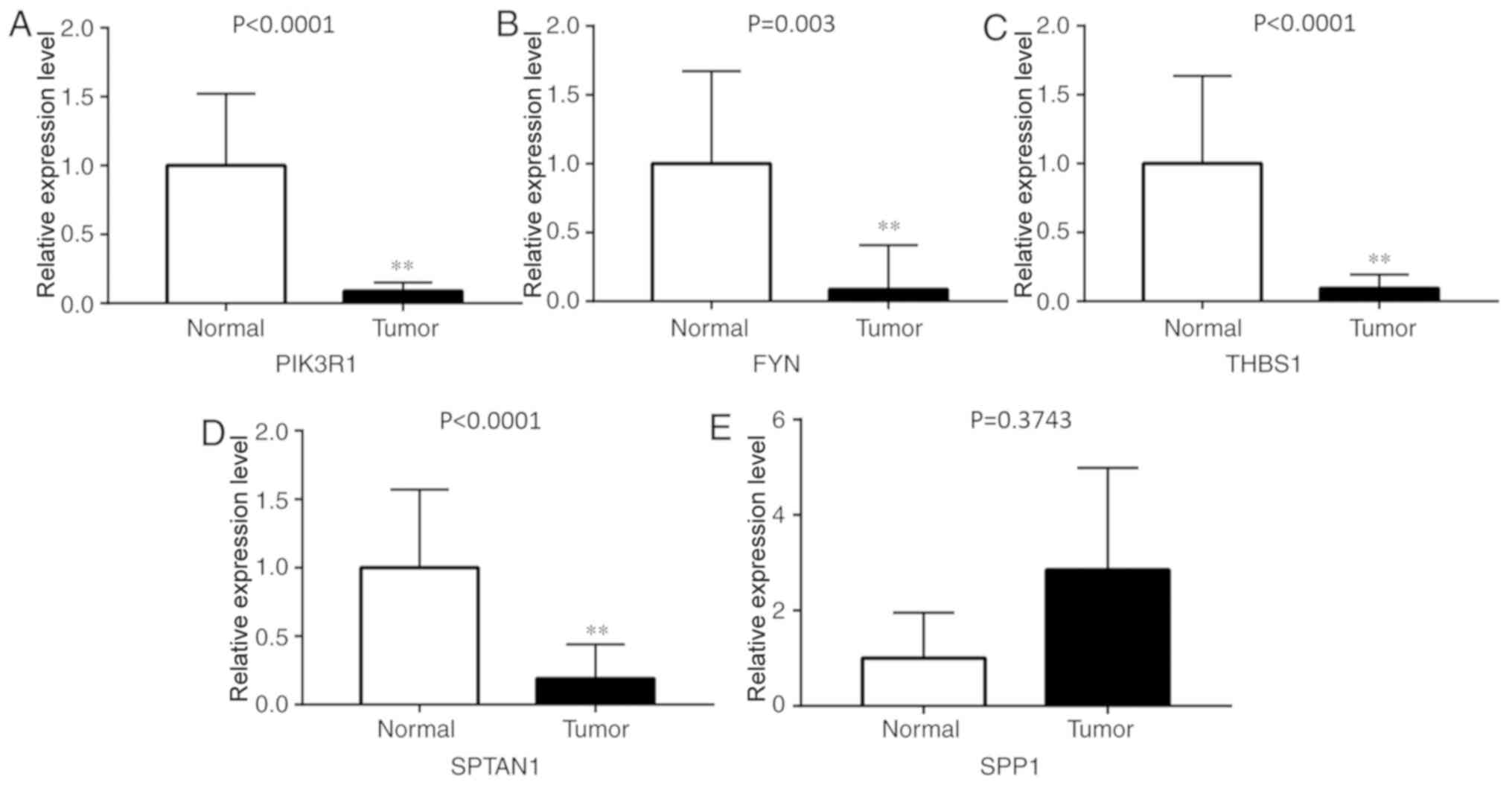
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Goldstraw P, Ball D, Jett JR, Le Chevalier
T, Lim E, Nicholson AG and Shepherd FA: Non-small-cell lung cancer.
Lancet. 378:1727–1740. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brody H: Lung cancer. Nature. 513
(Suppl):S12014. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Malone JH and Oliver B: Microarrays, deep
sequencing and the true measure of the transcriptome. BMC Biol.
9:342011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Allison DB, Cui X, Page GP and Sabripour
M: Microarray data analysis: From disarray to consolidation and
consensus. Nat Rev Genet. 7:55–65. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zequn N, Xuemei Z, Wei L, Zongjuan M,
Yujie Z, Yanli H, Yuping Z, Xia M, Wei W, Wenjing D, et al: The
role and potential mechanisms of LncRNA-TATDN1 on metastasis and
invasion of non-small cell lung cancer. Oncotarget. 7:18219–18228.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Győrffy B, Surowiak P, Budczies J and
Lanczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8:e822412013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gene Ontology Consortium: Gene ontology
consortium: Going forward. Nucleic Acids Res. 43:D1049–D1056. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jamal-Hanjani M, Wilson GA, McGranahan N,
Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R,
Rosenthal R, et al: Tracking the evolution of non-small-cell lung
cancer. N Engl J Med. 376:2109–2121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hamilton G and Rath B:
Mesenchymal-epithelial transition and circulating tumor cells in
small cell lung cancer. Adv Exp Med Biol. 994:229–245. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kriplani N, Hermida MA, Brown ER and
Leslie NR: Class I PI 3-kinases: Function and evolution. Adv Biol
Regul. 59:53–64. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chagpar RB, Links PH, Pastor MC, Furber
LA, Hawrysh AD, Chamberlain MD and Anderson DH: Direct positive
regulation of PTEN by the p85 subunit of phosphatidylinositol
3-kinase. Proc Natl Acad Sci USA. 107:5471–5476. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen L, Yang L, Yao L, Kuang XY, Zuo WJ,
Li S, Qiao F, Liu YR, Cao ZG, Zhou SL, et al: Characterization of
PIK3CA and PIK3R1 somatic mutations in Chinese breast cancer
patients. Nat Commun. 9:13572018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Saito YD, Jensen AR, Salgia R and Posadas
EM: Fyn: A novel molecular target in cancer. Cancer. 116:1629–1637.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li X, Yang Y, Hu Y, Dang D, Regezi J,
Schmidt BL, Atakilit A, Chen B, Ellis D and Ramos DM:
Alphavbeta6-Fyn signaling promotes oral cancer progression. J Biol
Chem. 278:41646–41653. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Posadas EM, Al-Ahmadie H, Robinson VL,
Jagadeeswaran R, Otto K, Kasza KE, Tretiakov M, Siddiqui J, Pienta
KJ, Stadler WM, et al: FYN is overexpressed in human prostate
cancer. BJU Int. 103:171–177. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang X, Li F and Zhu PL: Fyn-related
kinase expression predicts favorable prognosis in patients with
cervical cancer and suppresses malignant progression by regulating
migration and invasion. Biomed Pharmacother. 84:270–276. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lewin B, Siu A, Baker C, Dang D, Schnitt
R, Eisapooran P and Ramos DM: Expression of Fyn kinase modulates
EMT in oral cancer cells. Anticancer Res. 30:2591–2596.
2010.PubMed/NCBI
|
|
23
|
Kim AN, Jeon WK, Lim KH, Lee HY, Kim WJ
and Kim BC: Fyn mediates transforming growth factor-beta1-induced
down-regulation of E-cadherin in human A549 lung cancer cells.
Biochem Biophys Res Commun. 407:181–184. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li Y, Turpin CP and Wang S: Role of
thrombospondin 1 in liver diseases. Hepatol Res. 47:186–193. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang T, Sun L, Yuan X and Qiu H:
Thrombospondin-1 is a multifaceted player in tumor progression.
Oncotarget. 8:84546–84558. 2017.PubMed/NCBI
|
|
26
|
Jeanne A, Schneider C, Martiny L and
Dedieu S: Original insights on thrombospondin-1-related
antireceptor strategies in cancer. Front Pharmacol. 6:2522015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pal SK, Nguyen CT, Morita KI, Miki Y,
Kayamori K, Yamaguchi A and Sakamoto K: THBS1 is induced by TGFB1
in the cancer stroma and promotes invasion of oral squamous cell
carcinoma. J Oral Pathol Med. 45:730–739. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bienes-Martinez R, Ordonez A,
Feijoo-Cuaresma M, Corral-Escariz M, Mateo G, Stenina O, Jiménez B
and Calzada MJ: Autocrine stimulation of clear-cell renal carcinoma
cell migration in hypoxia via HIF-independent suppression of
thrombospondin-1. Sci Rep. 2:7882012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hartwig JH: Actin-binding proteins 1:
Spectrin superfamily. Protein Profile. 1:706–778. 1994.PubMed/NCBI
|
|
30
|
Cianci CD, Zhang Z, Pradhan D and Morrow
JS: Brain and muscle express a unique alternative transcript of
alphaII spectrin. Biochemistry. 38:15721–15730. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ackermann A, Schrecker C, Bon D,
Friedrichs N, Bankov K, Wild P, Plotz G, Zeuzem S, Herrmann E,
Hansmann ML and Brieger A: Downregulation of SPTAN1 is related to
MLH1 deficiency and metastasis in colorectal cancer. PLoS One.
14:e02134112019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sheng G, Zhang J, Zeng Z, Pan J, Wang Q,
Wen L, Xu Y, Wu D and Chen S: Identification of a novel
CSF3R-SPTAN1 fusion gene in an atypical chronic myeloid leukemia
patient with t(1;9)(p34;q34) by RNA-Seq. Cancer Genet 216–217.
16–19. 2017. View Article : Google Scholar
|
|
33
|
Sun Z, Wang L, Eckloff BW, Deng B, Wang Y,
Wampfler JA, Jang J, Wieben ED, Jen J, You M and Yang P: Conserved
recurrent gene mutations correlate with pathway deregulation and
clinical outcomes of lung adenocarcinoma in never-smokers. BMC Med
Genomics. 7:322014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wei R, Wong JPC and Kwok HF: Osteopontin-a
promising biomarker for cancer therapy. J Cancer. 8:2173–2183.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fedarko NS, Jain A, Karadag A and Fisher
LW: Three small integrin binding ligand N-linked glycoproteins
(SIBLINGs) bind and activate specific matrix metalloproteinases.
FASEB J. 18:734–736. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cabiati M, Gaggini M, Cesare MM, Caselli
C, De Simone P, Filipponi F, Basta G, Gastaldelli A and Del Ry S:
Osteopontin in hepatocellular carcinoma: A possible biomarker for
diagnosis and follow-up. Cytokine. 99:59–65. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zeng B, Zhou M, Wu H and Xiong Z: SPP1
promotes ovarian cancer progression via Integrin beta1/FAK/AKT
signaling pathway. Onco Targets Ther. 11:1333–1343. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hu Z, Lin D, Yuan J, Xiao T, Zhang H, Sun
W, Han N, Ma Y, Di X, Gao M, et al: Overexpression of osteopontin
is associated with more aggressive phenotypes in human non-small
cell lung cancer. Clin Cancer Res. 11:4646–4652. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hao C, Cui Y, Hu MU, Zhi X, Zhang L, Li W,
Wu W, Cheng S and Jiang WG: OPN-a splicing variant expression in
non-small cell lung cancer and its effects on the bone metastatic
abilities of lung cancer cells in vitro. Anticancer Res.
37:2245–2254. 2017. View Article : Google Scholar : PubMed/NCBI
|