furthermore,
cell-cell adhesion (P=3.99×10−4), cell-cell adherens
junction (P=0.002) and cadherin binding involved in cell-cell
adhesion (P=0.001) were notably enriched in biological process,
cellular component and molecular function analysis, respectively. A
recently published paper suggested that MIR210HG facilitates cancer
cell invasion and metastasis in osteosarcoma (25), which is consistent with the findings
of the present study. These findings suggest that MIR210HG may
exert its effects in colon cancer by modulating energy metabolism
and cell adhesion, which are closely associated with the prognosis
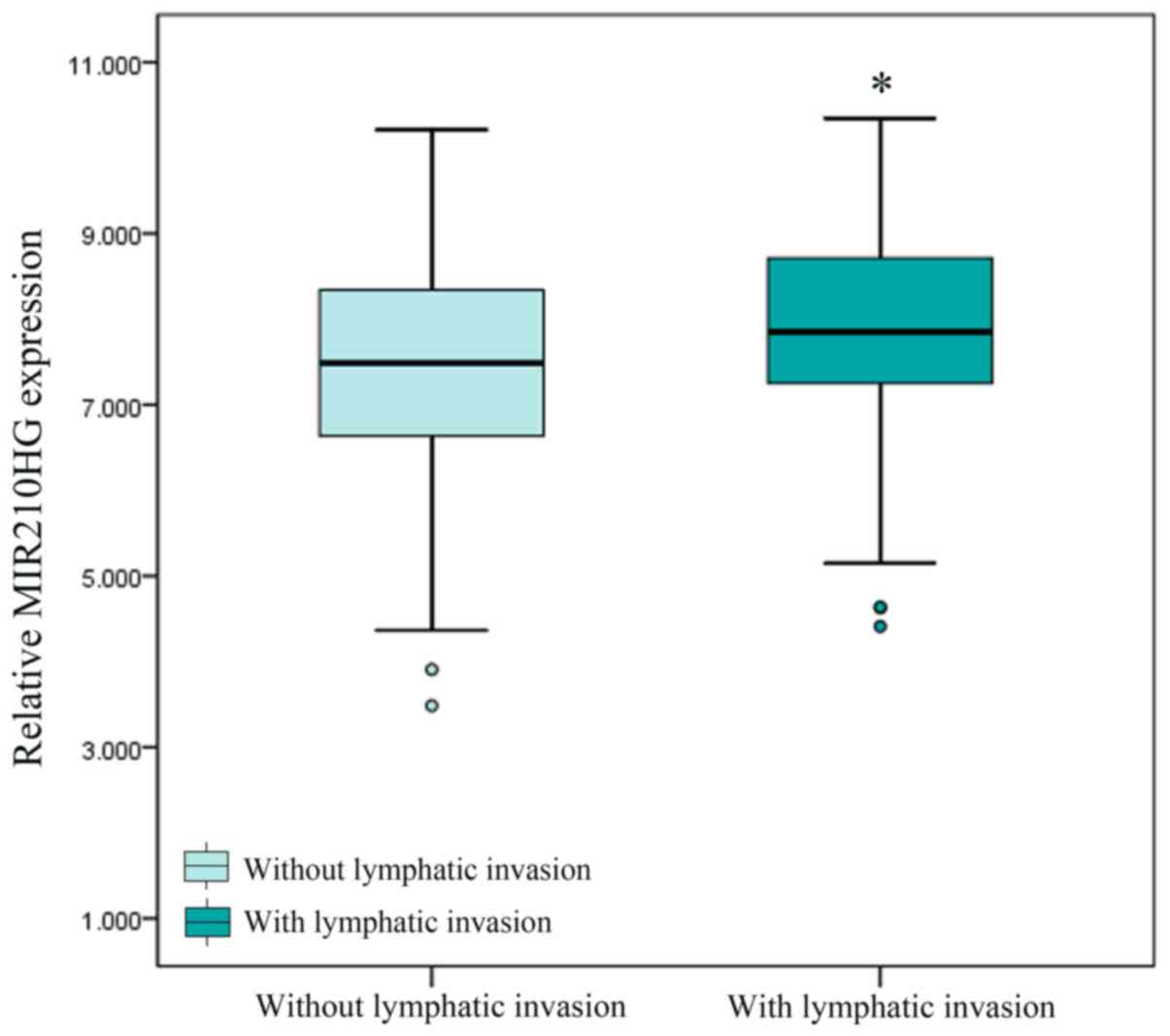
of patients with colon cancer. Furthermore, a Mann Whitney U test
was performed to verify the association between lymphatic
metastasis and MIR210HG expression in colon cancer, and a strong
association was identified (P=0.02; Fig.
6). These results suggest that MIR210HG may be a modulator of
colon cancer cell metastasis, and are consistent with the results
of the GO and KEGG enrichment analyses.
MIR210HG may influence colon cancer
via transcription and post-transcriptional processing
To further investigate the mechanism of MIR210HG in
colon cancer, various possible interactions between MIR210HG and
screened MIR210HG candidates were predicted. Known MIR210HG-protien
interactions were summarized using starBase v2.0. Specific genes
associated with RNA splicing and degradation were detected
(Table IV), including FUS RNA
binding protein (FUS). These genes may therefore serve as
post-transcriptional regulators in colon cancer. Novel
lncRNA-protein interactions were also predicted using RPISeq. Since
transcription factors are considered to be important regulators of
gene expression, the focus was on their association with MIR210HG
(Table V). Predictions for Mediator
of RNA polymerase II transcription (MED)15, MED16 and B-cell
lymphoma 3 protein (BCL3; probabilities >0.5) were considered to
be ‘positive’. It was observed that MED16 and BCL3 were connected
to hub genes within the MIR210HG regulatory network, suggesting
that they may modulate the network by stimulating these genes.
 | Table IV.Known MIG210HG-protien interactions
via starBase v2.0. |
Table IV.
Known MIG210HG-protien interactions
via starBase v2.0.
| Name | lncRNA | Target sites | BioComplex | ClipReadNum |
|---|
| eIF4AIII | MIR210HG | 12 | 2 | 31 |
| FUS | MIR210HG | 1 | 1 | 5 |
| SFRS | MIR210HG | 1 | 1 | 800 |
| U2AF65 | MIR210HG | 1 | 1 | 2 |
 | Table V.Prediction of lncRNA-protein
interactions between MIR210HG and candidate transcription factors
through RPIseq. |
Table V.
Prediction of lncRNA-protein
interactions between MIR210HG and candidate transcription factors
through RPIseq.
| Transcription
factors | Isoform | Gene names | RF classifier | SVM classifier |
|---|
| MED15 | 1 | Mediator complex
subunit 15 | 0.9 | 0.57 |
| MED15 | 2 | Mediator complex
subunit 15 | 0.9 | 0.59 |
| MED15 | 3 | Mediator complex
subunit 15 | 0.9 | 0.62 |
| MED16 | 1 | Mediator complex
subunit 16 | 0.8 | 0.61 |
| MED16 | 2 | Mediator complex
subunit 16 | 0.75 | 0.61 |
| MED16 | 3 | Mediator complex
subunit 16 | 0.75 | 0.6 |
| MED16 | 4 | Mediator complex
subunit 16 | 0.8 | 0.54 |
| MED16 | 5 | Mediator complex
subunit 16 | 0.8 | 0.48 |
| CDK9 | 1 | Cyclin dependent
kinase 9 | 0.85 | 0.55 |
| CDK9 | 2 | Cyclin dependent
kinase 9 | 0.85 | 0.51 |
| BCL3 | 1 | B-cell CLL/lymphoma
3 | 0.75 | 0.53 |
| PPP1R13L | 1 | Protein phosphatase
1 regulatory subunit 13 like | 0.85 | 0.49 |
lncRNAs also interacted with other cis acting
elements of promoters or co-expressed genes, thus regulating gene
expression via transcription or post-transcriptional processes. We
therefore investigated neighboring mRNAs located within 1 Mbps of
the MIR210HG locus to predict possible cis target genes. A
total of 13 genes were identified (Table VI), 5 of which were members of the
MIR210HG regulatory network. One of these genes was HRAS, which is
known to be involved in central carbon metabolism in cancer. HRAS
is a member of the Ras superfamily of GTPases, which are involved
in the regulation of cell division in response to growth factor
stimulation. Activated mutant HRAS (Hras-V12) is able to initiate
irreversible cell cycle arrest and increase the expression of the
tumor suppressors p16 and p53 (26).
 | Table VI.Prediction of nearby cis target genes
of MIR210HG. |
Table VI.
Prediction of nearby cis target genes
of MIR210HG.
| Nearby gene
names | Whether in PPI
network (1=yes, 0=no) |
|---|
| RNH1 | 0 |
| LOC101059906 | 0 |
| HRAS | 1 |
| LRRC56 | 0 |
| LMNTD2 | 1 |
| RASSF7 | 1 |
| MIR210 | 1 |
| LOC143666 | 0 |
| PHRF1 | 0 |
| IRF7 | 1 |
| CDHR5 | 0 |
| SCT | 0 |
| DRD4 | 0 |
Discussion
Lymphatic metastasis is one of the principal factors
to affect the prognosis of patients with colon cancer (27). The survival rates of patients with
lymphatic metastasis is significantly lower compared with that of
patients without lymphatic metastasis (28). Furthermore, it has been identified
that metastasis-related factors may be used as prognostic
biomarkers or potential therapeutic targets for improving the
survival rates of patients with colon cancer (29–31).
In the present study, candidate lncRNAs were
screened based on the effects of lncRNA expression on the survival
rate of patients with colon cancer. The employed screening method
may be more suitable for clinical practice compared with
traditional selection methods, and is more likely to be applied to
treatment. The same method was used to screen survival-related
lncRNAs in breast cancer, and a co-expressed gene network of the
lncRNA with the greatest effect, LINC00704 (data not shown), was
also constructed. The results of GO and KEGG analyses suggested
that LINC00704-related genes are associated with the regulation of
cell proliferation, which is in agreement with a previous report
(32).
Glycolysis is the primary method by which tumor
cells generate energy, even under normoxic conditions (33). Glycolysis provides energy for tumor
cells, and so targeting glycolysis may have potential as a method
to control tumor proliferation. Changes in glycolysis in tumor
cells are associated with resistance to anticancer drugs. For
instance, Song et al (34)
reported that increased glycolytic activity and low phosphorylation
efficiency increases chemo-resistance in patients with acute
myeloid leukemia. This may be regulated by glycolysis-related
molecules, including hypoxia-inducible factor 1α, hexokinase II,
glucose transport 1 and lactate dehydrogenase (34).
In the present study, a number of survival-related
lncRNAs in colon cancer were identified. Notably, the upregulation
of a tumor promoter lncRNA, MIR210HG, was associated with shorter
overall patient survival. To the best of our knowledge, current
investigations into the association between MIR210HG and colon
cancer prognosis are limited. Here, linear regression analysis was
performed and an MIR210HG-related regulatory network was
constructed. Specific MIR210HG-related candidates have been
reported to be associated with colon cancer cell survival and
metastasis, suggesting a possible link between MIR210HG expression
and these genes that affects overall survival in patients with
colon cancer. MIR210HG also influences osteosarcoma metastasis
(25); furthermore, it was revealed
that patients with high MIR210HG expression levels were more likely
to have lymphatic metastasis. Genes that were not reported may be
regarded as novel colon cancer-specific hubs in the MIR210HG
regulatory network. Two notable modules and the top 10 hub genes
were screened out using modularity analysis and MCC. A number of
these hub genes, including fructose-bisphosphate aldolase A (ALDOA)
and pyruvate kinase M (PKM)2, were associated with glycolysis, cell
migration and apoptosis in colon cancer cells. These hub genes have
been demonstrated to affect colon cancer prognosis. For instance,
ALDOA was reported to be a key promoting element in colon cancer;
ALDOA acts as a significant regulator of hypoxic adaptation in
colon cancer cells (35), whilst it
has also been reported to contribute to metastasis of colon cancer.
Another key hub gene, PKM2 was able to disturb the Warburg effect
and increase the sensitivity of cancer cells to tumor necrosis
factor-related apoptosis-inducing ligand-induced cell death. Yang
et al (36) reported that
PKM2 overexpression was able to modulate STAT3 signaling, thereby
facilitating the migration of colorectal cancer cells (36). Of these genes, ALDOA was found to
have the strongest correlation with MIR210HG
(adj.r2=0.4296; P=7.450×10−55), which
suggests that it may be a direct target of MIR210HG.
The possible mechanism of interaction between
MIR210HG and its co-expressed genes was also predicted. Reports
have indicated that MIR210HG may affect colon cancer via
transcription and post-transcriptional processing. Certain genes
associated with RNA splicing and degradation, including FUS, were
reported to interact with MIR210HG, suggesting that they may serve
as post-transcriptional regulators in colon cancer. mRNA splicing
is a vital step in post-transcriptional regulation, as it can
trigger translation and functionally diversify proteins. In part of
a study by Zarnack et al (37) experimental verification was carried
out and reported a correlation between MIR210HG and U2AF65
(supplementary material; summary by starBase v2.0 database:
http://starbase.sysu.edu.cn/index.php). While,
according to the present study, MIR210HG and U2AF65 also showed
associations (probabilities >0.5 using RPISeq: http://pridb.gdcb.iastate.edu/RPISeq/index.html),
which is consistent with Zarnack's experimental results. However,
in order to develop novel treatment methods to improve the
prognosis of patients with colon cancer, we must improve our
understanding of the molecular mechanisms of lncRNAs. In the
present study, novel lncRNA-protein interactions were detected
using RPISeq, and numerous transcription factors (including MED15,
MED16 and BCL3) were considered to be positive candidates. In
particular, MED16 and BCL3 were connected to hub genes within the
MIR210HG regulatory network, reinforcing our prediction that
MIR210HG serves as a transcriptional modulator in colon cancer.
Although there is a lack of evidence indicating the specific
molecular association between the selected transcription factors
and candidates, it was revealed that some transcription factors
(including MED15 and Angiopoietin-related protein 4, BCL3 and
GAPDH, Cyclin-dependent kinase 9 and HRAS) interact with the hub
genes and are strongly correlated with MIR210HG. A total of 5
selected transcription factors were associated with cancer
metastasis. Decreased MED15 expression can suppress the
transforming growth factor-β/Smad signaling pathway, thereby
downregulating the metastasis of highly invasive breast cancer
cells (38). BCL3 can also reduce
cancer cell metastasis by influencing cell motility (39). Furthermore, neighboring mRNAs located
within 1 Mbps of the MIR210HG loci were investigated to predict
possible cis element target genes of MIR210HG. A total of 13
genes were identified, 5 of which, including HRAS, were members of
the MIR210HG regulatory network. These results provide new
information as how MIR210HG regulates the regulatory network and
affects survival in patients with colon cancer.
In summary; in the present study survival-related
lncRNAs in colon cancer were screened and bioinformatics analyses
were performed to investigate the prognostic potential of MIR210HG
and its related genes in colon cancer. The findings indicated that
MIR210HG may exert its effects in colon cancer by modulating energy
metabolism and cell adhesion. Further predictions suggested that
MIR210HG may also affect colon cancer via transcription and
post-transcriptional processing. Together, these results provide
evidence of the transcriptional regulatory network of MIR210HG in
colon cancer, and suggest that MIR210HG may serve a potential role
as a novel biomarker and therapeutic target. Despite the multiple
bioinformatic and statistical methods used in the present study, it
does have some limitations, and further studies are required to
verify these results. Experimental evidence will be provided in
future studies; for example, the high expression level of MR210HG
may regulate the expression levels of screened MIR210HG-related
genes, or induce the phenotype obtained with bioinformatics
analyses (such as the metastasis of colon cancer cells), which will
be assessed using cell-based assays with colon cancer cell
lines.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant nos. 81760094
and 31602111), the Science and Technology Research Project for
Youth of Educational Commission in Jiangxi Province (grant no.
GJJ160238), the Science and Technology Foundation for Youths of
Jiangxi province (grant no. 20171BAB215021) and the Science and
Technology Project of Health and Family Planning commission of
Jiangxi Province (grant. no. 20175081).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YL and ZR designed the study. ZR, ZX and ZL
performed the data analyses. All authors contributed to the
conception of the study and drafted the manuscript. All authors
contributed significantly in writing the manuscript and read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
lncRNA
|
long non-coding RNA
|
|
DHFR
|
dihydrofolate reductase
|
|
TCGA
|
The Cancer Genome Atlas
|
|
ALDOA
|
fructose-bisphosphate aldolase A
|
|
PKM
|
pyruvate kinase M
|
|
DAVID
|
Database for Annotation,
Visualization, and Integrated Discovery
|
|
GO
|
gene ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
PPI
|
protein-protein interaction
|
|
STRING
|
Search Tool for the Retrieval of
Interacting Genes/Proteins
|
|
ClusterONE
|
Clustering with Overlapping
Neighborhood Expansion
|
References
|
1
|
Centers for Disease and Control Prevention
(CDC), . Vital Signs: Colorectal cancer screening, incidence and
mortality-United States, 2002–2010. MMWR Morb Mortal Wkly Rep.
60:884–889. 2011.PubMed/NCBI
|
|
2
|
Okugawa Y, Grady WM and Goel A: Epigenetic
alterations in colorectal cancer: Emerging biomarkers.
Gastroenterology. 149:1204–1225. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Balaguer F, Link A, Lozano JJ, Cuatrecasas
M, Nagasaka T, Boland CR and Goel A: Epigenetic silencing of
miR-137 is an early event in colorectal carcinogenesis. Cancer Res.
70:6609–6618. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang LM, Wang P, Liu XM and Zhang YJ:
LncRNA SUMO1P3 drives colon cancer growth, metastasis and
angiogenesis. Am J Transl Res. 9:5461–5472. 2017.PubMed/NCBI
|
|
6
|
Dianatpour A and Ghafouri-Fard S: The role
of long non coding RNAs in the repair of DNA double strand breaks.
Int J Mol Cell Med. 6:1–12. 2017.PubMed/NCBI
|
|
7
|
Espinoza CA, Goodrich JA and Kugel JF:
Characterization of the structure, function and mechanism of B2
RNA, an ncRNA repressor of RNA polymerase II transcription. RNA.
13:583–596. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Engreitz JM, Haines JE, Perez EM, Munson
G, Chen J, Kane M, McDonel PE, Guttman M and Lander ES: Local
regulation of gene expression by lncRNA promoters, transcription
and splicing. Nature. 539:452–455. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reinius B, Shi C, Hengshuo L, Sandhu KS,
Radomska KJ, Rosen GD, Lu L, Kullander K, Williams RW and Jazin E:
Female-biased expression of long non-coding RNAs in domains that
escape X-inactivation in mouse. BMC Genomics. 11:6142010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cancer Genome Atlas Research Network, .
Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA,
Ellrott K, Shmulevich I, Sander C and Stuart JM: The cancer genome
atlas pan-cancer analysis project. Nat Genet. 45:1113–1120. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang DW, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nepusz T, Yu H and Paccanaro A: Detecting
overlapping protein complexes in protein-protein interaction
networks. Nature Methods. 9:471–472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Muppirala UK, Honavar VG and Drena D:
Predicting RNA-protein interactions using only sequence
information. BMC Bioinformatics. 12:4892011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Y, Li Y, Chen W, He F, Tan Z, Zheng J,
Wang W, Zhao Q and Li J: NEAT expression is associated with tumor
recurrence and unfavorable prognosis in colorectal cancer.
Oncotarget. 6:27641–27650. 2015.PubMed/NCBI
|
|
21
|
Meng J, Li P, Zhang Q, Yang Z and Fu S: A
four-long non-coding RNA signature in predicting breast cancer
survival. J Exp Clin Cancer Res. 33:842014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shen Z, Li Q, Deng H, Lu D, Song H and Guo
J: Long non-coding RNA profiling in laryngeal squamous cell
carcinoma and its clinical significance: Potential biomarkers for
LSCC. PLoS One. 9:e1082372014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Min W, Dai D, Wang J, Zhang D, Zhang Y,
Han G, Zhang L, Chen C, Li X, Li Y and Yue Z: Long noncoding RNA
miR210HG as a potential biomarker for the diagnosis of glioma. PLoS
One. 11:e01604512016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xia W, Ren X, Li B, Yue J and Long L:
Applying modularity analysis of PPI networks to sequenced
organisms. Virulence. 3:459–463. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li J, Wu QM, Wang XQ and Zhang CQ: Long
noncoding RNA miR210HG sponges miR-503 to facilitate osteosarcoma
cell invasion and metastasis. DNA Cell Biol. 36:1117–1125. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Serrano M, Lin AW, Mccurrach ME, Beach D
and Lowe SW: Oncogenic ras provokes premature cell senescence
associated with accumulation of p53 and p16INK4a. Cell. 88:593–602.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Harris GJ, Church JM, Senagore AJ, Lavery
IC, Hull TL, Strong SA and Fazio VW: Factors affecting local
recurrence of colonic adenocarcinoma. Dis Colon Rectum.
45:1029–1034. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Baxter NN, Virnig DJ, Rothenberger DA,
Morris AM, Jessurun J and Virnig BA: Lymph node evaluation in
colorectal cancer patients: A population-based study. J Natl Cancer
Inst. 97:219–225. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin CW, Li XR, Zhang Y, Hu G, Guo YH, Zhou
JY, Du J, Lv L, Gao K, Zhang Y and Deng H: TAp63 suppress
metastasis via miR-133b in colon cancer cells. Br J Cancer.
110:2310–2320. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tan X, He X, Jiang Z, Wang X, Ma L, Liu L,
Wang X, Fan Z and Su D: Derlin-1 is overexpressed in human colon
cancer and promotes cancer cell proliferation. Mol Cell Biochem.
408:205–213. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yue B, Qiu S, Zhao S, Liu C, Zhang D, Yu
F, Peng Z and Yan D: LncRNA-ATB mediated E-cadherin repression
promotes the progression of colon cancer and predicts poor
prognosis. J Gastroenterol Hepatol. 31:595–603. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tracy KM, Tye CE, Ghule PN, Malaby HLH,
Stumpff J, Stein JL, Stein GS and Lian JB: Mitotically-associated
lncRNA (MANCR) affects genomic stability and cell division in
aggressive breast cancer. Mol Cancer Res. 16:587–598. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Song K, Li M, Xu X, Xuan LI, Huang G and
Liu Q: Resistance to chemotherapy is associated with altered
glucose metabolism in acute myeloid leukemia. Oncol Lett.
12:334–342. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kawai K, Uemura M, Munakata K, Takahashi
H, Haraguchi N, Nishimura J, Hata T, Matsuda C, Ikenaga M, Murata
K, et al: Fructose-bisphosphate aldolase A is a key regulator of
hypoxic adaptation in colorectal cancer cells and involved in
treatment resistance and poor prognosis. Int J Oncol. 50:525–534.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang P and Li Z, Fu R, Wu H and Li Z:
Pyruvate kinase M2 facilitates colon cancer cell migration via the
modulation of STAT3 signalling. Cell Signal. 26:1853–1862. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zarnack K, Konig J, Tajnik M, Martincorena
I, Eustermann S, Stévant I, Reyes A, Anders S, Luscombe NM and Ule
J: Direct competition between hnRNP C and U2AF65 protects the
transcriptome from the exonization of Alu elements. Cell.
152:453–466. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhao M, Yang X, Fu Y, Wang H, Ning Y, Yan
J, Chen YG and Wang G: Mediator MED15 modulates transforming growth
factor beta (TGFβ)/Smad signaling and breast cancer cell
metastasis. J Mol Cell Biol. 5:57–60. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wakefield A, Soukupova J, Montagne A,
Ranger J, French R, Muller WJ and Clarkson RW: Bcl3 selectively
promotes metastasis of ERBB2-driven mammary tumors. Cancer Res.
73:745–755. 2013. View Article : Google Scholar : PubMed/NCBI
|