Introduction
Gastric cancer has the sixth highest incidence of
cancer worldwide (1). Surgical
resection can be effective for the treatment of patients with early
gastric cancer (2). However, ~50% of
patients are diagnosed with advanced-stage disease; at which point
surgical treatment alone is not effective (2). Therefore, chemotherapy is one of the
most common treatment options for these cancer patients (3). 5-Fluorouracil (5-FU)-based chemotherapy
is the first line treatment for advanced gastric cancer; however,
its effectiveness is limited by drug resistance (3). Therefore, there is a requirement for
elucidating the molecular mechanisms underlying resistance to 5-FU,
as well as the development of novel treatment strategies, for
patients with advanced gastric cancer.
The N-myc downstream-regulated gene (NDRG) protein
family includes 4 members, NDRG1, NDRG2, NDRG3 and NDRG4. NDRG
proteins participate in multiple cellular processes, including cell
proliferation, differentiation and the stress response (4). In addition, NDRG family members are
tumor-associated proteins and their dysregulation may result in
tumorigenesis (5). NDRG3 has been
reported to promote colorectal cancer metastasis by activating Src
phosphorylation (6), and is
associated with poor survival in non-small cell lung cancer
(7). However, the role of NDRG3 and
regulatory mechanisms of NDRG3 in gastric cancer remain
unclear.
The present study investigates the role of NDRG3 in
gastric cancer. The clinical relevance of NDRG3 in gastric cancer
was examined and it was revealed that NDRG3 was upregulated in
tumor tissues obtained from patients with gastric cancer. The
biological role of NDRG3 in gastric cancer cell lines was
investigated and the results demonstrated that NDRG3 increased the
proliferation of these cells. The long non-coding (lncRNA) small
nucleolar RNA host gene 20 (SNHG20)/microRNA (miR)-140-5p signaling
pathway was subsequently identified to regulate the expression of
NDRG3. The SNHG20/miR-140-5p/NDRG3 axis was revealed to be
implicated in resistance to 5-FU in gastric cancer cell lines, and
may therefore present a potential therapeutic target for overcoming
5-FU-associated drug resistance in gastric cancer.
Materials and methods
Cell culture
The gastric cancer cell lines BGC-823 (cat. no.
C023) and AGS (cat. no. C015) were purchased from Wuhan Fine
Biotech Co., Ltd. All cells were cultured in RPMI-1640 medium (cat.
no. 11875093; Thermo Fisher Scientific, Inc.) containing 5% fetal
bovine serum (Thermo Fisher Scientific, Inc.). The cells were
maintained in a humidified atmosphere at 37°C and 5%
CO2.
Cell transfection
The miR-140-5p mimic and miR-inhibitor
(anti-miR-140-5p) used in this study were purchased from Shanghai
Gene Pharma Co., Ltd. The sequences were as follows: miR mimic
negative control (miR-NC), 5′-UUCUCCGAACGUGUCAACGUTT-3′; miR-140-5p
mimic, 5′-CAGUGGUUUUACCCUAUGGUAGACCAUAGGGUAAAACCACUGUU-3;
anti-miR-NC, 5′-CAGUACUUUGUGUAGUACAA-3′; anti-miR-140-5p,
5′-AACCCAUGGAAUUCAGUUCUCA-3′. The small interfering (si)RNA
sequences used in this study were purchased from Shanghai Gene
Pharma Co., Ltd., and their sequences were as follows: si-NC,
5′-CACTGACGGTGACCAGAACAAAGAT-3′; si-SNHG20,
5′-GAAUCGAUAGGUCGAGGGGTT-3′. AGS and BGC-823 cells were transfected
with miR-140-5p mimic or inhibitor and their respective negative
controls (100 nM), si-NC (50 nM) or si-SNHG20 (50 nM) sequences
using a lipid-based method (Lipofectamine® 2000; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
Gene and protein expression analyses were performed at 48 h
following transfection.
Lentivirus-based negative control and gene-specific
short hairpin (sh)RNAs were purchased from Sigma-Aldrich; Merck
KGaA. Transfections were performed using Lipofectamine®
2000 (Thermo Fisher Scientific, Inc.). A total of 2 µg
gene-specific shRNA or shControl were transfected into 293T cells
(5×105 cells; cat. no. C004; Wuhan Fine Biotech Co.,
Ltd.). At 48 h following transfection, the culture medium of 293T
cells was collected (5,000 viral particles/µl) and 10 µl viral
medium was applied to 100,000 BGC-823 and AGS gastric cancer cells
to get a multiplicity of infection of 0.5. Gastric cancer cells
were cultured in 5% CO2 at 37°C for 48 h, before
puromycin (0.75 µg/ml; Sigma-Aldrich; Merck KGaA) was added. Cells
were collected at 72 h post-transfection. The knockdown efficiency
was confirmed using western blotting analysis. The shRNA sequences
used were as follows: shControl,
5′-CCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGG-3′; shNDRG3#1,
5′-CCGGCCACTCCATAATATAACATTTCTCGAGAAATGTTATATTATGGAGTGGTTTTTG-3′;
shNDRG3#2,
5′-CCGGTTCCCGCCTGAACCCTATAAACTCGAGTTTATAGGGTTCAGGCGGGAATTTTTG-3′.
NDRG3 expression and correlation
analysis using the Gene Expression Profiling Interactive Analysis
(GEPIA) web tool
The GEPIA (gepia.cancerpku.cn/index.html) (8) online database was used to analyze RNA
sequencing expression data downloaded from the The Cancer Genome
Atlas (TCGA; http://portal.gdc.cancer.gov/) and the Genotype-Tissue
Expression (GTEx; http://www.gtexportal.org/home/) projects. GEPIA
performed NDRG3 expression analysis based on gene expression
levels; NDRG3 expression was compared between gastric cancer
(n=408) and normal stomach tissues (n=211). Tboxplot analysis used
log2 (transcripts per million + 1) for the log-scale, and was
conducted by the GEPIA web tool. In addition, GEPIA performed
pairwise gene correlation analysis for gastric cancer tissue
samples vs. normal gastric tissue samples of TCGA and/or GTEx
expression data using Pearson's correlation test.
Western blotting
Total protein was extracted from BGC-823 and AGS
gastric cancer cells using Cell Lysis Buffer for Western and
Immunoprecipitation (cat. no. P0013; Beyotime Institute of
Biotechnology) and centrifuged at 10,000 × g for 5 min at 4°C.
Protein concentrations were determined using a bicinchoninic acid
assay. Proteins were denatured at 100°C for 5 min in sample buffer.
An equal amount of protein (60 µg/lane) for each sample was
separated by 6–10% SDS-PAGE and transferred to nitrocellulose
membranes. Subsequently, membranes were incubated with primary
antibody overnight at 4°C. The membranes were then washed with 1X
Tris-buffered saline and Tween 20 and incubated with anti-rabbit
immunoglobulin G (IgG; cat. no. MR-R100; 1:3,000 dilution; Shanghai
MRbiotech, Co., Ltd.) and anti-mouse IgG (cat. no. MR-M100; 1:3,000
dilution; Shanghai MRbiotech, Co., Ltd.) horseradish
peroxidase-conjugated secondary antibodies for 1 h at room
temperature. The protein bands were visualized by chemiluminescence
using SuperSignal West Pico Stable Peroxide solution (Thermo Fisher
Scientific, Inc.). Image J software (ImageJ bundled with Java
version 1.8.0_112; National Institutes of Health) was used for
semi-quantification of protein expression levels. The primary
antibodies used were purchased from Santa Cruz Biotechnology, Inc.,
and were as follows: Anti-NDRG3 (dilution, 1:1,000; cat. no.
sc-514561) and anti-GAPDH (dilution, 1:5,000; cat. no.
sc-47724).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cultured cells using
TRIzol® reagent (Thermo Fisher Scientific, Inc.). A
total of 2 µg RNA was reverse transcribed into cDNA using a cDNA
reverse transcription kit (PrimeScript™ RT reagent kit; cat. no.
RR037A; Takara Bio, Inc.) according to the manufacturer's protocol.
qPCR analysis was performed using a PCR kit (TB Green™ Fast qPCR
Mix; cat. no. RR430A; Takara Bio, Inc.) according to the
manufacturer's protocol. The two kits were purchased from Takara
Bio Inc. The thermocycling conditions were as follows: Initial
denaturation at 95°C for 60 sec; denaturation at 95°C for 20 sec,
annealing at 58°C for 30 sec and extension at 72°C for 30 sec (43
cycles); and melting curve at 65–95°C with increments of 0.5°C for
5 sec. The following primer pairs were used for qPCR: NDRG3
forward, 5′-GCAGCTTCCAAACTCTCTGG-3′ and reverse,
5′-AGCTGCAGGTTGTCTTGGTT-3′; and GAPDH forward,
5′-TGTGGTCATGAGTCCTTCCA-3′ and reverse, 5′-CGAGATCCCTCCAAAATCAA-3′.
NDRG3 mRNA levels were quantified using the 2−ΔΔCq
method (9) and normalized to the
internal reference gene GAPDH.
Luciferase assay
Potential regulators of NDRG3 were identified using
the TargetScan (version 7.2; http://www.targetscan.org/vert_72/) and MicroRNA
(August 2010 Release; http://www.microrna.org/microrna/home.do) tools
according to the protocols of these web tools. A miR-140-5p
recognition site in the 3′-untranslated region (UTR) of NDRG3 was
identified The NDRG3 3′-UTR containing the putative miR-140-5p
binding site was cloned into a pMIR-REPORT plasmid (Promega
Corporation) to construct the reporter vector, pMIR-NDRG3-WT. The
GeneArt™ Site-Directed Mutagenesis system (Thermo Fisher
Scientific, Inc.) was used to the produce mutant-type NDRG3
reporter (NDRG3-Mut). The NDRG3-WT (50 nM) and Mut (50 nM)
reporters were co-transfected with miR-140-5p mimics (100 nM) into
the BGC-823 and AGS gastric cancer cells using a lipid-based method
(Lipofectamine® 2000; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. At 24 h post-infection,
luciferase activities were determined using a dual-luciferase assay
system (Promega Corporation) according to the manufacturer's
protocol. The red firefly luciferase signal was used as a
normalization control.
MTS assay
Cell viability was determined using a colorimetric
MTS assay kit (Abcam) according to the manufacturer's protocol.
Briefly, 10,000 cells/wells were plated into the 96-well plates. At
48 h after infection, cells were treated with increasing
concentrations (0, 0.625, 1.25, 2.5, 5, 10, 20, 40, 80 and 160
µg/ml) of 5-FU (cat. no. HY-90006; MedChemExpress) for a further 24
h at 37°C. MTS reagent was added to each well of the 96-well plate
[cells not treated with 5-FU were treated with a vehicle
(dimethylsulfoxide)]. After 60 min of incubation at 37°C in a cell
incubator, the absorbance at a wavelength of 490 nm was measured
using a plate reader.
Statistical analysis
Microsoft Excel software (Microsoft Excel 2013
version 15.19.1; Microsoft Corporation) was used for statistical
analysis. A two-sided paired Student's t-test was used for
comparisons between two groups, and one-way analysis of variance
followed by a Tukey's multiple comparisons test was used for
comparing multiple groups. All values were expressed as the mean ±
standard deviation. P<0.05 was considered to indicate a
statistically significant difference.
Results
Knockdown of NDRG3 inhibits gastric
cancer proliferation
Since aberrant expression of NDRG3 promotes
colorectal cancer metastasis and is associated with poor survival
in non-small cell lung cancer (7),
the role of NDRG3 in gastric cancer was investigated in the current
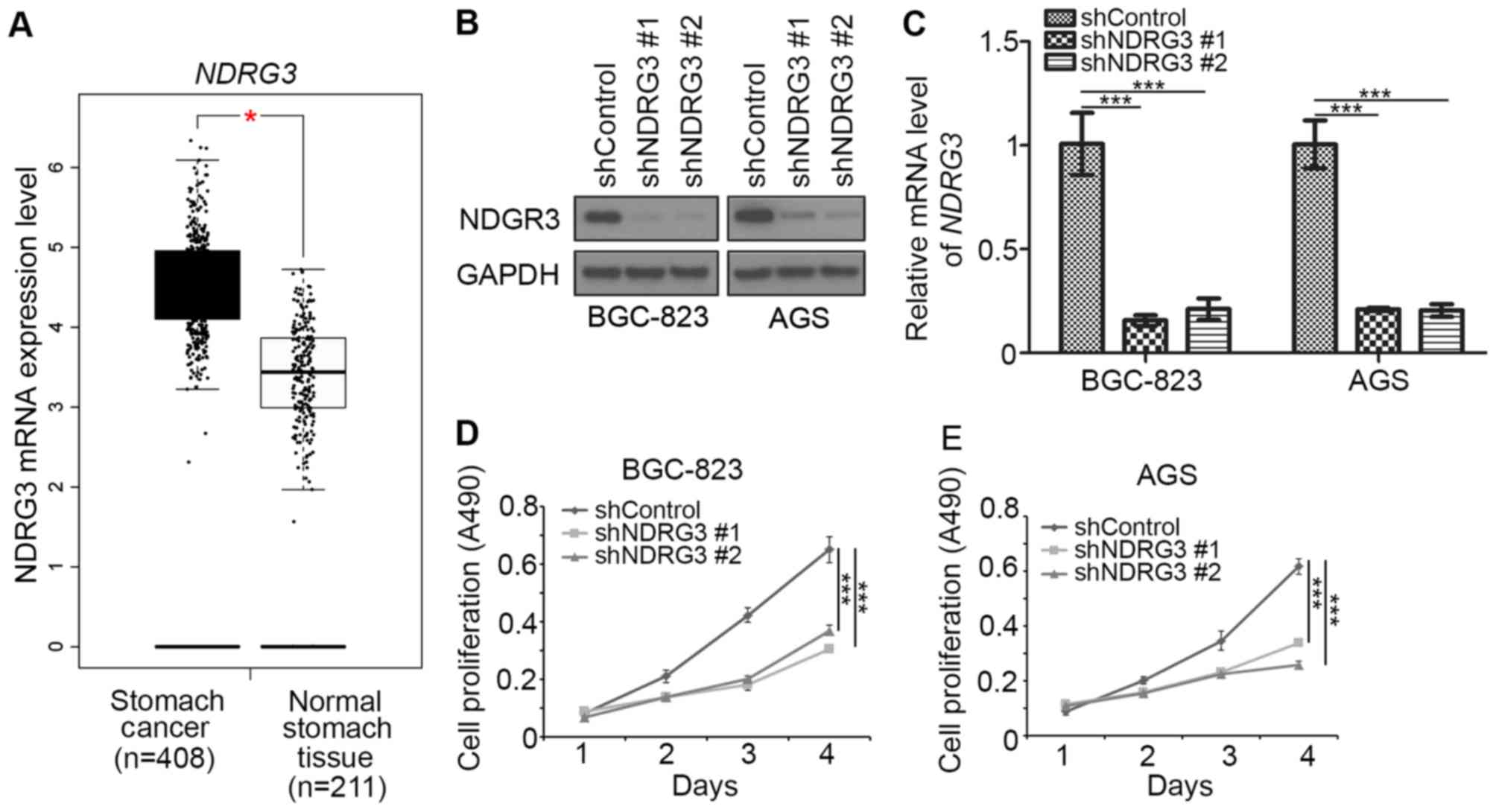
study. The expression of NDRG3 mRNA in gastric cancer and adjacent
normal gastric tissues was analyzed using the GEPIA web tool
(8). The mRNA level of NDRG3 in
gastric cancer was significantly increased when compared with
adjacent normal gastric tissues (Fig.
1A). Therefore, NDRG3 may serve a role in gastric cancer. To
examine the effect of NDRG3 on the growth of gastric cancer cells,
NDRG3 expression was silenced using two different shRNAs (shNDRG3-1
and shNDRG3-2) in BGC-823 and AGS gastric cancer cells (Fig. 1B and C). Knockdown of NDRG3
significantly decreased the cell growth rate of BGC-823 and AGS
cells when compared with controls (Fig.
1D and E). The two different shRNAs (shNDRG3-1 and shNDRG3-2)
could knockdown NDRG3 effectively, and shNDRG3-1 was used in
subsequent experiments. Together, these data suggest that NDRG3 may
be an important protein in gastric cancer.
miR-140-5p inhibits NDRG3 expression
in gastric cancer cells
Potential regulators of NDRG3 were identified using
the TargetScan and MicroRNA tools according to the protocols of
these web tools. A miR-140-5p recognition site in the 3′-UTR of
NDRG3 was identified (Fig. 2A).
Overexpression and knockdown of miR-140-5p in BGC-823 and AGS cells
were examined by RT-qPCR analysis (Fig.
2B). The interaction between NDRG3 and miR-140-5p was examined
using a luciferase assay in BGC-823 cells (Fig. 2C). The expression of NDRG3 was
increased following treatment with anti-miR-140-5p in BGC-823 and
AGS gastric cancer cells when compared with the controls (Fig. 2D and E). These results suggest that
the expression of NDRG3 in gastric cancer may be regulated by
miR-140-5p.
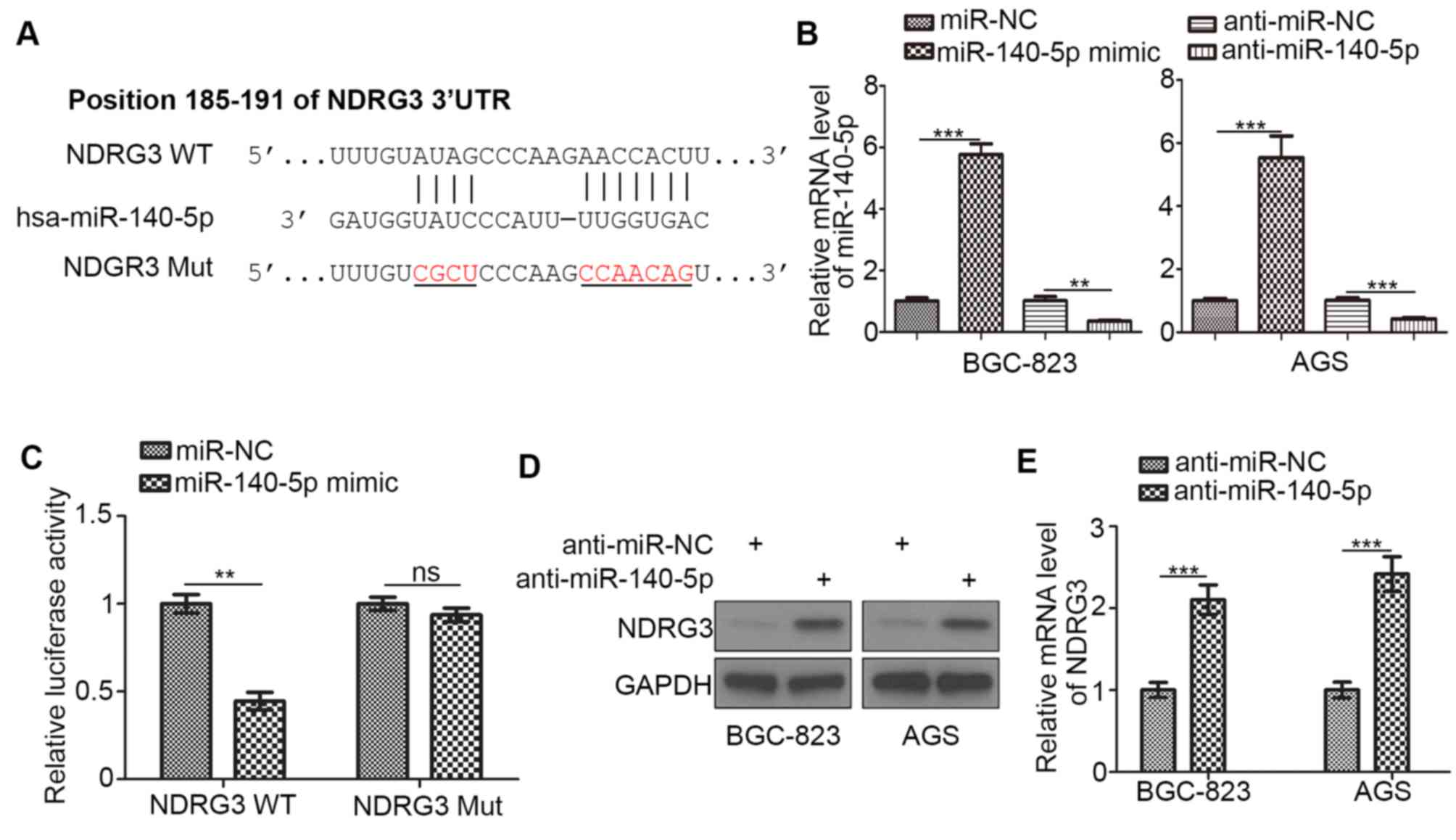 | Figure 2.miR-140-5p inhibits NDRG3 expression
in gastric cancer cells. (A) The predicted position of miR-140-5p
binding sites on the NDRG3 3′-UTR. BGC-823 cells were transfected
with NDRG3 (WT) or NDRG3 (Mut) reporters and miR-NC or miR-140-5p
mimics. (B) BGC-823 and AGS cells were transfected with miR-NC,
miR-140-5p mimic, anti-miR-NC or anti-miR-140-5p. At 48 h
post-transfection, cells were harvested for RT-qPCR analysis. (C) A
total of 48 h following transfection of BGC-823 cells, the relative
luciferase activity was detected using a dual-luciferase reporter
assay. BGC-823 and AGS cells were transfected with the indicated
constructs. A total of 48 h following transfection, cells were
harvested for (D) western blotting and (E) reverse
transcription-quantitative polymerase chain reaction analysis. Data
are presented as the mean ± standard deviation of three replicates.
**P<0.01 and ***P<0.001, as indicated. miR, microRNA; NDRG3,
N-myc downstream regulated gene, family member 3; UTR, untranslated
region; WT, wild type; Mut, mutant; NC, negative control; ns, not
significant. |
SNHG20 regulates NDRG3 expression via
miR-140-5p in gastric cancer cells
The SNHG20 lncRNA has been demonstrated to interact
with miR-140-5p and decrease its expression in cervical cancer
(10). In addition, SNHG20 promotes
gastric cancer progression by inhibiting the expression of cyclin
dependent kinase inhibitor 1A (CDKN1A) and by regulating the
glycogen synthase kinase-3β/β-catenin signaling pathway (11). To investigate the role of SNHG20 in
gastric cancer further, SNHG20 expression was silenced in BGC-823
and AGS gastric cancer cell lines in the current study. The
expression of NDRG3 was decreased following SNHG20 knockdown when
compared with the controls (Fig. 3A and
B). In addition, the correlation between NDRG3 and SNHG20 mRNA
levels in clinical specimens derived from patients with gastric
cancer was analyzed using the GEPIA web tool. Consistent with the
results observed in BGC-823 and AGS cells, NDRG3 was positively
correlated with SNHG20 (P=4×10−10, r=0.3; Fig. 3C). SNHG20 knockdown downregulated the
expression of NDRG3 in BGC-823 and AGS cells and this effect was
reduced following transfection of anti-miR-140-5p (Fig. 3D). Taken together, these results
suggest that the SNHG20/miR-140-5p axis may regulate the expression
of NDRG3 in gastric cancer.
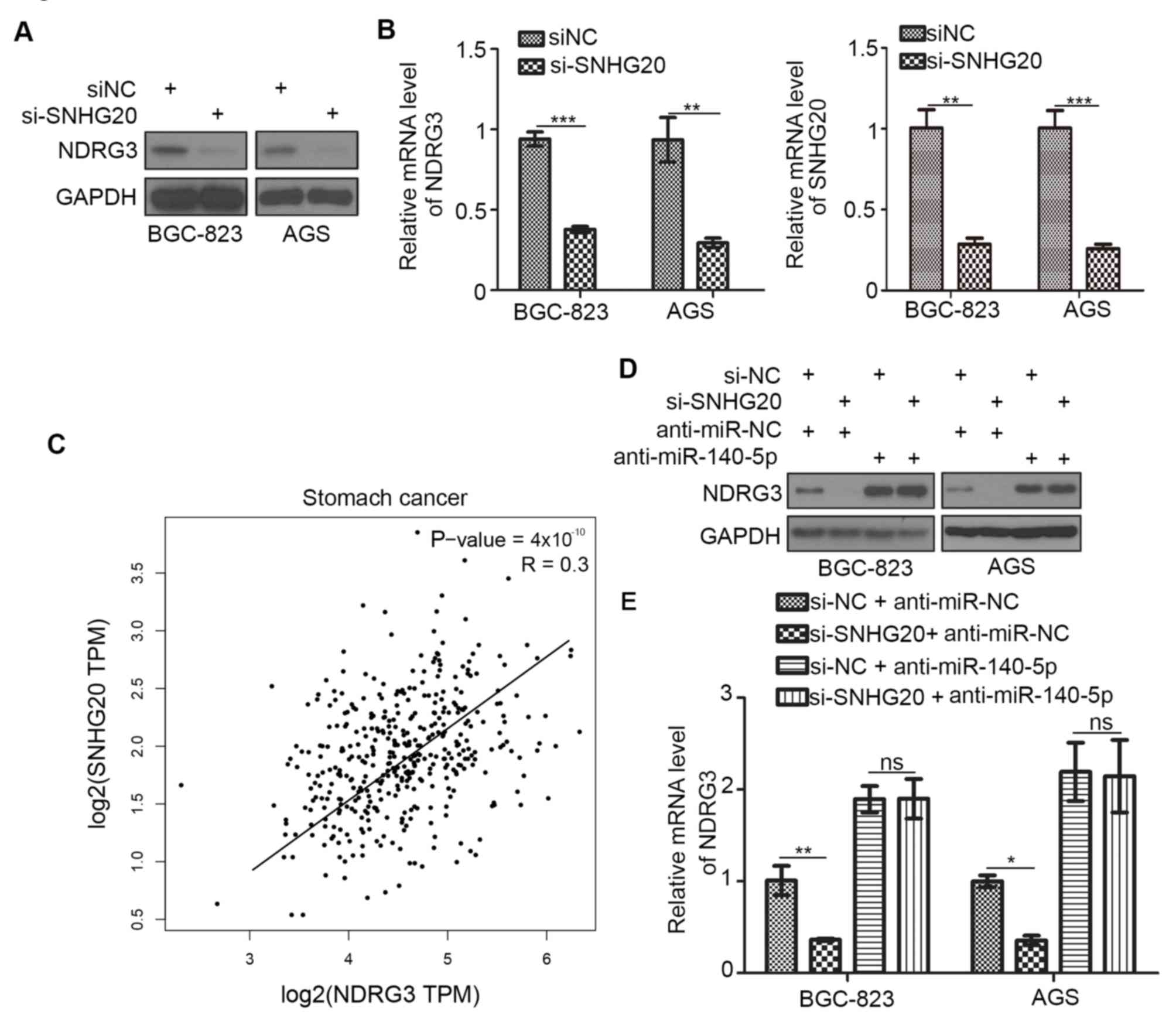 | Figure 3.SNHG20 regulates NDRG3 expression via
miR-140-5p in gastric cancer cells. BGC-823 and AGS cells were
transfected with the indicated constructs. A total of 48 h
following transfection, cells were harvested for (A) western
blotting and (B) RT-qPCR analyses. (C) The Gene Expression
Profiling Interactive Analysis web tool was used to determine the
correlation between the mRNA expression levels of NDRG3 and SNHG20
among patients with gastric cancer. BGC-823 and AGS cells were
transfected with indicated constructs. 48 h post transfection,
cells were harvested for (D) western blotting and (E) RT-qPCR
analyses. Data are presented as the mean ± standard deviation of
three replicates. *P<0.05, **P<0.01 and ***P<0.001, as
indicated. SNHG20; small nucleolar RNA host gene 20; NDRG3, N-myc
downstream regulated gene, family member 3; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; miR,
microRNA; ns, not significant; NC, negative control; TPM,
transcripts per million. |
SNHG20 contributes to 5-FU resistance
in gastric cancer cells by regulating the expression of NDRG3
The present study investigated whether the
SNHG20/NDRG3 axis may be responsible for mediating resistance to
5-FU in gastric cancer. The cell proliferation rate following
exposure to different concentrations of 5-FU following NDRG3
knockdown in BGC-823 and AGS cells was first measured (Fig. 4A and B). The half maximal inhibitory
concentration of 5-FU was decreased following knockdown of NDRG3 in
BGC-823 and AGS gastric cancer cells, and the viability of cells in
the shNDRG3 group was significantly decreased when compared with
that of the shControl group at a concentration of 5 µg/ml (Fig. 4A and B). Since Shen et al
(12) revealed that after 3 days of
treatment with 5-FU the viability of BGC-823 and AGS cells is
significantly decreased in the 5-FU group compared with the control
group, the difference in viability of cells transfected with
shControl and shNDRG3 was then compared at the same concentration
of 5-FU on day 3 (Fig. 4C and D).
Knockdown of NDRG3 significantly enhanced the sensitivity of
gastric cancer cell lines to 5-FU treatment. Knockdown of SNHG20
increased the sensitivity of the gastric cancer cell lines to 5-FU
treatment (Fig. 4C and D).
Co-knockdown of SNHG20 and NDRG3 did not further increase the
sensitivity to 5-FU compared with knockdown of either SNHG20 or
NDRG3 alone in the BGC-823 and AGS cells (Fig. 4C and D). The viability of cells in
the co-knockdown group was decreased when compared with the control
group in the two gastric cancer cell lines (Fig. 4C and D). Together, these data
indicated that SNHG20/NDRG3 axis may contribute to 5-FU resistance
in gastric cancer.
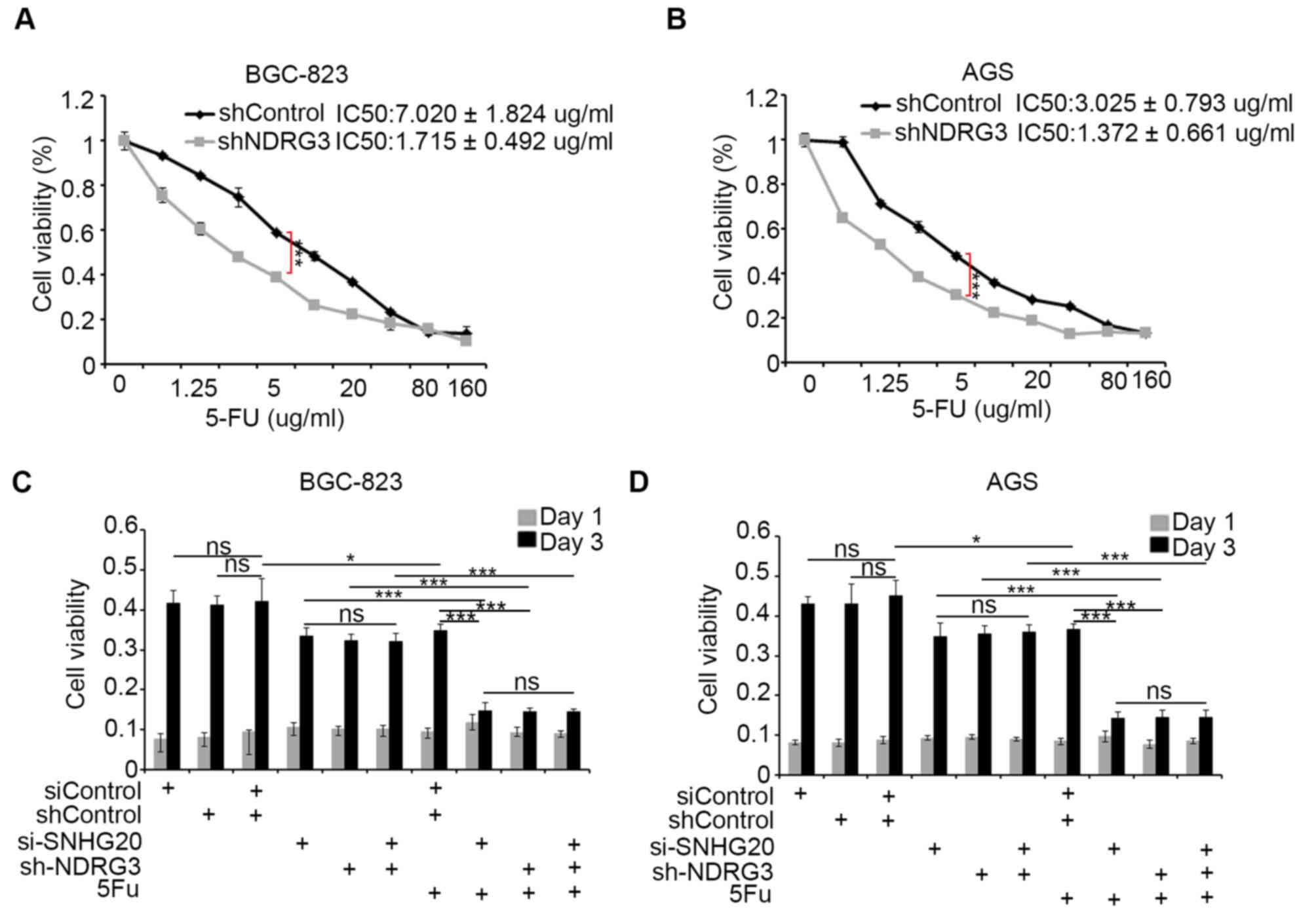 | Figure 4.SNHG20 may be involved in mediating
resistance to 5-FU in gastric cancer cells via NDRG3. (A) BGC-823
and (B) AGS cells were infected with lentivirus expressing control,
NDRG3-specific shRNAs. A total of 48 h following infection, cells
were treated with increasing concentrations of 5-FU for a further
24 h. Cells were subsequently harvested for the MTS assay. The cell
viability was compared at the concentration of 5 µg/ml in the two
gastric cancer cell lines. (C) BGC-823 and (D) AGS cells were
transfected with indicated constructs. A total of 48 h following
transfection, cells were treated with or without 2.5 µg/ml of 5-FU.
Cell viability was measured using the MTS assay. Data presented as
the mean ± standard deviation from five replicates; *P<0.05 and
***P<0.001, as indicated. SNHG20; small nucleolar RNA host gene
20; 5-FU, 5-fluorouracil; NDRG3, N-myc downstream regulated gene,
family member 3; shRNA, short hairpin RNA; IC50, half
maximal inhibitory concentration; ns, not significant. |
Discussion
The NDRG family serves important roles the
proliferation, invasion and metastasis of cells in solid tumors,
including breast, colon and prostate cancer (13,14).
However, the specific role of NDRG3 in gastric cancer remains
unclear. The results of the present study revealed that NDRG3 may
function as a tumor growth promoting protein in gastric cancer.
NDRG3 was previously reported to mediate drug resistance in
hepatocellular cancer (15).
Consistent with this aforementioned study, the results of the
present study suggest that NDRG3 may be involved in the development
of 5-FU resistance in gastric cancer.
lncRNAs are non-coding transcripts consisting of
>200 nucleotides (16). Aberrant
expression of lncRNAs is common in several types of cancer,
including gastric cancer, breast cancer and osteosarcoma, and may
be involved in chromatin remodeling and transcriptional and
post-transcriptional regulation (16–18). As
such, lncRNAs serve important roles in regulating the tumor cell
growth, metastasis and drug resistance of tumors (19). SNHG20 has been identified as an
oncogenic RNA in numerous types of cancer, including cervical
(10), breast (18) and colorectal cancer (20). SNHG20 decreased the expression of
CDKN1A and increased the proliferation and decreased apoptosis of
non-small cell lung cancer (21) and
gastric cancer cells (22). The
present study demonstrated that SNHG20 increased the expression
level of NDRG3 and that it may be involved in mediating resistance
to 5-FU in gastric cancer cell lines, indicating a novel role of
SNHG20 in tumorigenesis.
In conclusion, the present study revealed that NDRG3
was upregulated in gastric cancer samples compared with normal
healthy controls, and promoted the proliferation of gastric cancer
cell lines. Furthermore, the current study demonstrated that the
SNHG20/miR-140-5p signaling pathway may regulate the expression of
NDRG3. As different types of cancer may develop resistance to
traditional chemotherapies (23),
future studies investigating the mechanisms underlying drug
resistance may promote the development of novel treatment
strategies. The current study also revealed that the
SNHG20/miR-140-5p/NDRG3 axis may serve an important role in
mediating resistance to 5-FU in gastric cancer. Therefore,
therapeutic targeting of this axis may present a novel treatment
strategy to overcome 5-FU resistance in gastric cancer.
Acknowledgements
Not applicable.
Funding
The current study was supported by the National
Natural Science Foundation of Hubei, China (grant no.
2018CFC834).
Availability of data and materials
The datasets generated/analyzed in the present study
are available upon reasonable request from the corresponding
author.
Authors' contributions
JY, JS and XQ performed the experiments and wrote
the manuscript. LC, ZY, HY, CX, QZ and PW collected data. ZG wrote
the manuscript and analyzed the data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 scountries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen Y, Lian G, Ou G, Yang K, Chen J, Li
H, Chen S, Li J, Zeng L and Huang K: Inverse association between
Bmi-1 and RKIP affecting clinical outcome of gastric cancer and
revealing the potential molecular mechanisms underlying tumor
metastasis and chemotherapy resistance. Gastric Cancer. 19:392–402.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu ZY, Tang JN, Xie HX, Du YA, Huang L, Yu
PF and Cheng XD: 5-fluorouracil chemotherapy of gastric cancer
generates residual cells with properties of cancer stem cells. Int
J Biol Sci. 11:284–294. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Melotte V, Qu X, Ongenaert M, van
Criekinge W, de Bruïne AP, Baldwin HS and van Engeland M: The N-myc
downstream regulated gene (NDRG) family: Diverse functions,
multiple applications. FASEB J. 24:4153–4166. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cho Y, Yoon JH, Yoo JJ, Lee M, Lee DH, Cho
EJ, Lee JH, Yu SJ, Kim YJ and Kim CY: Fucoidan protects hepatocytes
from apoptosis and inhibits invasion of hepatocellular carcinoma by
up-regulating p42/44 MAPK-dependent NDRG-1/CAP43. Acta Pharm Sin B.
5:544–553. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li T, Sun R, Lu M, Chang J, Meng X and Wu
H: NDRG3 facilitates colorectal cancer metastasis through
activating Src phosphorylation. Onco Targets Ther. 11:2843–2852.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Luo X, Hou N, Chen X, Xu Z, Xu J, Wang L,
Yang S, Liu S, Xu L, Chen Y, et al: High expression of NDRG3
associates with unfavorable overall survival in non-small cell lung
cancer. Cancer Biomark. 21:461–469. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res 45 (W1).
W98–W102. 2017. View Article : Google Scholar
|
|
9
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo H, Yang S, Li S, Yan M, Li L and Zhang
H: LncRNA SNHG20 promotes cell proliferation and invasion via
miR-140-5p-ADAM10 axis in cervical cancer. Biomed Pharmacother.
102:749–757. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao L, Li C, Liu F, Zhao Y, Liu J, Hua Y,
Liu J, Huang J and Ge C: A blockade of PD-L1 produced antitumor and
antimetastatic effects in an orthotopic mouse pancreatic cancer
model via the PI3K/Akt/mTOR signaling pathway. Onco Targets Ther.
10:2115–2126. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shen X, Gu Y, Yu S, Gong P, Mao Y, Li Y,
Zheng Y, Qiao F, Zhao Z and Fan H: Silenced PITX1 promotes
chemotherapeutic resistance to 5-fluorocytosine and cisplatin in
gastric cancer cells. Exp Ther Med. 17:4046–4054. 2019.PubMed/NCBI
|
|
13
|
Kovacevic Z and Richardson DR: The
metastasis suppressor, Ndrg-1: A new ally in the fight against
cancer. Carcinogenesis. 27:2355–2366. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lin HC, Yeh CC, Chao LY, Tsai MH, Chen HH,
Chuang EY and Lai LC: The hypoxia-responsive lncRNA NDRG-OT1
promotes NDRG1 degradation via ubiquitin-mediated proteolysis in
breast cancer cells. Oncotarget. 9:10470–10482. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Du Z, Niu S, Xu X and Xu Q:
MicroRNA31-NDRG3 regulation axes are essential for hepatocellular
carcinoma survival and drug resistance. Cancer Biomark. 19:221–230.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang W, Luo P, Guo W, Shi Y, Xu D, Zheng H
and Jia L: LncRNA SNHG20 knockdown suppresses the osteosarcoma
tumorigenesis through the mitochondrial apoptosis pathway by
miR-139/RUNX2 axis. Biochem Biophys Res Commun. 503:1927–1933.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB,
Yin DD, Kong R, Xia R, Lu KH, Li JH, et al: Lnc RNA HOTAIR
functions as a competing endogenous RNA to regulate HER2 expression
by sponging miR-331-3p in gastric cancer. Mol Cancer. 13:922014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guan YX, Zhang MZ, Chen XZ, Zhang Q, Liu
SZ and Zhang YL: Lnc RNA SNHG20 participated in proliferation,
invasion, and migration of breast cancer cells via miR-495. J Cell
Biochem. 119:7971–7981. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li C, Zhou L, He J, Fang XQ, Zhu SW and
Xiong MM: Increased long noncoding RNA SNHG20 predicts poor
prognosis in colorectal cancer. BMC Cancer. 16:6552016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen Z, Chen X, Chen P, Yu S, Nie F, Lu B,
Zhang T, Zhou Y, Chen Q, Wei C, et al: Long non-coding RNA SNHG20
promotes non-small cell lung cancer cell proliferation and
migration by epigenetically silencing of P21 expression. Cell Death
Dis. 8:e30922017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu J, Liu L, Wan JX and Song Y: Long
noncoding RNA SNHG20 promotes gastric cancer progression by
inhibiting p21 expression and regulating the GSK-3β/β-catenin
signaling pathway. Oncotarget. 8:80700–80708. 2017.PubMed/NCBI
|
|
23
|
Housman G, Byler S, Heerboth S, Lapinska
K, Longacre M, Snyder N and Sarkar S: Drug resistance in cancer: An
overview. Cancers (Basel). 6:1769–1792. 2014. View Article : Google Scholar : PubMed/NCBI
|