Introduction
Colorectal cancer (CRC) is one of the most common
malignant tumors of the digestive system worldwide (1). In the United States in 2017, an
estimated 135,430 individuals were newly diagnosed with CRC, and
there were 50,260 mortalities due to the disease (2). According to latest tumor statistics,
CRC is the fourth most common form of cancer and the fifth most
frequent cause of cancer-related mortality in China (3). Tumor metastasis is the leading cause of
mortality; CRC has a high probability of invasion and metastasis,
which makes the prognosis of this disease very poor (4). Multiple genes and cellular pathways
have been demonstrated to participate in the process of metastasis
(5). To date, although there are
extensive studies relating to the molecular mechanisms of CRC onset
and progression, the precise mechanisms and potential targets for
therapy are not clear.
In CRC, lymph node metastasis, which is the first
step of distant metastasis, is a marker for prognosis. Firstly,
lymph node metastasis is a factor of the American Joint Committee
on Cancer/Union for International Cancer Control
tumor-node-metastasis (TNM) system, which is the most trusted
classification currently used to determine cancer treatment and
prognosis. In addition, lymph node metastasis is a critical
diagnostic indicator that the patient should receive adjuvant
chemotherapy after resection (6). As
lymph node metastasis promotes the malignant progression of
colorectal cancer, understanding the molecular mechanism is
critical and demanded in this field.
MicroRNAs (miRNAs) are small noncoding RNAs, ~22
nucleotides long, that regulate target gene expression by binding
to different regions of mRNAs, including 3′ untranslated regions
(3′UTRs), 5′UTR and protein-coding sequences (7,8). miRNA
expression is tissue-specific (9,10); one
miRNA can regulate multiple genes, and multiple miRNAs can also
regulate the same gene, which constitutes a complex network of
miRNAs and mRNAs in the development of disease (11,12). In
cancer, the miRNA-mRNA network can modulate cellular proliferation,
differentiation and metastasis (13)
Thus, the study of the miRNA-mRNA network is important to
understand the mechanism of CRC development.
The Cancer Genome Atlas (TCGA) project is a
large-scale effort, which aims to identify changes in each type of
cancer to understand how these changes interact with each other to
drive the disease pathogenesis. TCGA data, which contains clinical
information about participants and molecular information derived
from samples, such as mRNA and miRNA expression, protein
expression, copy number and methylation, are accessible to the
public and have been used widely in previous studies (14). Analyzing TCGA data comprehensively
may be a crucial step in improving cancer prevention, early
detection and treatment.
In the present study, the original CRC
RNA-sequencing data and miRNA isoform profiles were downloaded from
the TCGA database. The gene and miRNA signatures associated with
lymph node metastasis were analyzed, and a miRNA-mRNA network was
constructed, which revealed a mechanism involved in lymph node
metastasis in CRC.
Materials and methods
Identification of differentially
expressed genes (DEGs)
Public TCGA colon adenocarcinoma (TCGA-COAD) (v9.0;
http://portal.gdc.cancer.gov/repository) level-3
RNA-sequencing data repositories (including miRNA-sequencing) were
downloaded using the Genomics Data Commons data transfer tool
(https://gdc.cancer.gov/access-data/gdc-data-transfer-tool).
For RNA sequencing analysis, the samples were divided into two
groups based on clinical records: 157 samples with lymph node
metastasis and 230 samples with no lymph node metastasis. The
fragments per kilobase of transcript per million mapped
reads-normalized RNA sequencing data were analyzed using Ballgown
package in R (Bioconductor) (https://bioconductor.org/packages/release/bioc/html/ballgown.html)
to identify DEGs. Statistically significant DEGs were defined with
P<0.05 and fold change (FC)>1.2 or FC<0.8. DEGs were
presented as a volcano plot and a heatmap. For miRNA profiling, the
changes of miRNA isoform expression were compared between samples
with lymph node metastasis and no lymph node metastasis by Wilcoxon
rank-sum test. Differentially expressed (DE) miRNAs were defined as
absolute log2(FC) >0.5 and P<0.05.
Target genes of DE miRNAs and
transcription factors (TFs) for DEG screening
Two integrated online databases, TarBase v.8
(15) and miRTarBase (16), were used to evaluate the
miRNA-regulating mRNAs. In these databases, the associations
between miRNAs and mRNAs were validated experimentally (15–17). In
TarBase v8.0, the screening criteria for miRNA-regulated mRNA was
Argonaute (AGO) immunoprecipitation, luciferase reporter assay,
quantitative PCR (qPCR) or western blot. In miRTarBase, the
screening criterion was strong evidence (luciferase reporter assay,
qPCR or western blot). The association map between DEGs and TFs was
evaluated, constructed and visualized by Generadar (https://www.gcbi.com.cn/gcanalyze/html/generadar/index).
Pathway and process enrichment
analysis of DEGs or DE miRNA targets
Pathway and process enrichment analysis is a useful
method for annotating genes, identifying enriched biological themes
and manually drawn pathway maps that represent the knowledge of
molecular interactions, reactions and relations. To analyze the
DEGs or DE miRNA targets at the functional level, pathway and
process enrichment analyses were performed by Metascape (http://metascape.org) online tool. Log10
(P-value)<-2 was considered to indicate a statistically
significant difference, and q-value was used as a reference. For DE
miRNA targets and modules, the relationships among enriched terms
were analyzed and presented as a network plot, where terms with
similarity >0.3 were connected by edges. For DEGs, the network
was constructed by custom analysis, and the standard was Min
Overlap 10, P-value cutoff=0.001 and Min Enrichment=1.5. The
network was visualized using Cytoscape (3.7.0; http://cytoscape.org/).
Protein-protein interaction (PPI)
network analysis and module screening
To evaluate the interactive relationships among
DEGs, the STRING database (https://string-db.org) was used. Interaction networks
were constructed using the Cytoscape software. The plug-in
Molecular Complex Detection (MCODE; http://apps.cytoscape.org/apps/mcode) was used to
calculate the node degree. Central node genes were identified with
the filtering of degree >10. Furthermore, significant modules
were screened with the criteria of MCODE scores >3 and number of
nodes >4. Pathway and process enrichment analysis were performed
for DEGs in the identified modules.
Results
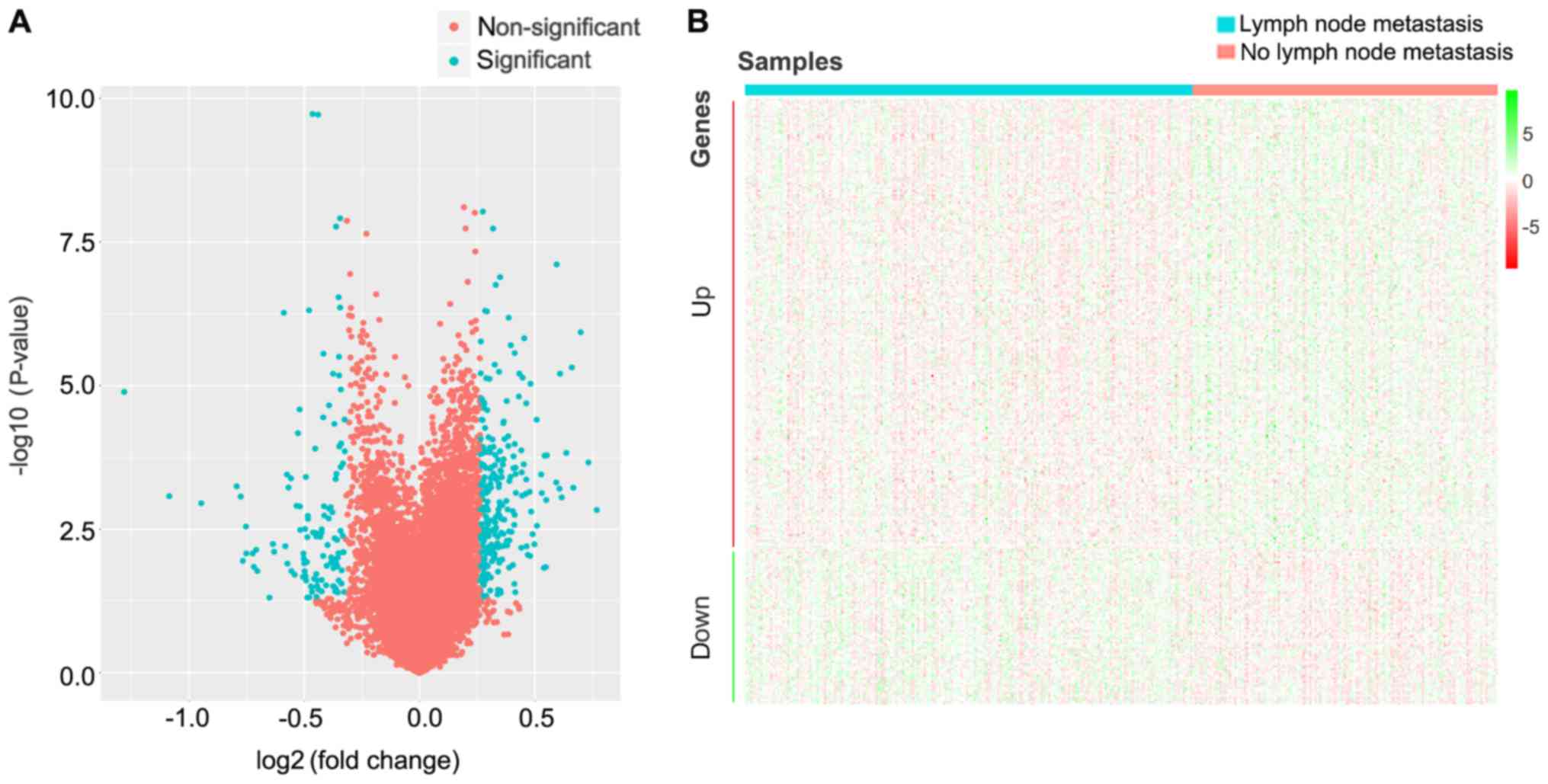
Identification of DEGs
The changes of gene expression levels associated
with lymph node metastasis were analyzed by Ballgown package in R.
Using P<0.05 and FC>1.2 (upregulated) or FC<0.8
(downregulated) as thresholds for significance, 305 DEGs were
screened, which included 227 upregulated and 78 downregulated genes
in patients with CRC lymph node metastasis compared with patients
with CRC without lymph node metastasis (Fig. 1; Table
I).
 | Table I.DEGs in patients with CRC lymph node
metastasis compared with patients with CRC without lymph node
metastasis (corresponding to heatmap in Fig. 1B from top to bottom). |
Table I.
DEGs in patients with CRC lymph node
metastasis compared with patients with CRC without lymph node
metastasis (corresponding to heatmap in Fig. 1B from top to bottom).
| DEG expression | Gene |
|---|
| Upregulated | DES, MSLN,
SFTA2, MIR675, COL9A3, WNT11, TACSTD2, C6orf15, GNG4, TNNC1, MYH11,
APOD, ACTG2, LY6G6D, MMP11, KRT23, CNN1, RBP4, PRAP1, PMEPA1,
TNNC2, SFRP4, PCSK1N, CACNG4, MGP, UCA1, CES1, TPM2, EDAR, TAGLN,
GRP, LMOD1, SERPINE2, MYL9, PRELP, COMP, CAPS, VIP, ACTA2, PKDCC,
CST1, CST2, GTF3A, MIR3131, NPTX2, CRABP2, GGT7, AQP1, QPRT, CHP2,
LGR6, PPL, PLA2G16, ELN, ASPN, BMP7, NKD2, DDAH2, CDKN2A, AMIGO2,
S100A4, SLCO4A1-AS1, MAP7D2, FAM127B, PPP1R14A, MOGAT3, ISLR,
PLEKHA4, S100A2, SQLE, RGCC, CARD11, PALM3, MFAP4, FABP3, CAB39L,
MXRA8, FADS2, SYNM, HSPB8, RAMP1, MIR936, HSPB1, FAM127A, NKD1,
NKILA, CDKN1C, AKAP12, SLC2A1, BGN, EPB41L1, HTRA1, PDX1, TNS1,
FOXQ1, L1CAM, CTSF, MIR4649, COL10A1, AKR1C3, KRT17, HSPH1, FKBP10,
LEMD1, TIMP2, AOC3, CRYAB, GJB3, MLXIPL, FHL1, CDIP1, SYT7,
LINC00543, SLPI, DSG3, HTRA3, VSTM2L, ARHGDIG, FREM1, UCHL1, PTK7,
B3GALT4, SPARCL1, LAPTM4B, KISS1, ANTXR1, DPYSL3, TPPP3, LTBP3,
LTBP1, MEGF6, GPC1, ECHDC3, MFAP2, AHNAK2, EDN1, FOLR1, YPEL3,
GLIS2, PRSS8, TGFB1I1, MUC20, PLN, EPDR1, IGFBP6, MIR4758, MAB21L2,
HSPB7, OSER1-AS1, MLF1, GDPD3, FBLN1, THBS4, SLC22A3, MAPRE3,
PLEKHB1, FAM234A, MRGPRF, SYNPO2, MDFI, FAM127C, FNDC1, KRT80,
BCAM, CST6, LYPD3, SULT2B1, TSC22D3, PLTP, EEF1A2, PRR15, DHRS12,
UPK2, NES, MYLK, NTSR1, MIR647, BAIAP2, C15orf52, FXYD6, SLC39A4,
PPDPF, TM4SF20, HSPA1A, SLC14A1, HIST1H2AC, PBXIP1, AZGP1, EEPD1,
HIST2H2BE, FER1L4, PTK6, IHH, MTIF3, EMILIN1, COL8A1, FKBP9,
C1QTNF12, SERP2, ARHGEF25, DDX27, LBH, FLNA, NINL, FSTL3, SSC5D,
LINC01006, HIST1H2BD, MSRB3, SYT1, CYB5B, IGFL1, C2orf54, MMP14,
TMEM139, ISM2, RGL2, NR1D1, MOSPD3, ANXA9, CALB2, DKK1, SLC35D3,
COL5A1, MAGEA6, VPS37D, PDLIM4 |
| Downregulated | FAM46C, RPL22L1,
C8orf4, LRRC26, CCL4L2, SLC12A2, IL1B, NAT1, INTS10, LOC101928100,
TYMS, FAS, CDCA2, CXCL8, FOS, RARRES3, B3GNT6, CD74, FAM26F, MT1E,
TNFRSF10A, IDO1, GBP1, ALDOB, EPHB3, PLAC8, CCL4, TNFRSF11A, FOXA1,
CES3, SLC39A8, GBP4, SLC6A14, HLA- DRA, IGLL5, CXCL3, CCL5, GNLY,
HLA-DRB5, GZMB, CXCL1, ST6GALNAC1, AGPAT5, ATOH1, CCL25, GZMA, GSR,
PBK, UBD, HEPACAM2, CA2, JCHAIN, KIAA1324, NOS2, MUC5B, L1TD1,
CXCL9, C2CD4A, CXCL2, SERPINA1, CXCL11, CXCL10, MMP12, CASP1,
REG1B, OLFM4, REG3A, ITLN1, DEFA5, DEFA6, SPINK4, REG4, HULC,
FCGBP, CLCA1, REG1A, PIGR |
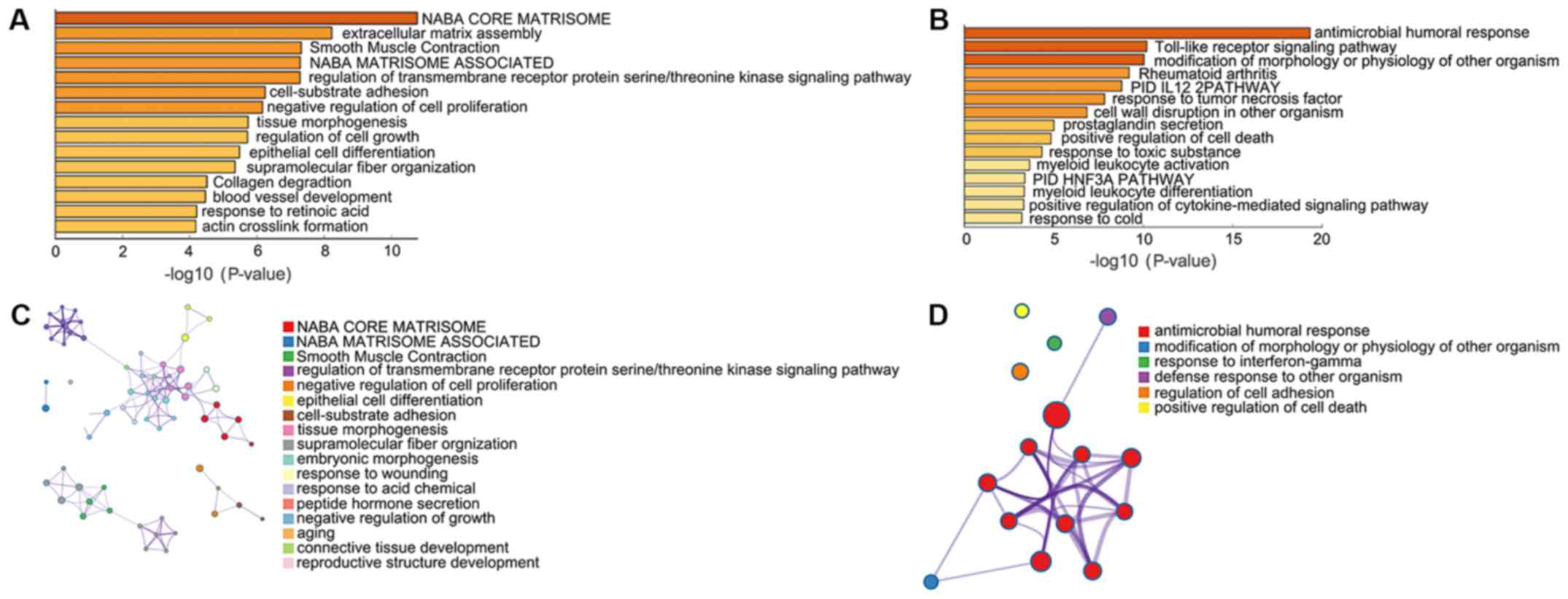
Pathway and process enrichment
analysis
Pathway and process enrichment analyses were
performed by Metascape, including Kyoto Encyclopedia of Genes and
Genomes pathway analysis, Gene Ontology terms in biological
processes, Reactome Gene Sets, Canonical Pathways and Comprehensive
resource of mammalian protein complexes (CORUM). Upregulated DEGs
were significantly enriched in ‘NABA CORE MATRISOME’,
‘extracellular matrix assembly’, ‘Smooth Muscle Contraction’ ‘NABA
MATRISOME ASSOCIATED’, ‘regulation of transmembrane receptor
protein serine/threonine kinase signaling pathway’ and
‘cell-substrate adhesion’ (Fig. 2A and
B). Downregulated DEGs were significantly enriched in
‘antimicrobial humoral response’, ‘Toll-like receptor signaling
pathway’, ‘modification of morphology or physiology of other
organism’, ‘Rheumatoid arthritis’, ‘PID IL12 2PATHWAY’ and
‘response to tumor necrosis factor’ (Fig. 2C and D).
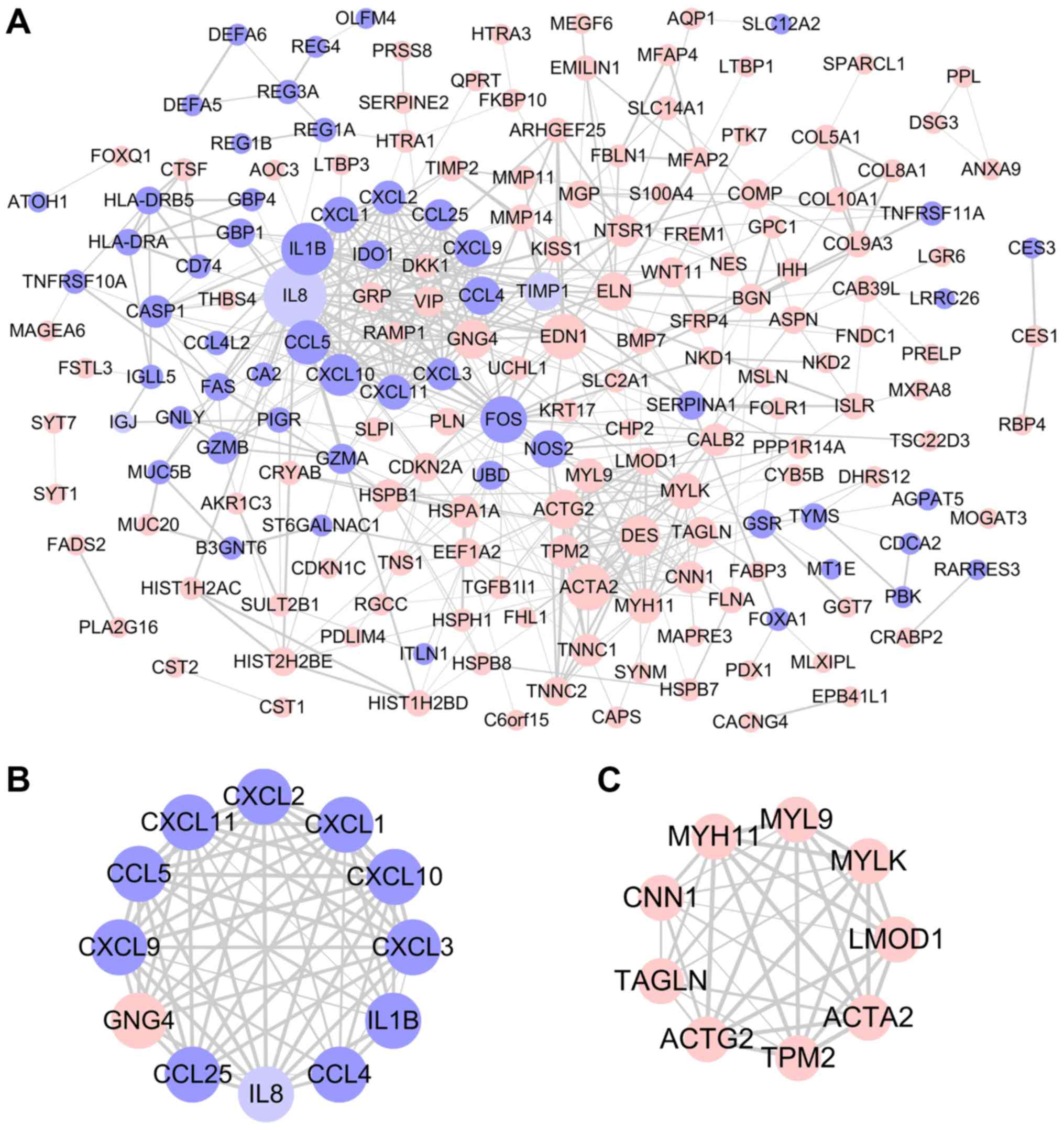
Key candidate genes and pathways
identified by PPI network
Using the STRING database and Cytoscape software, a
total of 183 DEGs were filtered into the PPI network complex,
including 186 nodes and 507 edges (Fig.
3A). With the filtering of degree >10, a total of 29 central
node genes were identified, of which the 10 most significant hub
genes were interleukin 1β (IL1B), actin α2, smooth muscle
(ACTA2), Fos proto-oncogene, AP-1 transcription factor
subunit, endothelin 1, C-C motif chemokine ligand 5 (CCL5),
C-X-C motif chemokine ligand 10 (CXCL10), desmin
(DES), actin γ, smooth muscle (ACTG2), G protein
subunit γ4 and CCL4. In addition, the top two significant
modules from the PPI network were selected for pathway and process
enrichment analyses. Module 1, consisting of 12 nodes and 62 edges,
was mainly associated with ‘neutrophil chemotaxis’ and ‘cellular
response to lipopolysaccharide’ (Figs.
3B and S1A). Module 2,
consisting of 9 nodes and 35 edges, was mainly associated with
‘smooth muscle contraction’ (Figs.
3C and S1B).
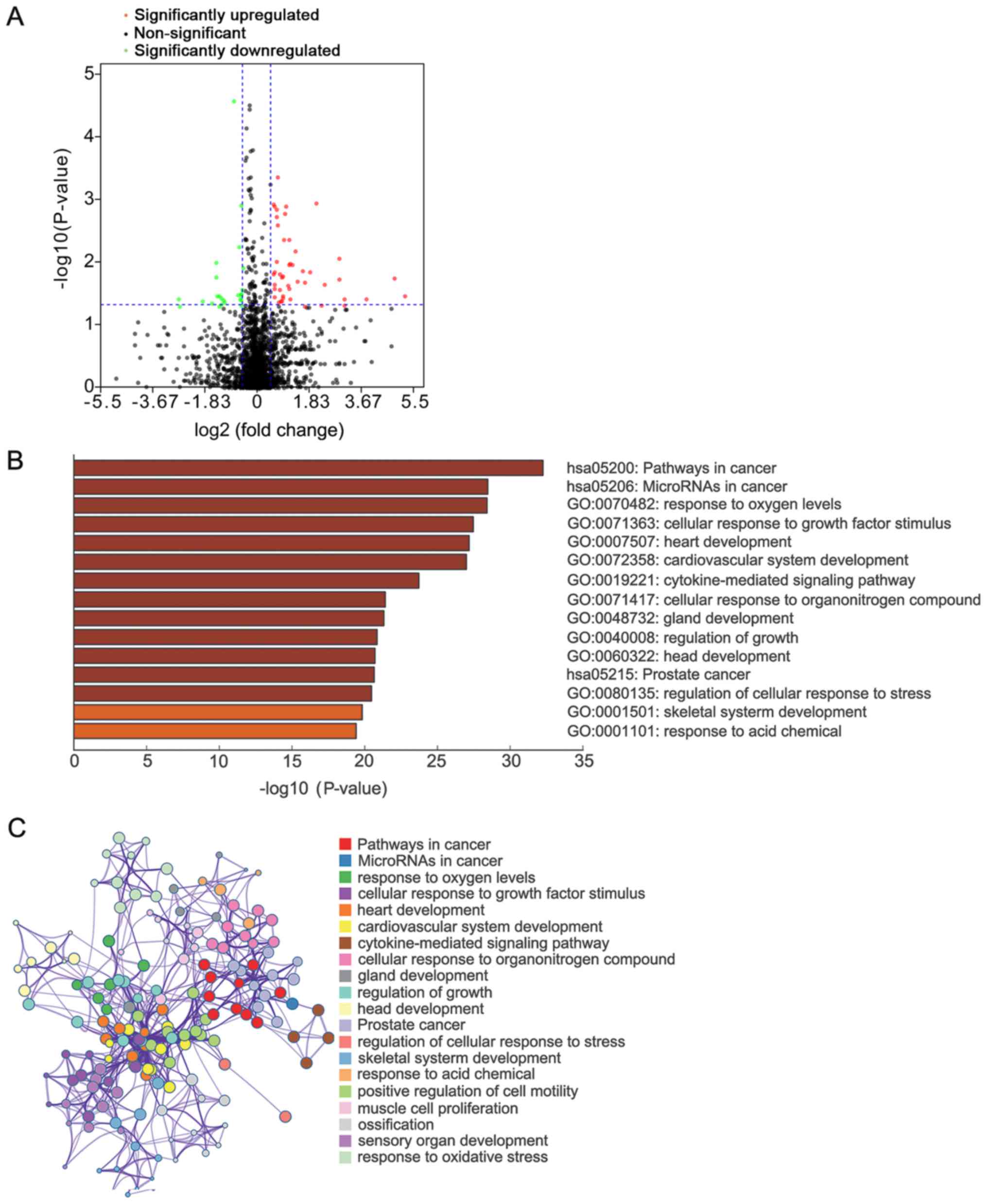
Identification of DE miRNAs
miRNAs have a regulatory role by guiding AGO
proteins to target mRNAs. To discover upstream miRNAs that regulate
DEG expression, the expression profiles of miRNA isoforms from the
TCGA database were analyzed, and miRNA signatures associated with
lymph node metastasis were screened. A total of 73 mature miRNAs
were discovered, of which 48 were upregulated and 25 were
downregulated (Fig. 4A). The targets
of mature DE miRNAs with strong experimental evidence were screened
through TarBase v.8 and miRTarBase databases (Fig. S2). These targets were significantly
enriched in ‘pathways in cancer’, ‘microRNAs in cancer’, ‘response
to oxygen levels’ and ‘cellular response to growth factor stimulus’
(Fig. 4B and C).
miRNA-mRNA network
There are two distinct ways of miRNA regulation:
Silencing expression of target mRNAs by binding to 3′untranslated
region (UTR) or protein-coding sequences, or upregulation of
targeted mRNAs by binding to 5′UTR (7,8,18). Therefore, changes in DEG expression
may be induced by the regulation of miRNAs. Nine pairs of DE
miRNA-DEG regulatory networks were identified (Table II). In mammals, translation
repression is the main mode of miRNA regulation. Changes in protein
levels of certain TFs may occur through translational regulation by
miRNAs, which triggers differences in expression levels of
downstream genes. TFs that regulate DEG expression were analyzed
using PubMed and Transfac databases. The relationship between TFs
and DEGs in the PubMed and Transfac databases is documented and is
credible. Subsequently, a TF-DEG network was constructed by
Generadar (Fig. 5A). By analyzing
the relationship between these TFs and DE miRNAs, a DE miRNA-TF-DEG
network related to lymph node metastasis of CRC was constructed
(Fig. 5B). In this network, miRNA
(miR)-612, miR-1-3p, miR-133b and miR-133a-3p jointly inhibited
translational regulation of the TF specificity protein 1
(SP1), and Sp1 further induced changes in the expression of
downstream DEGs. In addition, tumor protein p53 (TP53), a
well-known suppressor gene, was also demonstrated to be involved in
the regulation of DEGs.
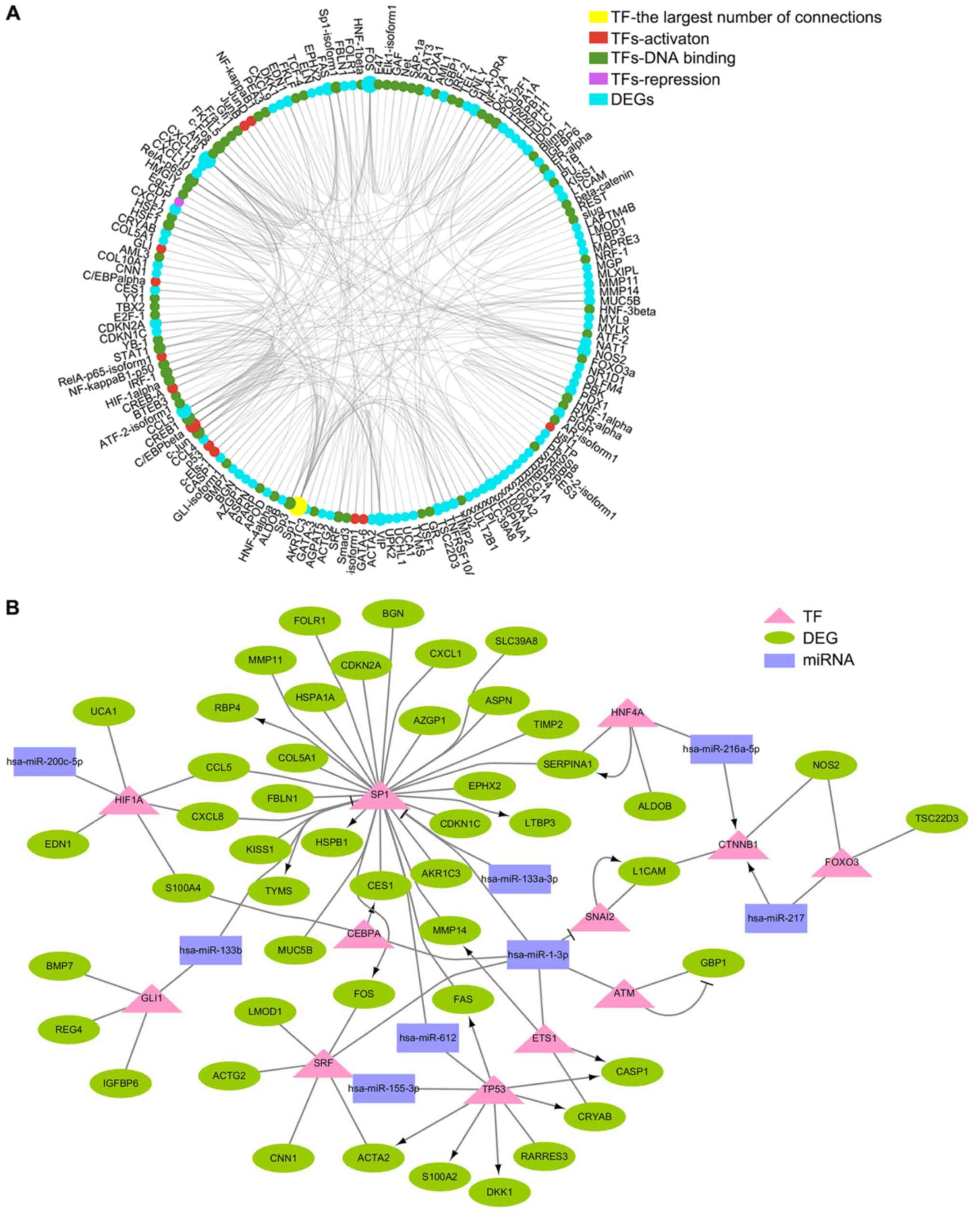 | Figure 5.DE miRNA-TF-DEG network associated
with lymph node metastasis of colorectal cancer. (A) Network
between TFs and DEGs was constructed by Generadar. Red, activation
TF; green, DNA binding TF; magenta, repression TF; cyan, DEG. (B)
DE miRNA-TF-DEs network related to lymph node metastasis of
colorectal cancer was constructed by Cytoscape. Purple rectangle,
DE miRNA; pink triangle, TF; green ellipse, DEG. DE, differentially
expressed; DEG, DE gene; miRNA, microRNA; TF, transcription
factor. |
 | Table II.miRNA-mRNA network associated with
lymph node metastasis in CRC. |
Table II.
miRNA-mRNA network associated with
lymph node metastasis in CRC.
| miRNA | Gene |
|---|
| hsa-miR-767-5p | COL10A1 |
|
hsa-miR-487b-5p | NES |
| hsa-miR-217 | CXCL2 |
| hsa-miR-1-3p | RPL22L1, EDN1,
FABP3, |
| hsa-miR-133b | FAS, MMP14 |
Discussion
CRC is a complex disease caused by genetic,
epigenetic and somatic aberrations (19). Understanding the molecular mechanisms
and finding biomarkers of CRC progression are of importance for
improving patient survival rate. Potential therapeutic targets for
CRC may be predicted by developing high-throughput sequencing. In
the present study, key candidate genes and pathways that may serve
important roles in CRC metastasis were identified by bioinformatics
analysis. Additionally, a network between DEGs and miRNAs, which
may participate in lymph node metastasis in CRC, was
constructed.
In the present study, 305 DEGs were screened by
bioinformatics analysis. Due to the large sample size and
individual differences, the homogeneity within the groups is poor,
and the FC of DEGs was relatively small, but the results are
credible. For example, high expression of desmin (DES), one of
DEGs, as observed in the present study, was also found to be
associated with liver metastasis and decreased survival rate of CRC
patients in a previous study (20).
Another study also demonstrated a remarkably high DES expression in
patients with advanced CRC compared with patients with early stage
CRC (21). Polymeric immunoglobulin
receptor (PIGR) is another differently expressed gene identified in
the present study. Traicoff et al demonstrated that PIGR
levels were significantly lower in CRC tissues compared with
non-tumor tissues (22). Agesen
et al also indicated that metastatic colorectal cancer had
lower PIGR expression levels compared with stage I CRC (23). The consistency of the results between
the present and previous studies indicated the feasibility of the
study methods and reliability of the results of the present
study.
Lymph node metastasis is an indicator of distant
tumor metastasis and an important part of TNM staging. It has a
significant reference value for patient treatment and prognosis
(6). However, the mechanism of lymph
node metastasis remains largely unknown. In the present study,
upregulated genes were mainly enriched in ‘NABA CORE MATRISOME’,
‘extracellular matrix assembly’ and ‘Smooth Muscle Contraction’,
whereas downregulated genes were mainly enriched in ‘antimicrobial
humoral response’, ‘Toll-like receptor signaling pathway’ and ‘PID
IL12 2PATHWAY’. Extracellular matrix changes serve a vital role in
tumor migration and invasion by changing the cytoskeleton and
inducing epithelial-mesenchymal transition (EMT) (24,25).
DEGs that are located in the extracellular matrix are matrix
metalloproteinase 11 (MMP11) and MMP14. These
metalloproteinases induce tumor cell metastasis by degrading the
extracellular matrix (26). In CRC,
MMP11 and MMP14 also mediate cell invasion and
metastasis (27,28).
In the present study, the majority of the
downregulated genes were associated with chemokines, including
CXCL1, CXCL2, CXCL3, CXCL8, CXCL9, CCL4, CCL5 and CCL25. This
result was consistent with a previous study where CXCL3 was
downregulated in liver metastasis compared with primary colon
cancer (29). Genes that participate
in smooth muscle contraction are closely related to cytoskeleton
rearrangement and may induce tumor metastasis (25,30). In
addition, ACTG2 and ACTA2 are two members of the
actin family and serve an important role in cell motility (31,32).
Downregulated DEGs were significantly enriched in
‘antimicrobial humoral response’. In the gut, Salmonella is
a common cause of bacterial infection (33). A previous study has demonstrated that
Salmonella infection induces proliferation of epithelial
cells in the small intestine and colon (34). In addition, the S. enterica
effector avirulence protein A promotes colonic tumorigenesis
(34); therefore,
bacterial-infection related genes may be upregulated in the
occurrence of CRC, whereas the expression of these genes may be
decreased in lymph node metastasis.
A total of 29 central node genes were identified by
PPI analysis, and two significant modules were chosen. The first
module consisted of 12 genes, including IL1B, CCL4, CXCL9,
CXCL11, CXCL3, CXCL10 and CCL25. These genes are
associated with inflammation-related signaling pathways (35). Inflammatory bowel disease is a risk
factor for colon cancer (36). A
large number of microorganisms in the intestine affect gene
expression in human intestinal cells. For example,
Bifidobacterium, Lactobacillus and certain other strains
affect toll-like receptor (TLR) gene expression in
macrophages and dendritic cells (37). TLR signaling initiates an immune
defense mechanism that prevents invasion of microorganisms
primarily through the production of proinflammatory cytokines and
the promotion of the barrier function (38). Continued cascade of inflammatory
signals leads to proliferation, angiogenesis, apoptosis inhibition
and growth factor secretion, which initiate the occurrence of CRC
(39). In the present study, the
majority of these genes were downregulated, which suggested
differential gene expression patterns of metastatic carcinoma
compared with primary CRC.
Another module consisted of 9 genes, including
ACTA2, calponin 1, myosin heavy chain 11 and leiomodin 1.
The upregulation of these genes was associated with ‘smooth muscle
contraction’ and ‘vascular smooth muscle contraction,’ indicating
potential cytoskeleton changes. As cytoskeleton remodeling induces
EMT (24,26), and EMT is a means for tumor cells to
acquire invasive and metastatic abilities (40), the results of the present study
indicate that CRC may promote lymph node metastasis through
EMT.
MiRNAs are involved in tumorigenesis (41). In the present study, 73 DE miRNAs
were identified. ‘Response to oxygen levels’ was one of the
pathways associated with target genes of DE miRNAs. Tumors are
multicellular, heterogeneous entities composed of interacting tumor
cells and mesenchymal cells (42).
In addition, soluble molecules, such as oxygen, are involved in the
development of tumors (42,43). Environmental oxygen levels impact
cancer cell metabolism, resulting in changes in the expression of
several genes at the organism and cellular level (43). Hypoxia is a common phenomenon in
solid tumors, which triggers many hypoxic stress reactions. The
discovery of the hypoxia inducible transcription factor (HIF) has
promoted the understanding of the hypoxia response (44). The hypoxia response induced by HIFs
promotes cell survival and energy conservation (44). In addition, hypoxia promotes reactive
oxygen species (ROS) release (45).
Inokuma et al demonstrated that serum ROS levels are
associated with tumor size and lymph node metastasis (46). Lin et al indicated that ROS
levels involved in tumor lymphangiogenesis are mediated by
lysophosphatidic acid receptor (LPA)1/LPA3 signaling (47). In addition, high oxygen levels can
kill tumor cells in the lungs (48).
Under this pressure, tumor cells may reduce the damage caused by
oxygen through N-ethyl-maleimide sensitive fusion protein (49). The results of the present study
indicated that the response to oxygen levels may be an important
step in lymph node metastasis.
miRNAs regulate the expression of target genes in
multiple ways (7,8,18). In
the present study, nine pairs of DE miRNA-DEG mRNA associated with
lymph node metastasis in CRC were identified; others may form DE
miRNA-TF-DEG networks through translational inhibition of TFs,
which is consistent with other studies (49). In the present study, SP1 was
at the center of the DE miRNA-TF-DEG network. TF Sp1 is a zinc
finger TF that binds to GC-rich motifs of a number of promoters.
The results of the present study indicated that miR-612, miR-1-3p,
miR-133b and miR-133a-3p may synergistically regulate SP1
expression at the translational level, and subsequently Sp1 may
further adjust MMP11 and MMP14 expression at the
transcriptional level to promote the invasion and metastasis of
CRC. TP53 is a tumor suppressor gene, and its expression
changes promote occurrence and progression of CRC (50). The data from the present study
suggested that miR-155-3p and miR-612 may be involved in the
regulation of TP53 expression, and may participate in the
malignant development of colorectal cancer through
TP53-mediated ACTA2 and S100 calcium-binding protein
A2. Therefore, the networks between miRNAs and mRNAs may serve
important roles in lymph node metastasis of CRC.
In summary, using the TCGA database and integrated
bioinformatics analysis, the present study identified 305 DEGs
during lymph node metastasis in CRC, filtered 183 gene nodes in
DEGs to construct a PPI network complex, and identified two of the
most significant modules in the PPI network. Lymph node
metastasis-related DE miRNAs were further analyzed, and a
miRNA-mRNA network was constructed, which partially revealed the
mechanism of lymph node metastasis of CRC. However, molecular
biology experiments are required to confirm the results of the
present study.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by The National Science
Foundation for Young Scientists of China (grant no. 81802415 to
YZ), Shandong Provincial Natural Science Foundation (grant no.
ZR2018PH025 to YZ), The Doctoral Scientific Fund Project of the
Affiliated Hospital of Qingdao University (grant no. 2796 to QJ),
Clinical Medicine + X Project, Medical College, Qingdao
University.
Availability of data and materials
CRC level-3 RNA-sequencing data repositories
(including microRNA-sequencing) are available for download from the
The Cancer Genome Atlas dataset (http://cancergenome.nih.gov).
Authors' contributions
BS designed and supervised the study, interpreted
the results and wrote parts of the manuscript. QJ and YJZ managed
the project, supervised statistical analyses and drafted the
initial manuscript. YD designed the study. CC constructed the
network between miRNA and mRNA. SZ and YY performed statistical
analyses and PPI analyses. PL and DG screened the miRNA regulatory
mRNAs and transcription factors for DEGs. All authors read and
approved the final manuscript.
Ethics approval and consent for
participation
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu K, Yao H, Wen Y, Zhao H, Zhou N, Lei S
and Xiong L: Functional role of a long non-coding RNA
LIFR-AS1/miR-29a/TNFAIP3 axis in colorectal cancer resistance to
pohotodynamic therapy. Biochim Biophys Acta Mol Basis Dis.
1864:2871–2880. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RGS, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stintzing S, Tejpar S, Gibbs P, Thiebach L
and Lenz HJ: Understanding the role of primary tumour localisation
in colorectal cancer treatment and outcomes. Eur J Cancer.
84:69–80. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shinagawa T, Tanaka T, Nozawa H, Emoto S,
Murono K, Kaneko M, Sasaki K, Otani K, Nishikawa T, Hata K, et al:
Comparison of the guidelines for colorectal cancer in Japan, the
USA and Europe. Ann Gastroenterol Surg. 2:6–12. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang K, Zhang X, Cai Z, Zhou J, Cao R,
Zhao Y, Chen Z, Wang D, Ruan W, Zhao Q, et al: A novel class of
microRNA-recognition elements that function only within open
reading frames. Nat Struct Mol Biol. 25:1019–1027. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Long JM, Maloney B, Rogers JT and Lahiri
DK: Novel upregulation of amyloid-β precursor protein (APP) by
microRNA-346 via targeting of APP mRNA 5′-untranslated region:
Implications in Alzheimer's disease. Mol Psychiatry. 24:345–363.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ludwig N, Leidinger P, Becker K, Backes C,
Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stahler C, Meese E
and Keller A: Distribution of miRNA expression across human
tissues. Nucleic Acids Res. 44:3865–3877. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chipman LB and Pasquinelli AE: miRNA
targeting: Growing beyond the seed. Trends Genet. 35:215–222. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Uhlmann S, Mannsperger H, Zhang JD, Horvat
EA, Schmidt C, Kublbeck M, Henjes F, Ward A, Tschulena U, Zweig K,
et al: Global microRNA level regulation of EGFR-driven cell-cycle
protein network in breast cancer. Mol Syst Biol. 8:5702012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sells E, Pandey R, Chen H, Skovan BA, Cui
H and Ignatenko NA: Specific microRNA-mRNA regulatory network of
colon cancer invasion mediated by tissue kallikrein-related
peptidase 6. Neoplasia. 19:396–411. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hanauer DA, Rhodes DR, Sinha-Kumar C and
Chinnaiyan AM: Bioinformatics approaches in the study of cancer.
Curr Mol Med. 7:133–141. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Karagkouni D, Paraskevopoulou MD,
Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou
D, Kavakiotis I, Maniou S, Skoufos G, et al: DIANA-TarBase v8: A
decade-long collection of experimentally supported miRNA-gene
interactions. Nucleic Acids Res. 46:D239–D245. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chou CH, Shrestha S, Yang CD, Chang NW,
Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al: miRTarBase
update 2018: A resource for experimentally validated
microRNA-target interactions. Nucleic Acids Res. 46:D296–D302.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ozawa T, Kandimalla R, Gao F, Nozawa H,
Hata K, Nagata H, Okada S, Izumi D, Baba H, Fleshman J, et al: A
MicroRNA signature associated with metastasis of T1 colorectal
cancers to lymph nodes. Gastroenterology. 154:844–848. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gebert LFR and MacRae IJ: Regulation of
microRNA function in animals. Nat Rev Mol Cell Biol. 20:21–37.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sakai E, Fukuyo M, Ohata K, Matsusaka K,
Doi N, Mano Y, Takane K, Abe H, Yagi K, Matsuhashi N, et al:
Genetic and epigenetic aberrations occurring in colorectal tumors
associated with serrated pathway. Int J Cancer. 138:1634–1644.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ma Y, Peng J, Liu W, Zhang P, Huang L, Gao
B, Shen T, Zhou Y, Chen H, Chu Z, et al: Proteomics identification
of desmin as a potential oncofetal diagnostic and prognostic
biomarker in colorectal cancer. Mol Cell Proteomics. 8:1878–1890.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Arentz G, Chataway T, Price TJ, Izwan Z,
Hardi G, Cummins AG and Hardingham JE: Desmin expression in
colorectal cancer stroma correlates with advanced stage disease and
marks angiogenic microvessels. Clin Proteomics. 8:162011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Traicoff JL, De Marchis L, Ginsburg BL,
Zamora RE, Khattar NH, Blanch VJ, Plummer S, Bargo SA, Templeton
DJ, Casey G and Kaetzel CS: Characterization of the human polymeric
immunoglobulin receptor (PIGR) 3′UTR and differential expression of
PIGR mRNA during colon tumorigenesis. J Biomed Sci. 10:792–804.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Agesen TH, Sveen A, Merok MA, Lind GE,
Nesbakken A, Skotheim RI and Lothe RA: ColoGuideEx: A robust gene
classifier specific for stage II colorectal cancer prognosis. Gut.
61:1560–1567. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li Y, Kuscu C, Banach A, Zhang Q,
Pulkoski-Gross A, Kim D, Liu J, Roth E, Li E, Shroyer KR, et al:
miR-181a-5p inhibits cancer cell migration and angiogenesis via
downregulation of matrix metalloproteinase-14. Cancer Res.
75:2674–2685. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Przybyla L, Muncie JM and Weaver VM:
Mechanical control of epithelial-to-mesenchymal transitions in
development and cancer. Annu Rev Cell Dev Biol. 32:527–554. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gonzalez-Avila G, Sommer B, Mendoza-Posada
DA, Ramos C, Garcia-Hernandez AA and Falfan-Valencia R: Matrix
metalloproteinases participation in the metastatic process and
their diagnostic and therapeutic applications in cancer. Crit Rev
Oncol Hematol. 137:57–83. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Weng MT, Tsao PN, Lin HL, Tung CC, Change
MC, Chang YT, Wong JM and Wei SC: Hes1 increases the invasion
ability of colorectal cancer cells via the STAT3-MMP14 pathway.
PLoS One. 10:e01443222015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Andarawewa KL, Motrescu ER, Chenard MP,
Gansmuller A, Stoll I, Tomasetto C and Rio MC: Stromelysin-3 is a
potent negative regulator of adipogenesis participating to cancer
cell-adipocyte interaction/crosstalk at the tumor invasive front.
Cancer Res. 65:10862–10871. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Doll D, Keller L, Maak M, Boulesteix AL,
Siewert JR, Holzmann B and Janssen KP: Differential expression of
the chemokines GRO-2, GRO-3, and interleukin-8 in colon cancer and
their impact on metastatic disease and survival. Int J Colorectal
Dis. 25:573–581. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yamanaka R, Abe E, Sato T, Hayano A and
Takashima Y: Secondary intracranial tumors following radiotherapy
for pituitary adenomas: A systematic review. Cancers (Basel);
9(pii): pp. E1032017, PubMed/NCBI
|
|
31
|
Jeon M, You D, Bae SY, Kim SW, Nam SJ, Kim
HH, Kim S and Lee JE: Dimerization of EGFR and HER2 induces breast
cancer cell motility through STAT1-dependent ACTA2 induction.
Oncotarget. 8:50570–50581. 2016.PubMed/NCBI
|
|
32
|
Wu Y, Liu ZG, Shi MQ, Yu HZ, Jiang XY,
Yang AH, Fu XS, Xu Y, Yang S, Ni H, et al: Identification of ACTG2
functions as a promoter gene in hepatocellular carcinoma cells
migration and tumor metastasis. Biochem Biophys Res Commun.
491:537–544. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pace-Asciak CR: Hepoxilins in cancer and
inflammation-Use of hepoxilin antagonists. Cancer Metastasis Rev.
30:493–506. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gagnaire A, Nadel B, Raoult D, Neefjes J
and Gorvel JP: Collateral damage: Insights into bacterial
mechanisms that predispose host cells to cancer. Nat Rev Microbiol.
15:109–128. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
McDermott AJ, Falkowski NR, McDonald RA,
Frank CR, Pandit CR, Young VB and Huffnagle GB: Role of
interferon-γ and inflammatory monocytes in driving colonic
inflammation during acute clostridium difficile infection in mice.
Immunology. 150:468–477. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Long AG, Lundsmith ET and Hamilton KE:
Inflammation and colorectal cancer. Curr Colorectal Cancer Rep.
13:341–351. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Raskov H, Burcharth J and Pommergaard HC:
Linking gut microbiota to colorectal cancer. J Cancer. 8:3378–3395.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bischoff SC, Barbara G, Buurman W,
Ockhuizen T, Schulzke JD, Serino M, Tilg H, Watson A and Wells JM:
Intestinal permeability-A new target for disease prevention and
therapy. BMC Gastroenterol. 14:1892014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bultman SJ: Emerging roles of the
microbiome in cancer. Carcinogenesis. 35:249–255. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nieto MA, Huang RY, Jackson RA and Thiery
JP: Emt: 2016. Cell. 166:21–45. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fang Y, Zhang L, Li Z, Li Y, Huang C and
Lu X: MicroRNAs in DNA damage response, carcinogenesis, and
chemoresistance. Int Rev Cell Mol Biol. 333:1–49. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kenneth NS and Rocha S: Regulation of gene
expression by hypoxia. Biochem J. 414:19–29. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ortmann B, Druker J and Rocha S: Cell
cycle progression in response to oxygen levels. Cell Mol Life Sci.
71:3569–3582. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang H, Jiang H, Van De Gucht M and De
Ridder M: Hypoxic radioresistance: Can ROS be the key to overcome
it. Cancers (Basel). 11(pii): E1122019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Inokuma T, Haraguchi M, Fujita F,
Torashima Y, Eguchi S and Kanematsu T: Suppression of reactive
oxygen species develops lymph node metastasis in colorectal cancer.
Hepatogastroenterology. 59:2480–2483. 2012.PubMed/NCBI
|
|
47
|
Lin YC, Chen CC, Chen WM, Lu KY, Shen TL,
Jou YC, Shen CH, Ohbayashi N, Kanaho Y, Huang YL and Lee H: LPA1/3
signaling mediates tumor lymphangiogenesis through promoting CRT
expression in prostate cancer. Biochim Biophys Acta Mol Cell Biol
Lipids. 1863:1305–1315. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Alvarez SW, Sviderskiy VO, Terzi EM,
Papagiannakopoulos T, Moreira AL, Adams S, Sabatini DM, Birsoy K
and Possemato R: NFS1 undergoes positive selection in lung tumours
and protects cells from ferroptosis. Nature. 551:639–643. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lizarbe MA, Calle-Espinosa J,
Fernandez-Lizarbe E, Fernandez-Lizarbe S, Robles MA, Olmo N and
Turnay J: Colorectal cancer: From the genetic model to
posttranscriptional regulation by noncoding RNAs. Biomed Res Int.
2017:73542602017. View Article : Google Scholar : PubMed/NCBI
|