Introduction
Cervical cancer is one of the most common malignant
gynecological tumors, and squamous cell carcinoma of the cervix
(SCC) is the most common histological subtype (1). According to a 2015 report by the
Chinese Cancer Registry, cervical cancer has an incidence rate of
9.89 per 100,000 and a mortality rate of 2.6 per 100,000 (2). Further clarification of the molecular
mechanisms underlying cervical cancer may lead to the development
of improved treatment options for this disease.
Heat shock factor 1 (HSF1) was first identified as a
classical transcriptional factor activated in the heat shock
response (3). Upon activation by a
variety of stimuli such as heat shock, infection and heavy metal
toxicity, HSF1 forms trimers, translocates into the nucleus and
induces various heat shock proteins (HSPs) by binding to heat shock
elements (HSEs) on HSP promoters (4,5).
Accumulating evidence has suggested that HSF1 is a powerful
modifier of carcinogenesis; HSF1 levels are elevated in several
cancers, including breast (6),
ovarian (7) and cervical cancer
(8). Activated or elevated HSF1 is
often associated with resistance to chemotherapy drugs or poor
prognosis (9). A previous study
revealed that HSF1−/− mice are resistant to chemically
induced tumors, and that HSF1−/− mouse embryonic
fibroblasts resist oncogene-induced transformation (10). However, the underlying mechanisms of
the resistance remain to be determined.
Several studies have reported that HSF1 promotes a
specific transcriptional program in highly malignant cells
(11), as well as distinct
transcriptional programs in cancer-associated fibroblasts and
adjacent cancer cells (12). It is
currently unknown why HSF1 regulates the transcription of different
genes under different conditions. HSF1 has been reported to
interact with numerous protein factors and serves multiple roles in
various physiological and pathological processes, such as nuclear
factor of interleukin 6 (13),
14-3-3 epsilon (14), heat shock
transcription factor binding protein 1 (15) and 26S subunit, non-ATPase 10/gankyrin
(16). Interactions with different
proteins under various conditions may enable HSF1 to perform
multiple functions. Therefore, clarifying the protein interaction
profile of HSF1 in SCC may help identify the functions of HSF1.
In the present study, proteins that bind to HSF1 in
the cervical tissues of patients with SCC and control subjects were
obtained by using immunoprecipitation (IP) and separated by
SDS-PAGE. The gel bands were analyzed by matrix-assisted laser
desorption/ionization-time of flight mass spectrometry
(MALDI-TOF-MS) to identify the proteins. Based on these results,
interaction networks and potential molecular functions of the
identified proteins were analyzed by bioinformatic methods.
Materials and methods
Patients and tissue samples
Cervical tissue samples were collected from four
patients diagnosed with squamous cell carcinoma (SCC) of the
cervix, who underwent total hysterectomy, as well as four control
(Ctrl) patients diagnosed with hysteromyoma or adenomyosis with
unaffected cervical tissue in Affiliated Hospital of Guangdong
Medical University (Zhanjiang, China) between February and August
2013 (Table SI). All patients in
the SCC group were not complicated with other types of pelvic
cancer and did not receive chemoradiotherapy within 3 months before
surgery. Disease staging was performed following the International
Federation of Gynecology and Obstetrics (FIGO) classification. In
addition, two groups met three major criteria: i) Higher HSF1
expression in SCC compared to Ctrl; ii) matched for age in the two
groups (43±2.94 years in SCC; 45.25±3.69 years in Ctrl); iii)
similar clinical stage for SCC patients (three were in stage IIa
and one in stage Ib2). The study was approved by the Medical Ethics
Committee of the Affiliated Hospital of Guangdong Medical
University (Zhanjiang, China; reference no. PJ2013046) and written
informed consent was obtained from all patients before surgery.
Patients were not treated with chemotherapy or radiotherapy for 3
months prior to surgery.
Protein extraction
Total protein from each sample (200 mg) was
extracted in radioimmunoprecipitation assay (RIPA) lysis buffer
(EMD Millipore, Billerica, MA, USA) plus 0.01% phenylmethylsulfonyl
fluoride and protease/phosphatase inhibitor cocktail (Cell
Signaling Technology, Inc., Danvers, MA, USA) in a homogenizer on
ice, and insoluble debris was removed by centrifugation for 10 min
at 10,800 × g at 4°C. Protein concentrations were determined using
a bicinchoninic acid assay kit (Beyotime Institute of
Biotechnology, Beijing, China).
IP and electrophoresis
PureProteome protein G/A beads and a magnetic stand
(EMD Millipore) were used to immunoprecipitate the proteins that
bound to HSF1, following the manufacturer's protocol. Briefly, 100
µl G/A beads were incubated with rabbit polyclonal HSF1 antibody
(1:50, cat. no. ab2923; Abcam, Cambridge, UK) at 25°C for 10 min
with continuous mixing. Following washing with PBS containing 0.1%
Tween 20 (pH 7.4), the immobilized capture antibody was incubated
with pooled protein (500 µg, in equal amounts from 4 samples) at
4°C overnight with continuous mixing. The immunoprecipitates were
washed with PBS containing 0.1% Tween 20 (pH 7.4), eluted by
boiling in RIPA buffer and resolved by 10% SDS-PAGE.
Gel staining
The gels were stained with Coomassie Brilliant Blue
R250 (0.1% R250, 40% methanol, 10% glacial acetic acid) at 25°C for
20 min and then washed 4 times with washing buffer (10% methanol,
10% glacial acetic acid) for 10 min each time. To avoid
interference from heavy/light antibody chains in MS analysis, 10
pairs of bands distant from the antibody chains exhibiting
differences between the SCC and Ctrl groups were cut from the
gel.
In-gel digestion
In-gel digestions were prepared as described by
Ostling et al (17). Briefly,
gel sections were minced into 1 mm3 pieces and washed 4
times with 100 mM ammonium bicarbonate
(NH4HCO3) in 50% acetonitrile (ACN; Thermo
Fisher Scientific, Waltham, MA, USA). The gel fragments were
subjected to reduction using 10 mM dithiothreitol (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) in 100 mM
NH4HCO3 buffer at 60°C for 1 h. Alkylation
was performed in a solution of 55 mM iodoacetamide (Sigma-Aldrich;
Merck KGaA) in 100 mM NH4HCO3 for 30 min at
25°C in the dark followed by in-gel digestion with 10 µl of trypsin
(25 ng/µl; Promega Corporation, Madison, WI, USA) in 50 mM
NH4CO3 at 37°C for 14–16 h. Subsequently, the
supernatant was transferred to a new microcentrifuge tube and 100
µl of 60% ACN containing 0.1% trifluoroacetic acid (TFA;
Sigma-Aldrich; Merck KGaA) was added to the remaining debris, which
was then treated by sonication for 15 min to collect the
supernatant at room temperature. The two parts of the supernatant
were combined and vacuum-dried to a volume of 10 µl. The
vacuum-dried supernatant was concentrated with a
ZipTips®C18 (EMD Millipore) and washed 3 times with 0.1%
TFA. The samples were subsequently eluted with buffer (0.1% TFA and
50% ACN) and 1 µl α-cyano-4-hydroxycinnamic acid (5 mg/ml) was
added before spotting onto the target plate.
MALDI-TOF-MS
Peptide mass fingerprinting (PMF) and sequence
analysis were carried out on a MALDI-TOF/TOF mass spectrometer
(4800 Proteomics Analyzer; Applied Biosystems; Thermo Fisher
Scientific, Inc.). Internal standard calibration of the PMF spectra
was performed using the autolysis products of trypsin. Peptide mass
maps were acquired in positive reflection mode at an accelerating
voltage of 25 kV and laser intensity of 6,000, averaging 1,200
laser shots per MALDI-TOF spectrum and 2,400 shots per TOF/TOF
spectrum (the resolution was 20,000). The top 10 precursor ion mass
peaks with a mass range of 800–4,000 Da were selected for tandem
TOF/TOF analysis. Two analysis types, combined MS and MS/MS, were
used to examine the human NCBInr (ftp.ncbi.nlm.nih.gov/blast/db/FASTA/nr.gz) and
Swissprot (ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/complete/)databases
using Mascot software (version 2.2; Matrix Science Ltd.) to
identify proteins that bind to HSF1. Searches were performed to
allow for carbamidomethylation, oxidation and a maximum of one
missed trypsin cleavage. Peptide tolerance and MS/MS tolerance were
both 0.2 Da. Confidently identified proteins had a statistically
significant protein score (P<0.05 based on combined MS and MS/MS
spectra).
Bioinformatics analysis
The interactions of MS-identified proteins from SCC
and Ctrl samples were preliminarily confirmed using the STRING
(version 10.5) online database (https://string-db.org). Functional categories and the
Gene Ontology (GO) terms biological process, cellular component and
molecular function of MS-identified proteins were analyzed using
the Database for Annotation, Visualization and Integrated Discovery
(DAVID 6.8; http://david.ncifcrf.gov).
Results
Proteins binding to HSF1
In order to investigate the effect of HSF1, four SCC
samples with higher HSF1 expression levels than the Ctrl samples
were used in the current study (Fig.
S1). A total of 392 different proteins were identified by MS,
with 241 and 220 proteins in SCC and Ctrl samples, respectively.
The proteins were distributed as follows: 172 proteins only
occurred in SCC (termed Pro-S), 151 proteins were identified only
in Ctrl (termed Pro-C), and 69 proteins were identified in the two
groups (termed Pro-B) and were removed from further analysis
(Table I). Furthermore, the
immunoprecipitated proteins binding to HSF1 yielded a different
SDS-PAGE electropherogram (Fig.
S2).
 | Table I.Proteins binding to heat shock factor
1 identified by mass spectrometry. |
Table I.
Proteins binding to heat shock factor
1 identified by mass spectrometry.
| Set | Proteins |
|---|
| Pro-C (n=151) | ACP7, AEBP1, AGO3,
AKNAD1, ANKLE1, ARIH2OS, ASPM, ATP5I, ATPAF1, BICDL2, C10orf90,
C15orf56, C17orf58, C20orf96, C6orf223, C9orf3, CALCB, CALHM2,
CATSPERG, CCT3, CCT8, CDK10, CEP250, CEP57, CEP97, CHD2, CHD8,
CHST9, COL18A1, CPNE8, CSNK1G2-AS1, CSPP1, CXorf23, DAOA, DIP2C,
DKFZP434L187, DNAH10, DNAH11, DNAH6, DPCD, DRP2, DSTYK, DUSP1,
ECHDC1, FANK1, FAT2, FAT4, FAU, FMN2, FNIP1, FRA10AC1, GAB1, GLI3,
GMFB, GOLGA4, HACE1, HECW1, HRG, IGF1R, IGHA1, IGHG4, IGHM, IGSF21,
IL21, IPMK, IPPK, ITPR1, KCNH8, KDM2A, KIF13A, KLHL34, KRT10, KRT9,
LDOC1, LOXL2, LRRC27, LTA, MCEE, MGAT3, MICALL2, MIR4697HG, MRPL49,
MRPS7, MTCL1, MTIF2, MTO1, MX2, MYCBP, NEB, NEBL, NEU3, NR1I3,
NRAP, NUP160, PCDHB15, PCDHGC4, PDCD7, PGK2, PHLDA3, PLEKHA5,
PPARGC1A, PPP4R1, PRTG, PTPN23, RASSF3, RBM12B, RCAN2, RD3, RFC3,
RFX2, RIPK2, RND1, RPL13, RPL36AL, RPS20, RPS28, RTL1, S100P, SBDS,
SCAPER, SCGB1D2, SH2D1B, SHOX2, SLC15A2, SLC4A1AP, SMARCA5, SMC5,
SNAPC4, SNX31, STAT4, TAF6, TAP1, TENM2, TLN2, TMEM266, TMEM59L,
TP53TG1, TSGA13, TTN, TUBGCP2, TXNDC8, UBD, USP28, WFDC11, ZC2HC1B,
ZC3H18, ZMYND10, ZNF148, ZNF536, ZNF770, ZNF837 |
| Pro-S (n=172) | AASS, ABCG4, ALK,
ALPL, AMIGO2, AMPD3, AP2A1, AP3D1, ARHGAP32, ARHGEF18, ATN1,
ATP13A4, ATP5G3, ATXN7, AURKC, BAHCC1, BCL2L11, BIRC6, BLZF1,
BTN3A3, C7orf13, CAB39L, CACNA1H, CASP5, CCDC105, CCL17, CCT2,
CFAP57, CHAT, CHRNB4, CIC, CLASP2, CNTN1, COA4, COL4A3BP, CTNNBIP1,
CUL4B, CUX2, DDX41, DDX56, DEF8, DLEC1, DMWD, DNAJC22, DVL3,
DYNC1LI1, ENO1, ENPP3, FAM46C, FBXO42, FDXACB1, FGFRL1, FHOD3,
FLJ42569, FMNL2, FREM2, GAREM2, GEMIN4, GLRB, GLTP, GRAP, GUSB,
HDAC11, HECW2, HELLS, HIST1H1T, HLA-DPA1, ICAM3, IGHV3-30,
IGHV3-33, IKZF4, ITIH5, KAT2A, KDM2B, KDM5B, KIF14, KLHL25, KRR1,
KRT16, LMNB1, LRP4, MAP4K3, MDH1B, MESP1, MRM1, MRPS18A, MS4A10,
MYO15A, MYO1A, MYT1, MYT1L, NEDD8, NPC1L1, NPHP3, NRXN1, NSD1,
NXF3, OR1S2, OR2T7, OR6T1, OTUD4, OTUD7A, PADI6, PATE2, PCNX3,
PDGFRL, PDK4, PHB2, PHRF1, PKD1L1, PLXNB3, PNLIPRP3, PNN, PPP1R42,
PRDX3, PRPF4, PSMB10, PSMB3, PSMC3, RALGDS, RBBP5, RCN3, RGL1,
RHPN1, RIMKLA, RIMS2, RIPK4, RNF139, RNF168, RPGRIP1L, RTTN, RYR2,
SAG, SERBP1, SETD1A, SH2B3, SH2D4B, SHISA7, SLC25A30, SLFN11,
SMARCA5, SMIM21, SORD, SOWAHC, SPECC1L, SQRDL, SRRM1, ST20, STAR,
SUMO1, TBC1D10C, TBC1D4, TENM1, TENM3, TERT, TESK2, TFAP4, TGM7,
TMEM215, TNK1, TRABD, TSSK6, TXNRD2, VPS13A, WNT2B, YIF1A, ZDHHC2,
ZFP64, ZNF185, ZNF438, ZNF506, ZNF790 |
| Pro-B (n=69) | ACSM3, ANKRD6,
ARFGEF3, ASTE1, ATP5O, BCLAF1, BDP1, BRWD1, BTN3A2, C2CD2L, CCDC81,
CCDC88A, CDKN2AIP, CLIP3, COX6A2, DIO3, DNAJC19, DPF1, DPH5,
EFCAB10, GALP, GRIK2, GRIN1, HAX1, HSPE1, HYPK, IDH1, IFNA2, IGHA2,
IGHG1, IGHG2, IGHG3, IGHM, IGHV2-70, IGHV3-23, IGKC, IL4, KRT1,
MAST4, MBLAC2, MMEL1, NBR1, NPFFR1, NPHS1, PCBD2, PFDN6, PHKG1,
POLL, POLR2G, PRL, PROSC, PTH2, PYDC1, RAB6B, RPAP1, RPL10, RYR3,
SART1, SDR42E2, SLC46A3, SLIRP, SYT11, TAS2R14, TRIM32, UBE3D,
UMPS, URI1, ZBTB10, ZNF292 |
Interaction analysis
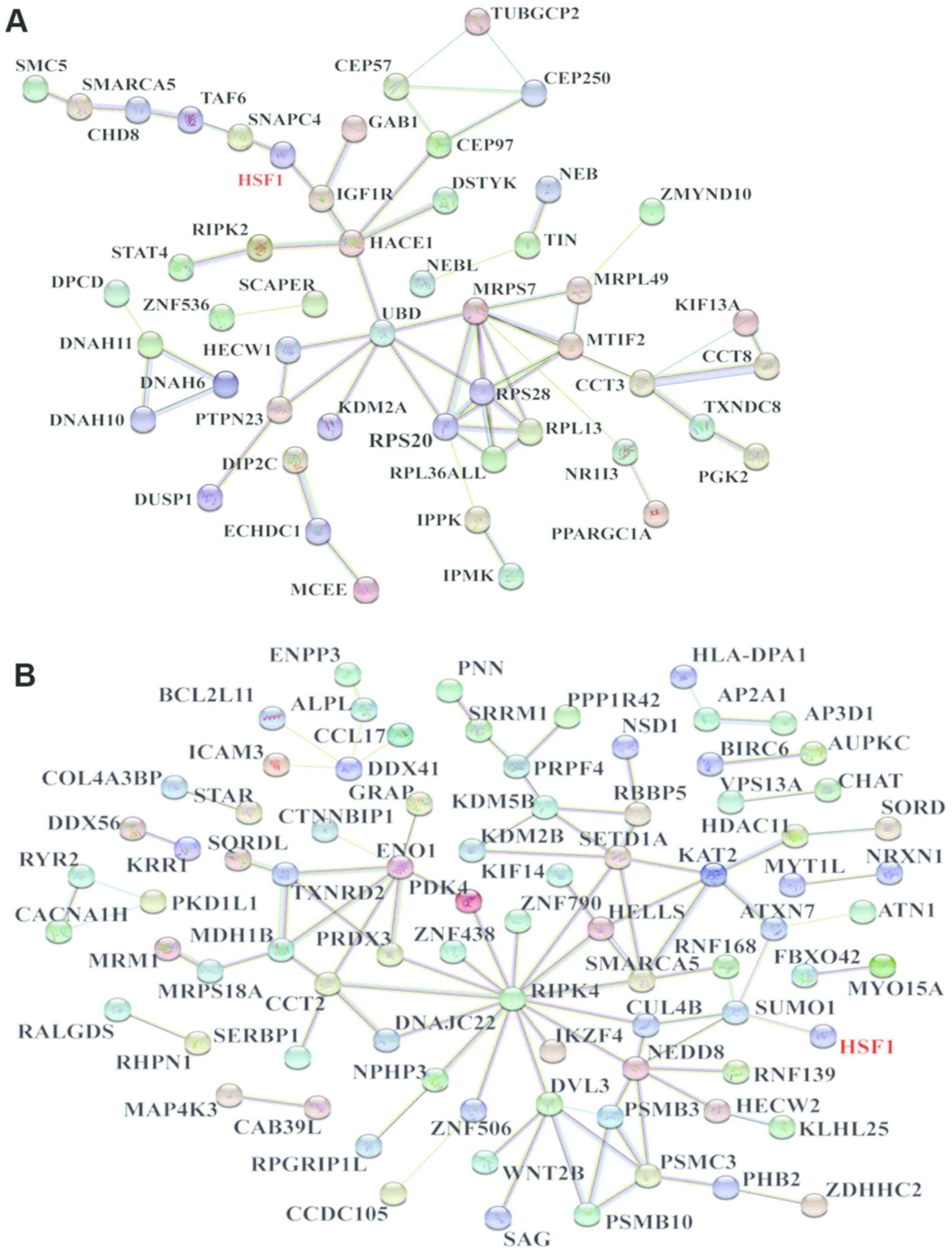
The STRING website was used to determine whether the
proteins identified by MS bound to HSF1. A total of 151 proteins
from the Pro-C set were entered along with HSF1, which produced a
network consisting of 144 nodes and 61 edges. In this network, HSF1
was indicated to interact with insulin-like growth factor 1
receptor and small nuclear RNA-activating protein complex subunit
4; these proteins interacted with others to form a highly complex
network (Fig. 1A).
HSF1 and the 172 proteins from the Pro-S set were
also entered, and a network consisting of 167 nodes and 92 edges
was obtained. The results identified an interaction between HSF1
and small ubiquitin-related modifier 1 (SUMO1). Additionally, SUMO1
was indicated to interact with other proteins to construct a highly
complicated network, which included neural precursor cell-expressed
developmentally downregulated protein 8 (NEDD8), cullin-4B,
receptor-interacting serine-threonine kinase 4 (RIPK4), and
disheveled segment polarity protein 3 (DVL3) from the Pro-S set
(Fig. 1B).
Functional classification
Subsequently, 151 proteins from the Pro-C set and
172 proteins from the Pro-S set were entered into DAVID
bioinformatics resources. Two of these proteins, immunoglobulin
heavy constant µ and caspase 5, were not recognized by the database
and removed from further analysis. Therefore, 150 proteins from
Pro-C and 171 proteins from Pro-S were subjected to functional
classification according to the UniProtKB Keywords original
database in DAVID. Following this analysis, 130/150 proteins from
Pro-C and 147/171 proteins from Pro-S were present in the output.
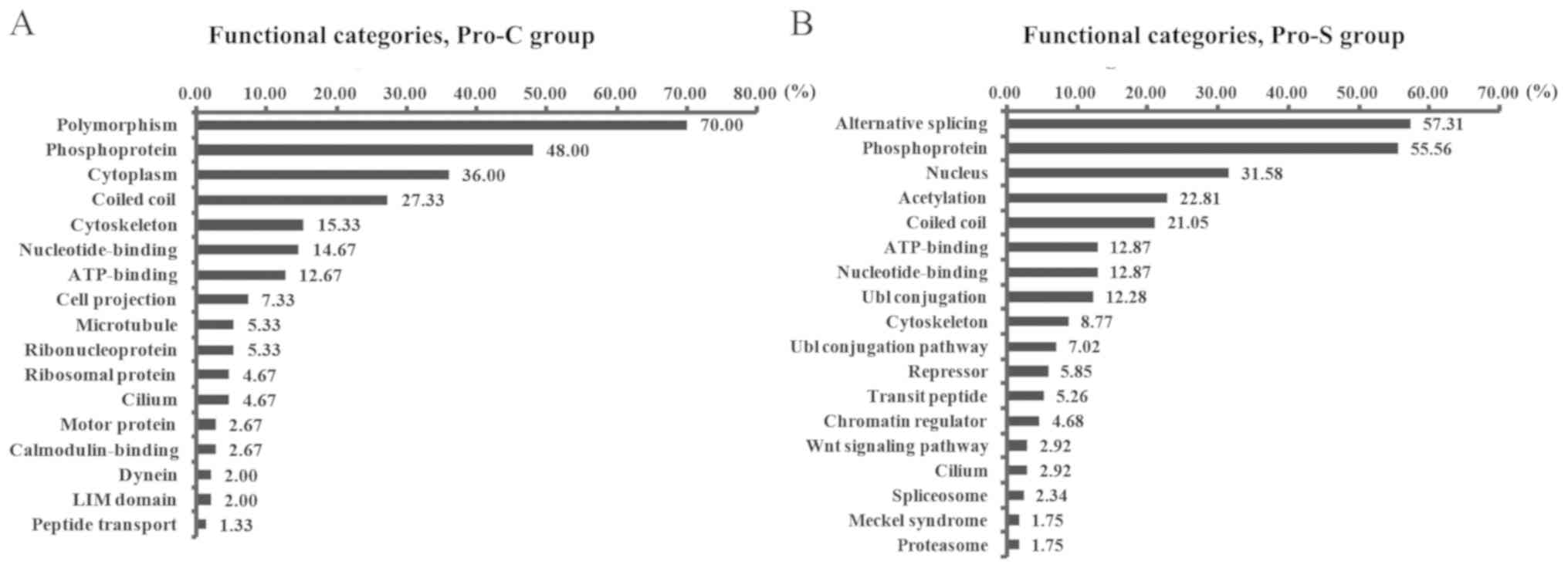
The results revealed the distribution of Pro-S proteins into
functional categories that differed from those of Pro-C (Fig. 2). The predominant category in Pro-S
was ‘alternative splicing’ (57.31%), whereas the largest category
in Pro-C was ‘polymorphism’ (70.00%). ‘Phosphoprotein’ was the
second largest category in both groups, representing 55.56% of
proteins in Pro-S and 48.00% of proteins in Pro-C. The third
largest category was related to cellular localization in both sets;
however, this category was ‘nuclear localization’ (31.58%) in Pro-S
and ‘cytoplasmic localization’ (36.00%) in Pro-C. Furthermore, five
categories were present in the two groups at comparable
frequencies: ‘Coiled coil’, ‘ATP binding’, ‘nucleotide binding’,
‘cytoskeleton’ and ‘cilium’. A considerable fraction of the
proteins indicated to bind HSF1 in Pro-S were associated with
protein modifications, including ‘acetylation’ (22.81%),
‘ubiquitin-like (Ubl) conjugation’ (12.28%) and ‘Ubl conjugation
pathway’ (7.02%).
GO term analysis
The protein sets obtained for Pro-S and Pro-C were
subjected to GO term analysis. For the cellular component category,
79/150 proteins in Pro-C and 82/171 proteins in Pro-S were
identified. The results revealed that the proteins were mainly
located in the cytoplasm and nucleoplasm in Pro-C, whereas the
proteins were in the cytosol, nucleoplasm and membrane in Pro-S
(Fig. 3A). For the molecular
function category, 49/150 proteins in Pro-C and 31/171 proteins in
Pro-S were analyzed. The largest category was ‘ATP binding’ in the
two sets and the category ‘poly (A) RNA binding’ also has large
proportion in Pro-C (Fig. 3B). For
the biological process category, 39/150 proteins in Pro-C and
51/171 proteins in Pro-S were analyzed. The largest two categories
were ‘protein phosphorylation’ and ‘negative regulation of the
canonical Wnt signaling pathway’ in Pro-S, and ‘translation’ and
‘positive regulation of apoptotic process’ in Pro-C (Fig. 3C).
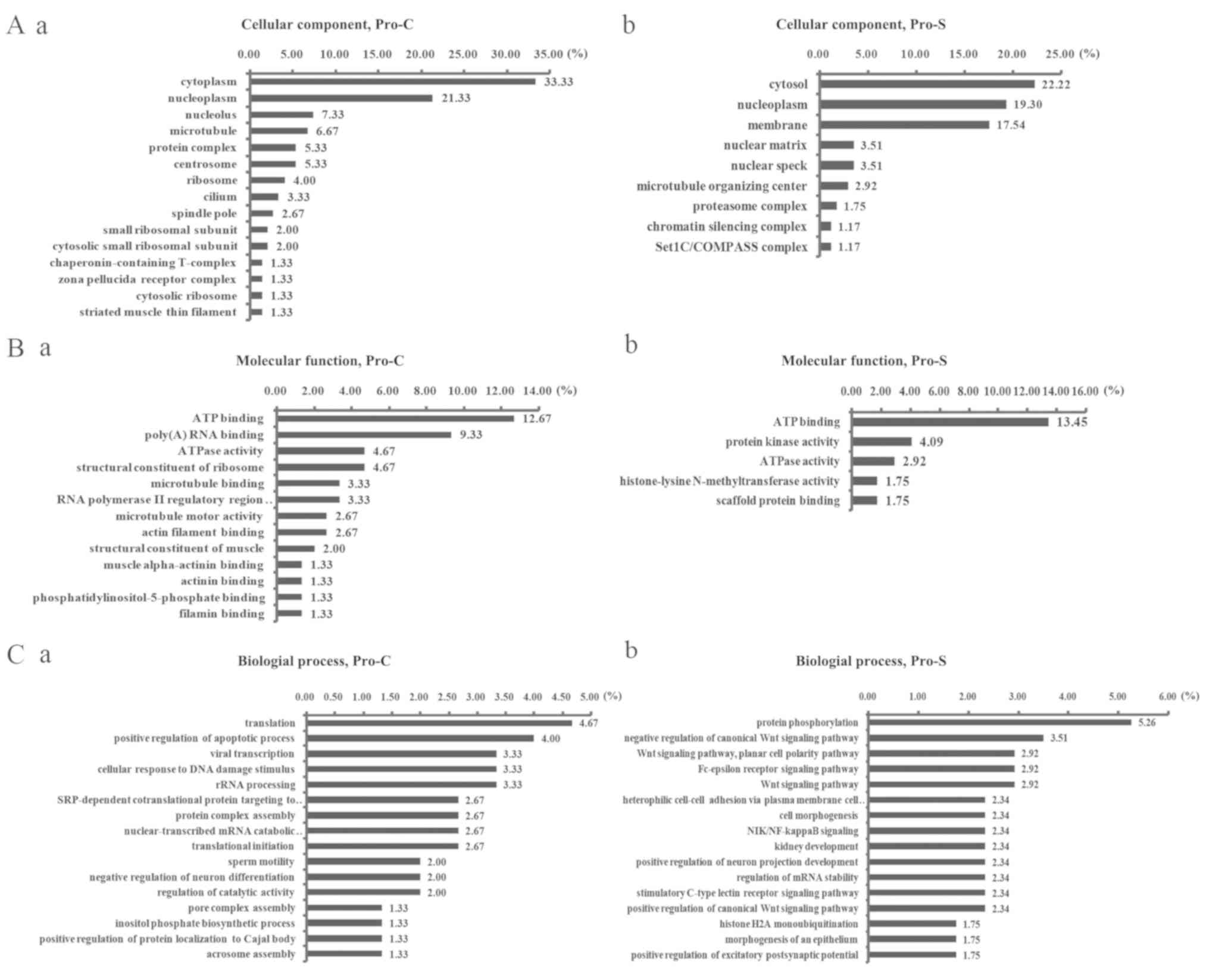 | Figure 3.GO term analysis of proteins
identified by MS. (A) The cellular component, (B) molecular
function and (C) biological process of GO analysis. GO analysis of
proteins occurred only in (A-a, B-a, C-a) Pro-C or (A-b, B-b, C-b)
Pro-S. GO, Gene Ontology; HSF1, heat shock factor 1; MS, mass
spectrometry; Pro-C, proteins in the control group only; Pro-S,
proteins in the cervical squamous cell carcinoma group only. |
Discussion
HSF1 may bind to distinct proteins in
SCC
Tumorigenesis is a complex process involving various
protein factors and molecular pathways. HSF1 is a transcription
factor involved in multiple diseases through interactions with
numerous proteins, including nuclear factor-interleukin-6 and
14-3-3ε (14,18). This is the first study, to the best
of our knowledge, to examine the function of HSF1 at the
interactional proteome level in SCC. Clear differences in the
proteins that bind to HSF1 were observed between cervical SCC
tissue and Ctrl tissue, which indicated that the proteins binding
to HSF1 were distinct in the two tissue types.
HSF1 may participate in alternative
splicing in SCC
The proteins identified by MS as only present in SCC
or Ctrl tissues were classified using the online functional
annotation tool DAVID. The largest category of proteins binding to
HSF1 in Pro-S were involved in alternative splicing. Alternative
splicing enables the production of different combinations of exons
from the same genomic template and therefore increases protein
complexity (19). In cervical
cancer, alternative splicing of numerous genes, including Numb
(20), survivin Dex3 (21) and RAB1B, member RAS oncogene family
(22), has been reported.
Additionally, the serine/arginine rich family of proteins dictate
splice site recognition (23). HSF1
is a serine-rich protein (24),
which suggests that HSF1 may interact with certain proteins to
participate in alternative splicing of genes in SCC. However, the
association between HSF1, alternative splicing and cervical cancer
has not been widely examined.
HSF1 may have unique phosphorylation
in SCC
Phosphoproteins was the second largest category in
Pro-C and Pro-S. HSF1 is a serine-rich constitutively
phosphorylated mediator of the stress response (24) and can be phosphorylated at multiple
sites, including Ser230 (25),
Ser303 and Ser307 (26). The
inducible phosphorylation of HSF1 is correlated with its
transcriptional activation (27).
piR-823 may influence whether HSF1 phosphorylation functions as a
tumor promoter (28). Therefore,
HSF1 phosphorylation status may serve a role in the genesis and
progression of cervical cancer. Among the phosphoproteins in Pro-S,
two common kinases were detected: RIPK4 and mitogen-activated
protein kinase kinase kinase kinase 3 (MAP4K3; Table I). RIPK4 functions as a node in the
protein network for the Pro-S set. Increased RIPK4 expression is
associated with the progression and favorable prognosis of cervical
cancer (29); RIPK4 promotes
canonical Wnt signaling by phosphorylating disheveled segment
polarity protein 2 (30). HSF1
Ser326 is a substrate of p38 mitogen-activated protein kinases
(31). These data suggested that
distinct kinases may be activated and interact with HSF1 in
cervical cancer.
SUMO1 was identified in the ‘alternative splicing’
and ‘phosphoproteins’ categories (data not shown) and was confirmed
by String to interact with HSF1. SUMO1 was demonstrated to regulate
HSF1 DNA-binding activity through sumoylation at Lys298 (32). No previously published studies have
investigated the correlation of SUMO1, HSF1 and cervical cancer.
Therefore, whether the interaction between SOMU1 and HSF1 is
involved in the occurrence and development of cervical cancer
deserves attention.
HSF1 may exhibit greater translocation
into the nucleus in SCC
The third largest functional category was ‘nucleus’
in Pro-S and ‘cytoplasm’ in Pro-C. Although GO cellular component
analysis revealed that the cytoplasm was the largest and
nucleoplasm the second largest term in both Pro-S and Pro-C, the
ratio of protein in the nucleoplasm vs. cytoplasm was higher in
Pro-S (0.87) compared with in Pro-C (0.64). HSF1 is activated and
translocates from the cytoplasm into the nucleus under stress
conditions (4). The results of the
present study indicated that more HSF1 translocated to the nucleus
in cervical SCC compared with normal tissue. Therefore, blocking
the translocation of HSF1 during malignant transformation, for
example, in human papilloma virus infection, may represent a
therapeutic strategy.
In the molecular function GO analysis, ‘ATP binding’
was the largest category in Pro-C and Pro-S. In this category,
dynein light intermediate chain 1 (DYNC1LI1) was identified in ATP
binding and nucleotide binding categories (data not shown).
DYNC1LI1 is a transporter in eukaryotic cells that serves important
roles in the development and function of the mammalian nervous
system (33) and malignancy of
glioma (34). HSF1 has been reported
to reinforce cell death resistance in glioma (35). The translocation of HSF1 under stress
is not well-understood; further studies of the role of DYNC1LI1 in
SCC may clarify the mechanism of HSF1 translocation.
HSF1 may influence acetylation in
SCC
The fourth largest functional category in the Pro-S
group was acetylation. Acetylation introduces an acetyl functional
group into a chemical compound, which is an important modification
of proteins at co-translational and post-translational level.
Studies of histone acetylation in cervical cancer have revealed
that histone H3 acetyl K9 was correlated with low grading and low
FIGO staging scores (36). Col et
al reported that HSF1 regulates the acetylation of pericentric
chromatin such as histone H3K9 or H3K4 under heat stress (37). These data suggested that HSF1 may
promote the development of cervical cancer by influencing the
acetylation of H3K9.
HSF1 may bind to a distinct ubiquitin
protein ligase in SCC
The fifth largest functional category in the Pro-S
group was ‘coiled coil’. Coiled coil is a structural motif in
proteins in which 2–7 a-helices are coiled together resembling
strands of a rope (38). In the
present study, two coiled-coil proteins were identified in the same
E3 ubiquitin protein ligase family: HECT, C2 and WW
domain-containing E3 ubiquitin protein ligase (HECW) 1 and HECW2;
HECW1 was identified in Pro-C and HECW2 was identified in Pro-S
(Table I). HECW1 interacts with
RNF43 to enhance pro-apoptotic activity through p53 (39). HECW2 enhances endothelial
cell-to-cell junctions, and its deficiency impairs angiogenesis
(40). This suggested that HSF1 may
interact with different members of the E3 ubiquitin protein ligase
family under pathological and normal conditions.
HSF1 and Wnt signaling pathway
Although the Wnt signaling pathway appeared very low
in the functional categories (2.92%, Fig. 2B), the biological process related to
Wnt signaling pathway appeared four times in GO analysis for Pro-S
(Fig. 3C b). The reason may be there
were fewer members of this pathway. Wnt signaling pathway is
involved in the development and metastasis of cervical cancer
(41,42). HSF1 increases Wnt/β-catenin pathway
activation (43), which suggested
that HSF1 may be involved in the carcinogenesis of cervical cancer
by regulating the Wnt signaling pathway. Interestingly, in the
present study, the biological processes included not only positive
but also negative regulation of canonical Wnt signaling pathway. It
is possible that HSF1 may increase Wnt pathway activation
indirectly by antagonizing the effect of a protein which can
negative regulate the Wnt signaling pathway. The biological process
of ‘negative regulation of the canonical Wnt signaling pathway’
included six proteins: Proteasome subunit β (PSMB) 10; nephrocystin
3; DVL3; low-density lipoprotein receptor-related protein 4; PSMB3;
and the proteasome 26S subunit, ATPase 3 (PSMC3) (data not shown).
Among these proteins, PSMB10, PSMB3, PSMC3 and DVL3 formed a
quadrilateral network as part of a larger network with HSF1 through
NEDD8 and SUMO1 (Fig. 1B). Further
studies are needed to explore whether HSF1 is able to regulate the
Wnt signaling pathway by binding to this protein.
Although SUMO1 was the only protein identified in
Pro-S and interacting with HSF1 directly (Fig. 1B), the results of the present study
indicated possible indirect interactions between HSF1 and numerous
proteins in the context of cervical cancer. The current study
provided insight into the mechanism by which HSF1 influences the
development of cervical cancers. However, owing to its preliminary
nature, the study has some deficiencies such as small sample size
and incomprehensive selection of reagents. For example, the RIPA
buffer may disrupt weak non-covalent protein-protein interactions.
In addition, we only chose 10 paired bands with obvious difference
for MS. All these may possibly result in certain proteins, which
have been reported as interactional proteins of HSF1, not being
identified. Thus, future studies need to be conducted to confirm
these interactions and to explore their functions in cervical
SCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was funded by The National Natural
Science Foundation of China (grant nos. 81302246 and 81600445), The
Start-up Fund for Doctors of Guangdong Medical University, China
(grant no. B2012038) and The Support Fund for Key Discipline of
Guangdong Medical University, China (grant no. 301K20140002).
Availability of data and materials
All data used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
LZ contributed to design and drafting the
manuscript. ZH analyzed the data of mass spectrometry. YZ analyzed
and interpreted the patients' data. JH conducted IP. XY conducted
protein extraction and electrophoresis. JW performed the
bioinformatics analysis and gave final approval of the version to
be published. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was approved by The Medical Ethics
Committee of Affiliated Hospital of Guangdong Medical University
(Zhanjiang, China; reference no. PJ2013046) and all patients
provided informed written consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HSF1
|
heat shock factor 1
|
|
IP
|
immunoprecipitation
|
|
MS
|
mass spectrometry
|
|
SCC
|
squamous cell carcinoma of cervix
|
References
|
1
|
Hsu HC, Li X, Curtin JP, Goldberg JD and
Schiff PB: Surveillance epidemiology and end results analysis
demonstrates improvement in overall survival for cervical cancer
patients treated in the era of concurrent chemoradiotherapy. Front
Oncol. 5:812015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ritossa F: A new puffing pattern induced
by temperature shock and DNP in Drosophila. Experientia.
18:571–573. 1962. View Article : Google Scholar
|
|
4
|
Baler R, Dahl G and Voellmy R: Activation
of human heat shock genes is accompanied by oligomerization,
modification, and rapid translocation of heat shock transcription
factor HSF1. Mol Cell Biol. 13:2486–2496. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shi Y, Mosser DD and Morimoto RI:
Molecular chaperones as HSF1-specific transcriptional repressors.
Genes Dev. 12:654–666. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Santagata S, Hu R, Lin NU, Mendillo ML,
Collins LC, Hankinson SE, Schnitt SJ, Whitesell L, Tamimi RM,
Lindquist S and Ince TA: High levels of nuclear heat-shock factor 1
(HSF1) are associated with poor prognosis in breast cancer. Proc
Natl Acad Sci USA. 108:18378–18383. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Powell CD, Paullin TR, Aoisa C, Menzie CJ,
Ubaldini A and Westerheide SD: The heat shock transcription factor
hsf1 induces ovarian cancer epithelial-mesenchymal transition in a
3D spheroid growth model. PLoS One. 11:e01683892016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu S, Cheng M, Hou L, Zhang W, Liu B and
Linghu H: HSF1 expression in cervical carcinoma and its correlation
with clinical pathological characteristics and prognosis. J Third
Mil Med Univ. 36:560–563. 2017.(In Chinese).
|
|
9
|
Calderwood SK: Elevated levels of HSF1
indicate a poor prognosis in breast cancer. Future Oncol.
8:399–401. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dai C, Whitesell L, Rogers AB and
Lindquist S: Heat shock factor 1 is a powerful multifaceted
modifier of carcinogenesis. Cell. 130:1005–1018. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mendillo ML, Santagata S, Koeva M, Bell
GW, Hu R, Tamimi RM, Fraenkel E, Ince TA, Whitesell L and Lindquist
S: HSF1 drives a transcriptional program distinct from heat shock
to support highly malignant human cancers. Cell. 150:549–562. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Scherz-Shouval R, Santagata S, Mendillo
ML, Sholl LM, Ben-Aharon I, Beck AH, Dias-Santagata D, Koeva M,
Stemmer SM, Whitesell L and Lindquist S: The reprogramming of tumor
stroma by HSF1 is a potent enabler of malignancy. Cell.
158:564–578. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang L, Yang M, Wang Q, Liu M, Liang Q,
Zhang H and Xiao X: HSF1 regulates expression of G-CSF through the
binding element for NF-IL6/CCAAT enhancer binding protein beta. Mol
Cell Biochem. 352:11–17. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang X, Grammatikakis N, Siganou A,
Stevenson MA and Calderwood SK: Interactions between extracellular
signal-regulated protein kinase 1, 14-3-3epsilon, and heat shock
factor 1 during stress. J Biol Chem. 279:49460–49469. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Eroglu B, Min JN, Zhang Y, Szurek E,
Moskophidis D, Eroglu A and Mivechi NF: An essential role for heat
shock transcription factor binding protein 1 (HSBP1) during early
embryonic development. Dev Biol. 386:448–460. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Luo T, Fu J, Xu A, Su B, Ren Y, Li N, Zhu
J, Zhao X, Dai R, Cao J, et al: PSMD10/gankyrin induces autophagy
to promote tumor progression through cytoplasmic interaction with
ATG7 and nuclear transactivation of ATG7 expression. Autophagy.
12:1355–1371. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ostling P, Björk JK, Roos-Mattjus P,
Mezger V and Sistonen L: Heat shock factor 2 (HSF2) contributes to
inducible expression of hsp genes through interplay with HSF1. J
Biol Chem. 282:7077–7086. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xie Y, Chen C, Stevenson MA, Auron PE and
Calderwood SK: Heat shock factor 1 represses transcription of the
IL-1beta gene through physical interaction with the nuclear factor
of interleukin 6. J Biol Chem. 277:11802–11810. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang ET, Sandberg R, Luo S, Khrebtukova I,
Zhang L, Mayr C, Kingsmore SF, Schroth GP and Burge CB: Alternative
isoform regulation in human tissue transcriptomes. Nature.
456:470–476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rong C, Feng Y and Ye Z: Notch is a
critical regulator in cervical cancer by regulating Numb splicing.
Oncol Lett. 13:2465–2470. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gaytan-Cervantes J, Gonzalez-Torres C,
Maldonado V, Zampedri C, Ceballos-Cancino G and Melendez-Zajgla J:
Protein Sam68 regulates the alternative splicing of survivin DEx3.
J Biol Chem. 292:13745–13757. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nikoshkov A, Broliden K, Attarha S,
Sviatoha V, Hellström AC, Mints M and Andersson S: Expression
pattern of the PRDX2, RAB1A, RAB1B, RAB5A and RAB25 genes in normal
and cancer cervical tissues. Int J Oncol. 46:107–112. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bradley T, Cook ME and Blanchette M: SR
proteins control a complex network of RNA-processing events. RNA.
21:75–92. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xia W, Guo Y, Vilaboa N, Zuo J and Voellmy
R: Transcriptional activation of heat shock factor HSF1 probed by
phosphopeptide analysis of factor 32P-labeled in vivo. J Biol Chem.
273:8749–8755. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Holmberg CI, Hietakangas V, Mikhailov A,
Rantanen JO, Kallio M, Meinander A, Hellman J, Morrice N,
MacKintosh C, Morimoto RI, et al: Phosphorylation of serine 230
promotes inducible transcriptional activity of heat shock factor 1.
EMBO J. 20:3800–3810. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chu B, Soncin F, Price BD, Stevenson MA
and Calderwood SK: Sequential phosphorylation by mitogen-activated
protein kinase and glycogen synthase kinase 3 represses
transcriptional activation by heat shock factor-1. J Biol Chem.
271:30847–30857. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cotto JJ, Kline M and Morimoto RI:
Activation of heat shock factor 1 DNA binding precedes
stress-induced serine phosphorylation. Evidence for a multistep
pathway of regulation. J Biol Chem. 271:3355–3358. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yin J, Jiang XY, Qi W, Ji CG, Xie XL,
Zhang DX, Cui ZJ, Wang CK, Bai Y, Wang J and Jiang HQ: piR-823
contributes to colorectal tumorigenesis by enhancing the
transcriptional activity of HSF1. Cancer Sci. 108:1746–1756. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Azizmohammadi S, Azizmohammadi S, Safari
A, Kaghazian M, Sadrkhanlo M, Behnod V and Seifoleslami M:
High-Level Expression of RIPK4 and EZH2 contributes to lymph node
metastasis and predicts favorable prognosis in patients with
cervical cancer. Oncol Res. 25:495–501. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang X, McGann JC, Liu BY, Hannoush RN,
Lill JR, Pham V, Newton K, Kakunda M, Liu J, Yu C, et al:
Phosphorylation of Dishevelled by protein kinase RIPK4 regulates
Wnt signaling. Science. 339:1441–1445. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dayalan Naidu S, Sutherland C, Zhang Y,
Risco A, de la Vega L, Caunt CJ, Hastie CJ, Lamont DJ, Torrente L,
Chowdhry S, et al: Heat shock factor 1 Is a substrate for p38
mitogen-activated protein kinases. Mol Cell Biol. 36:2403–2417.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hietakangas V, Ahlskog JK, Jakobsson AM,
Hellesuo M, Sahlberg NM, Holmberg CI, Mikhailov A, Palvimo JJ,
Pirkkala L and Sistonen L: Phosphorylation of serine 303 is a
prerequisite for the stress-inducible SUMO modification of heat
shock factor 1. Mol Cell Biol. 23:2953–2968. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Banks GT, Haas MA, Line S, Shepherd HL,
Alqatari M, Stewart S, Rishal I, Philpott A, Kalmar B, Kuta A, et
al: Behavioral and other phenotypes in a cytoplasmic Dynein light
intermediate chain 1 mutant mouse. J Neurosci. 31:5483–5494. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Szeliga M, Obara-Michlewska M, Matyja E,
Łazarczyk M, Lobo C, Hilgier W, Alonso FJ, Márquez J and Albrecht
J: Transfection with liver-type glutaminase cDNA alters gene
expression and reduces survival, migration and proliferation of
T98G glioma cells. Glia. 57:1014–1023. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Antonietti P, Linder B, Hehlgans S,
Mildenberger IC, Burger MC, Fulda S, Steinbach JP, Gessler F, Rödel
F, Mittelbronn M and Kögel D: Interference with the HSF1/HSP70/bag3
pathway primes glioma cells to matrix detachment and BH3
mimetic-induced apoptosis. Mol Cancer Ther. 16:156–168. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Beyer S, Zhu J, Mayr D, Kuhn C, Schulze S,
Hofmann S, Dannecker C, Jeschke U and Kost BP: Histone H3 acetyl K9
and histone H3 tri methyl K4 as prognostic markers for patients
with cervical cancer. Int J Mol Sci. 18(pii): E4772017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Col E, Hoghoughi N, Dufour S, Penin J,
Koskas S, Faure V, Ouzounova M, Hernandez-Vargash H, Reynoird N,
Daujat S, et al: Bromodomain factors of BET family are new
essential actors of pericentric heterochromatin transcriptional
activation in response to heat shock. Sci Rep. 7:54182017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu J, Zheng Q, Deng Y, Cheng CS,
Kallenbach NR and Lu M: A seven-helix coiled coil. Proc Natl Acad
Sci USA. 103:15457–15462. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shinada K, Tsukiyama T, Sho T, Okumura F,
Asaka M and Hatakeyama S: RNF43 interacts with NEDL1 and regulates
p53-mediated transcription. Biochem Biophys Res Commun.
404:143–147. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Choi KS, Choi HJ, Lee JK, Im S, Zhang H,
Jeong Y, Park JA, Lee IK, Kim YM and Kwon YG: The endothelial E3
ligase HECW2 promotes endothelial cell junctions by increasing
AMOTL1 protein stability via K63-linked ubiquitination. Cell
Signal. 28:1642–1651. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bahrami A, Hasanzadeh M, ShahidSales S,
Yousefi Z, Kadkhodayan S, Farazestanian M, Joudi Mashhad M, Gharib
M, Mahdi Hassanian S and Avan A: Clinical significance and
prognosis value of Wnt signaling pathway in cervical cancer. J Cell
Biochem. 118:3028–3033. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang R, Lu H, Lyu YY, Yang XM, Zhu LY,
Yang GD, Jiang PC, Re Y, Song WW, Wang JH, et al:
E6/E7-P53-POU2F1-CTHRC1 axis promotes cervical cancer metastasis
and activates Wnt/PCP pathway. Sci Rep. 7:447442017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li G, Song Y, Zhang Y, Wang H and Xie J:
miR-34b Targets HSF1 to suppress cell survival in acute myeloid
leukemia. Oncol Res. 24:109–116. 2016. View Article : Google Scholar : PubMed/NCBI
|