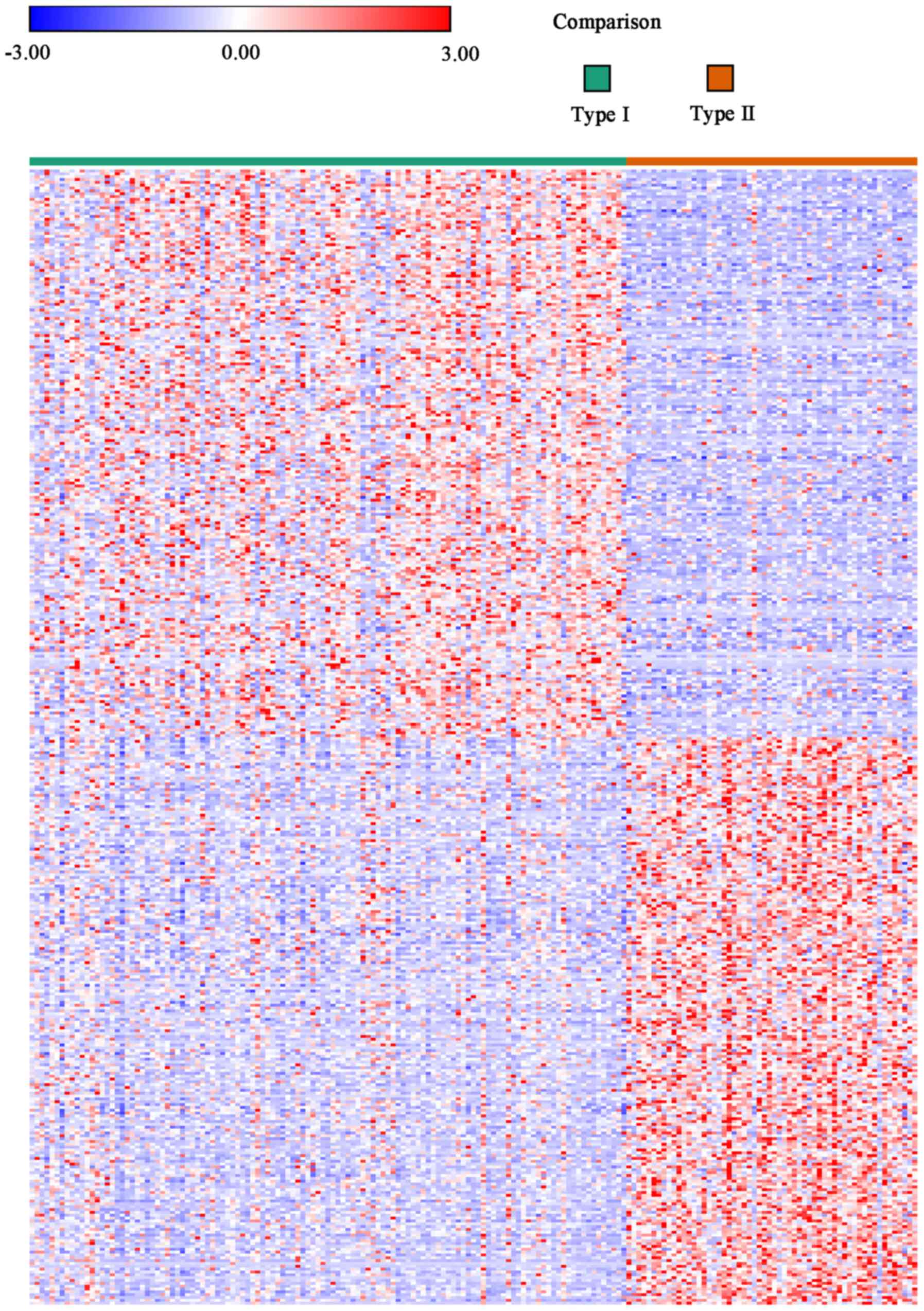
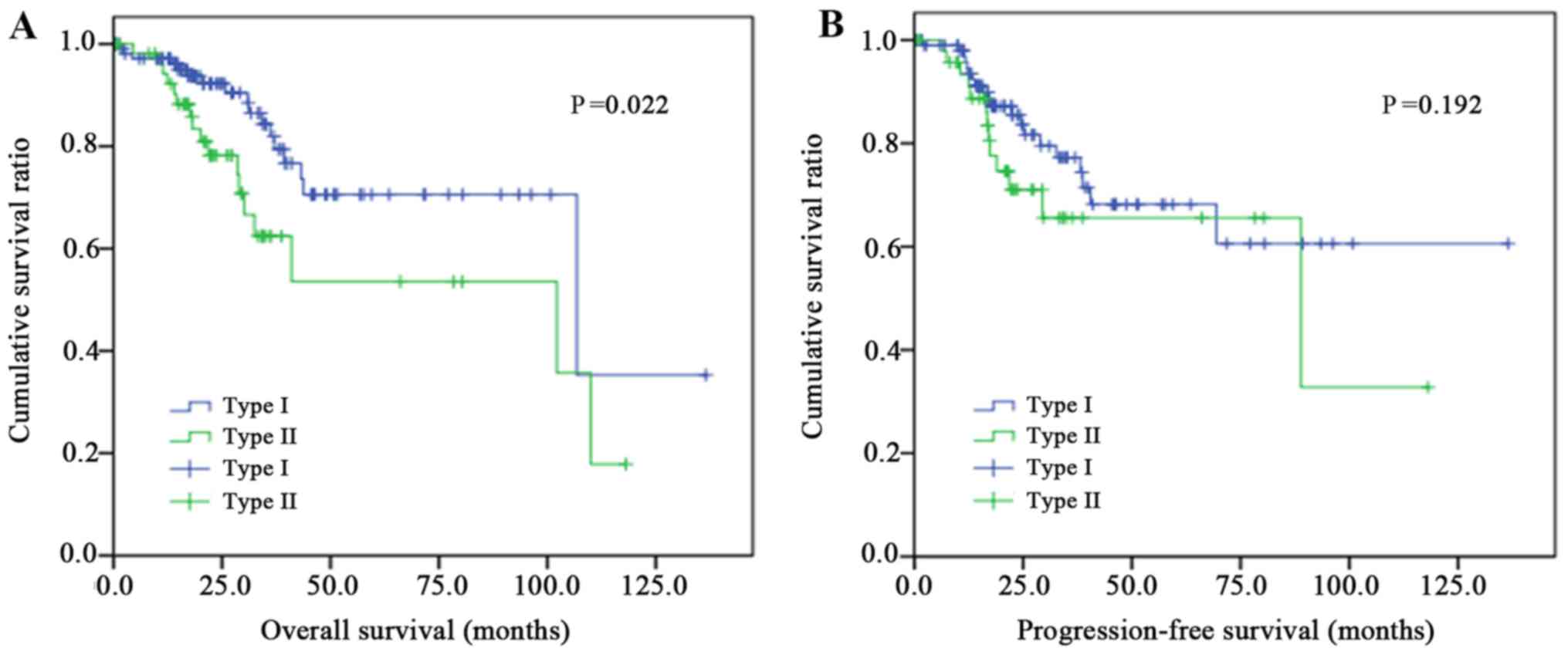
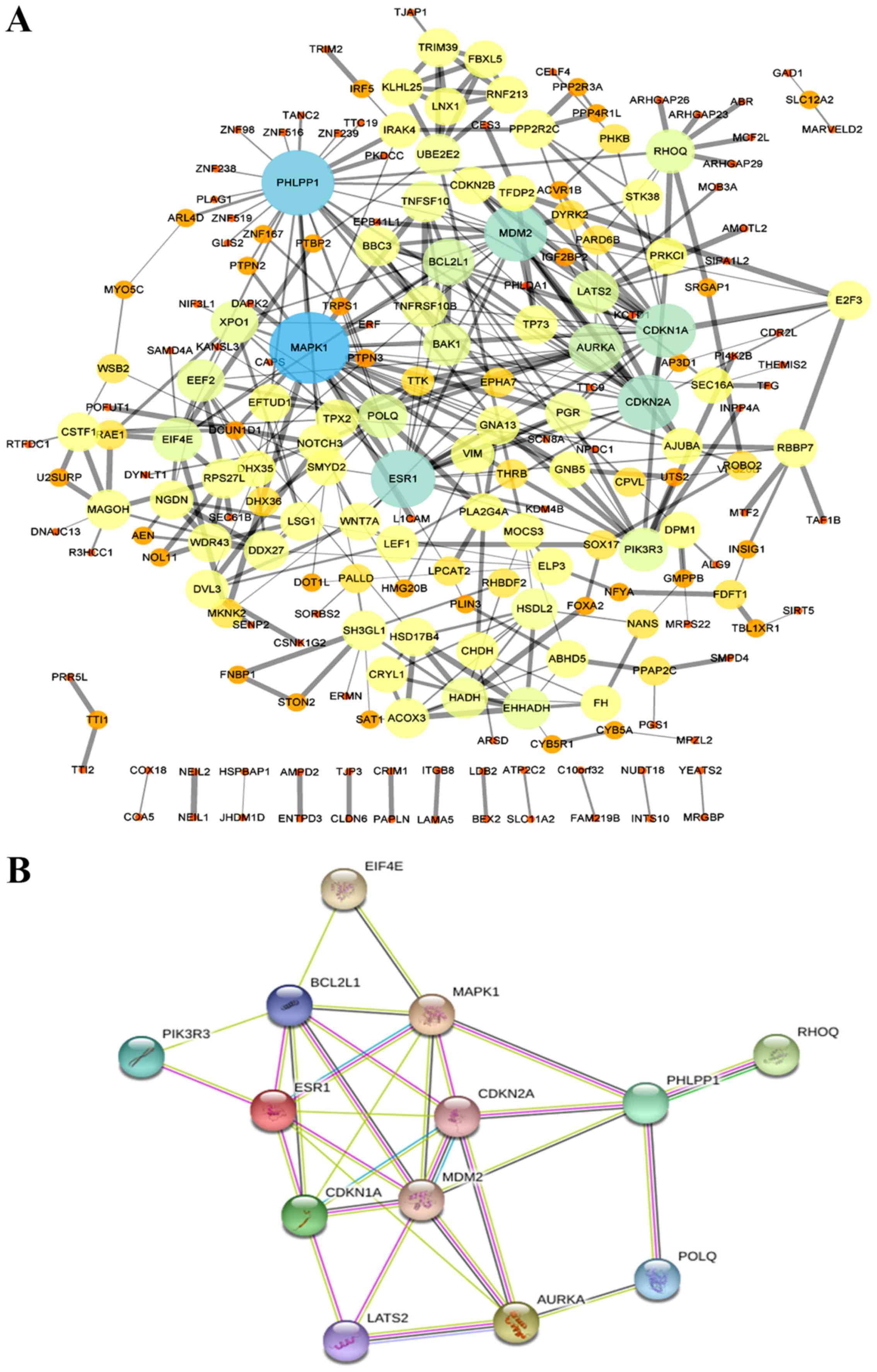
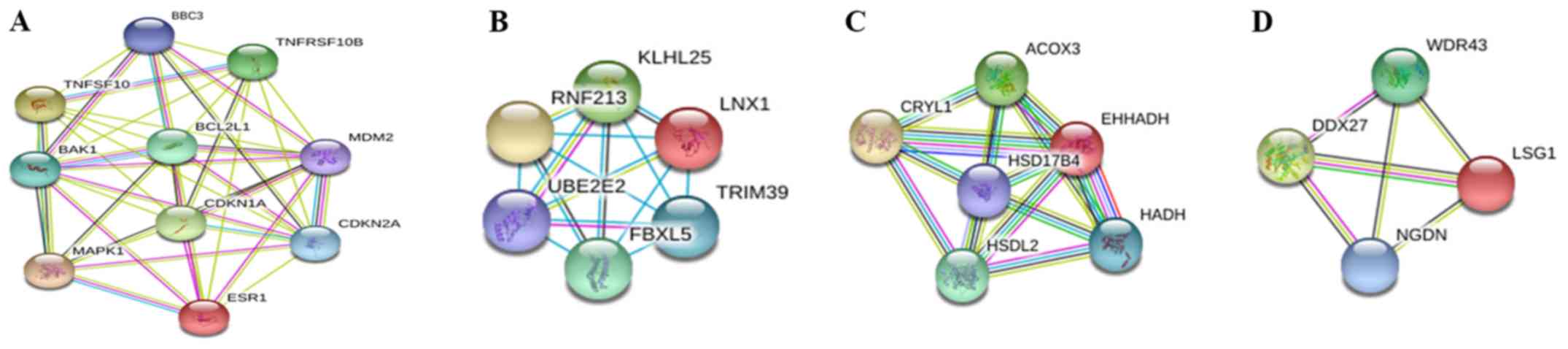
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bokhman JV: Two pathogenetic types of
endometrial carcinoma. Gynecol Oncol. 15:10–17. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Koh WJ, Abu-Rustum NR, Bean S, Bradley K,
Campos SM, Cho KR, Chon HS, Chu C, Cohn D, Crispens MA, et al:
Uterine neoplasms, version 1.2018, NCCN clinical practice
guidelines in oncology. J Natl Compr Canc Netw. 16:170–199. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Buhtoiarova TN, Brenner CA and Singh M:
Endometrial carcinoma: Role of current and emerging biomarkers in
resolving persistent clinical dilemmas. Am J Clin Pathol. 145:8–21.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Felix AS, Yang HP, Bell DW and Sherman ME:
Epidemiology of endometrial carcinoma: Etiologic importance of
hormonal and metabolic influences. Adv Exp Med Biol. 943:3–46.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mirakhor Samani S, Ezazi Bojnordi T,
Zarghampour M, Merat S and Fouladi DF: Expression of p53, Bcl-2 and
Bax in endometrial carcinoma, endometrial hyperplasia and normal
endometrium: A histopathological study. J Obstet Gynaecol.
38:999–1004. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen HX, Xu XX, Tan BZ, Zhang Z and Zhou
XD: MicroRNA-29b inhibits angiogenesis by targeting VEGFA through
the MAPK/ERK and PI3K/Akt signaling pathways in endometrial
carcinoma. Cell Physiol Biochem. 41:933–946. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Du J, Yuan Z, Ma Z, Song J, Xie X and Chen
Y: KEGG-PATH: Kyoto encyclopedia of genes and genomes-based pathway
analysis using a path analysis model. Mol Biosyst. 10:2441–2447.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cancer Genome Atlas Research Network, ;
Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA,
Ellrott K, Shmulevich I, Sander C and Stuart JM: The Cancer Genome
Atlas pan-cancer analysis project. Nat Genet. 45:1113–1120. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guan J, Xie L, Luo X, Yang B, Zhang H, Zhu
Q and Chen X: The prognostic significance of estrogen and
progesterone receptors in grade I and II endometrioid endometrial
adenocarcinoma: Hormone receptors in risk stratification. J Gynecol
Oncol. 30:e132019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tan W, Zhong Z, Carney RP, Men Y, Li J,
Pan T and Wang Y: Deciphering the metabolic role of AMPK in cancer
multi-drug resistance. Semin Cancer Biol. 56:56–71. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nadaraja S, Jørgensen TL, Matzen LE and
Herrstedt J: Impact of age, comorbidity, and FIGO stage on
treatment choice and mortality in older danish patients with
gynecological cancer: A retrospective register-based cohort study.
Drugs Real World Outcomes. 5:225–235. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lax SF: New features in the 2014 WHO
classification of uterine neoplasms. Pathologe. 37:500–511.
2016.(In German). View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kanehisa M, Goto S, Furumichi M, Tanabe M
and Hirakawa M: KEGG for representation and analysis of molecular
networks involving diseases and drugs. Nucleic Acids Res.
38:D355–D360. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ono K, Demchak B and Ideker T: Cytoscape
tools for the web age: D3.js and Cytoscape.js exporters. F1000Res.
3:1432014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Monterossi G, Ghezzi F, Vizza E, Zannoni
GF, Uccella S, Corrado G, Restaino S, Quagliozzi L, Casarin J,
Dinoi G, et al: Minimally invasive approach in type II endometrial
cancer: Is it wise and safe? J Minim Invasive Gynecol. 24:438–445.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu Y, Nan F, Lu K and Wang Y, Liu Y, Wei
S, Wu R and Wang Y: Identification of key genes in endometrioid
endometrial adenocarcinoma via TCGA database. Cancer Biomark.
21:11–21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Corbet C, Pinto A, Martherus R, Santiago
DJJ, Polet F and Feron O: Acidosis drives the reprogramming of
fatty acid metabolism in cancer cells through Changes in
mitochondrial and histone acetylation. Cell Metab. 24:311–323.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pan J, Cheng L, Bi X, Zhang X, Liu S, Bai
X, Li F and Zhao AZ: Elevation of omega-3 polyunsaturated fatty
acids attenuates PTEN-deficiency induced endometrial cancer
development through regulation of COX-2 and PGE2 production. Sci
Rep. 5:149582015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
de Lorgeril M and Salen P: New insights
into the health effects of dietary saturated and omega-6 and
omega-3 polyunsaturated fatty acids. BMC Med. 10:502012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zanoaga O, Jurj A, Raduly L,
Cojocneanu-Petric R, Fuentes-Mattei E, Wu O, Braicu C, Gherman CD
and Berindan-Neagoe I: Implications of dietary omega-3 and omega-6
polyunsaturated fatty acids in breast cancer. Exp Ther Med.
15:1167–1176. 2018.PubMed/NCBI
|
|
24
|
Edmondson RJ, Crosbie EJ, Nickkho-Amiry M,
Kaufmann A, Stelloo E, Nijman HW, Leary A, Auguste A, Mileshkin L,
Pollock P, et al: Markers of the p53 pathway further refine
molecular profiling in high-risk endometrial cancer: A TransPORTEC
initiative. Gynecol Oncol. 146:327–333. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nout RA, Bosse T, Creutzberg CL,
Jürgenliemk-Schulz IM, Jobsen JJ, Lutgens LC, van der Steen-Banasik
EM, van Eijk R, Ter Haar NT and Smit VT: Improved risk assessment
of endometrial cancer by combined analysis of MSI, PI3K-AKT,
Wnt/β-catenin and P53 pathway activation. Gynecol Oncol.
126:466–473. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Theisen ER, Gajiwala S, Bearss J, Sorna V,
Sharma S and Janat-Amsbury M: Reversible inhibition of lysine
specific demethylase 1 is a novel anti-tumor strategy for poorly
differentiated endometrial carcinoma. BMC Cancer. 14:7522014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gao X, Sun J, Huang C, Hu X, Jiang N and
Lu C: RNAi-mediated silencing of NOX4 inhibited the invasion of
gastric cancer cells through JAK2/STAT3 signaling. Am J Transl Res.
9:4440–4449. 2017.PubMed/NCBI
|
|
28
|
Shen Y, Bian R, Li Y, Gao Y, Liu Y, Xu Y,
Song X and Zhang Y: Liensinine induces gallbladder cancer apoptosis
and G2/M arrest by inhibiting ZFX-induced PI3K/AKT pathway. Acta
Biochim Biophys Sin (Shanghai). 607–614. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lo RC, Leung CO, Chan KK, Ho DW, Wong CM,
Lee TK and Ng IO: Cripto-1 contributes to stemness in
hepatocellular carcinoma by stabilizing Dishevelled-3 and
activating Wnt/beta-catenin pathway. Cell Death Differ.
25:1426–1441. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang J, Li Y, Dong M and Wu D: Long
non-coding RNA NEAT1 regulates E2F3 expression by competitively
binding to miR-377 in non-small cell lung cancer. Oncol Lett.
14:4983–4988. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu T, Jia W, An Q, Cao X and Xiao G:
Bioinformatic analysis of GLI1 and related signaling pathways in
chemosensitivity of gastric cancer. Med Sci Monit. 24:1847–1855.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang L, Feng T and Spicer LJ: The role of
tight junction proteins in ovarian follicular development and
ovarian cancer. Reproduction. 155:R183–R198. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Martin TA, Watkins G, Mansel RE and Jiang
WG: Loss of tight junction plaque molecules in breast cancer
tissues is associated with a poor prognosis in patients with breast
cancer. Eur J Cancer. 40:2717–2725. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Youakim A and Ahdieh M: Interferon-gamma
decreases barrier function in T84 cells by reducing ZO-1 levels and
disrupting apical actin. Am J Physiol. 276:G1279–G1288.
1999.PubMed/NCBI
|
|
35
|
Chen H, Wang Y and Xue F: Expression and
the clinical significance of Wnt10a and Wnt10b in endometrial
cancer are associated with the Wnt/β-catenin pathway. Oncol Rep.
29:507–514. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Han X, Cao Y, Wang K and Zhu G: HMGA1
facilitates tumor progression through regulating Wnt/beta-catenin
pathway in endometrial cancer. Biomed Pharmacother. 82:312–318.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang ZM, Wan XH, Sang GY, Zhao JD, Zhu QY
and Wang DM: miR-15a-5p suppresses endometrial cancer cell growth
via Wnt/β-catenin signaling pathway by inhibiting WNT3A. Eur Rev
Med Pharmacol Sci. 21:4810–4818. 2017.PubMed/NCBI
|
|
38
|
Wang T, Wang M, Fang S, Wang Q, Fang R and
Chen J: Fibulin-4 is associated with prognosis of endometrial
cancer patients and inhibits cancer cell invasion and metastasis
via Wnt/β-catenin signaling pathway. Oncotarget. 8:18991–19012.
2017.PubMed/NCBI
|
|
39
|
Chen QG, Zhou W, Han T, Du SQ, Li ZH,
Zhang Z, Shan GY and Kong CZ: MiR-378 suppresses prostate cancer
cell growth through downregulation of MAPK1 in vitro and in vivo.
Tumour Biol. 37:2095–2103. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wei WT, Nian XX, Wang SY, Jiao HL, Wang
YX, Xiao ZY, Yang RW, Ding YQ, Ye YP and Liao WT: miR-422a inhibits
cell proliferation in colorectal cancer by targeting AKT1 and
MAPK1. Cancer Cell Int. 17:912017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang Y, Gao C, Zhang Y, Gao J, Teng F,
Tian W, Yang W, Yan Y and Xue F: Visfatin stimulates endometrial
cancer cell proliferation via activation of PI3K/Akt and
MAPK/ERK1/2 signalling pathways. Gynecol Oncol. 143:168–178. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kanan Y, Matsumoto H, Song H, Sokolov M,
Anderson RE and Rajala RV: Serine/threonine kinase akt activation
regulates the activity of retinal serine/threonine phosphatases,
PHLPP and PHLPPL. J Neurochem. 113:477–488. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li X, Stevens PD, Liu J, Yang H, Wang W,
Wang C, Zeng Z, Schmidt MD, Yang M, Lee EY and Gao T: PHLPP is a
negative regulator of RAF1, which reduces colorectal cancer cell
motility and prevents tumor progression in mice. Gastroenterology.
146:1301–1312.e1-e10. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Huang YS, Chang CC, Lee SS, Jou YS and
Shih HM: Xist reduction in breast cancer upregulates AKT
phosphorylation via HDAC3-mediated repression of PHLPP1 expression.
Oncotarget. 7:43256–43266. 2016.PubMed/NCBI
|
|
45
|
Hribal ML, Mancuso E, Spiga R, Mannino GC,
Fiorentino TV, Andreozzi F and Sesti G: PHLPP phosphatases as a
therapeutic target in insulin resistance-related diseases. Expert
Opin Ther Targets. 20:663–675. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Toderow V, Rahmeh M, Hofmann S, Kirn V,
Mahner S, Jeschke U and von Schönfeldt V: Promotor analysis of ESR1
in endometrial cancer cell lines, endometrial and endometriotic
tissue. Arch Gynecol Obstet. 296:269–276. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jensen EV, Block GE, Smith S, Kyser K and
DeSombre ER: Estrogen receptors and breast cancer response to
adrenalectomy. Natl Cancer Inst Monogr. 34:55–70. 1971.PubMed/NCBI
|
|
48
|
Zhang CZ, Wang SX, Zhang Y, Chen JP and
Liang XM: In vitro estrogenic activities of Chinese medicinal
plants traditionally used for the management of menopausal
symptoms. J Ethnopharmacol. 98:295–300. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hall KL, Teneriello MG, Taylor RR, Lemon
S, Ebina M, Linnoila RI, Norris JH, Park RC and Birrer MJ: Analysis
of Ki-ras, p53, and MDM2 genes in uterine leiomyomas and
leiomyosarcomas. Gynecol Oncol. 65:330–335. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jeczen R, Skomra D, Cybulski M,
Schneider-Stock R, Szewczuk W, Roessner A, Rechberger T and Semczuk
A: P53/MDM2 overexpression in metastatic endometrial cancer:
Correlation with clinicopathological features and patient outcome.
Clin Exp Metastasis. 24:503–511. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wu JP and Luo X: The association between
murine double minute 2 (MDM2) rs2279744 and endometrial cancer risk
in a Chinese Han population. Cell Mol Biol (Noisy-le-grand).
63:128–130. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Dutto I, Tillhon M, Cazzalini O, Stivala
LA and Prosperi E: Biology of the cell cycle inhibitor p21(CDKN1A):
Molecular mechanisms and relevance in chemical toxicology. Arch
Toxicol. 89:155–178. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhao J, Shi L, Zeng S, Ma C, Xu W, Zhang
Z, Liu Q, Zhang P, Sun Y and Xu C: Importin-11 overexpression
promotes the migration, invasion, and progression of bladder cancer
associated with the deregulation of CDKN1A and THBS1. Urol Oncol.
36:11.e1–311.e13. 2018. View Article : Google Scholar
|
|
54
|
Seleznik GM, Reding T, Peter L, Gupta A,
Steiner SG, Sonda S, Verbeke CS, Dejardin E, Khatkov I, Segerer S,
et al: Development of autoimmune pancreatitis is independent of
CDKN1A/p21-mediated pancreatic inflammation. Gut. 67:1663–1673.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hayashi H, Kohno T, Ueno H, Hiraoka N,
Kondo S, Saito M, Shimada Y, Ichikawa H, Kato M, Shibata T, et al:
Utility of assessing the number of mutated KRAS, CDKN2A, TP53, and
SMAD4 genes using a targeted deep sequencing assay as a prognostic
biomarker for pancreatic cancer. Pancreas. 46:335–340. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Umene K, Banno K, Kisu I, Yanokura M,
Nogami Y, Tsuji K, Masuda K, Ueki A, Kobayashi Y, Yamagami W, et
al: Aurora kinase inhibitors: Potential molecular-targeted drugs
for gynecologic malignant tumors. Biomed Rep. 1:335–340. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tanaka Y, Ueda Y, Nakagawa S, Matsuzaki S,
Kobayashi E, Shiki Y, Nishio Y, Takemura M, Yamamoto T, Sawada K,
et al: A phase I/II study of GLIF combination chemotherapy for
taxane/platinum-refractory/resistant endometrial cancer (GOGO-EM2).
Cancer Chemother Pharmacol. 82:585–592. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Umene K, Yanokura M, Banno K, Irie H,
Adachi M, Iida M, Nakamura K, Nogami Y, Masuda K, Kobayashi Y, et
al: Aurora kinase A has a significant role as a therapeutic target
and clinical biomarker in endometrial cancer. Int J Oncol.
46:1498–1506. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Inoue-Yamauchi A, Jeng PS, Kim K, Chen HC,
Han S, Ganesan YT, Ishizawa K, Jebiwott S, Dong Y, Pietanza MC, et
al: Targeting the differential addiction to anti-apoptotic BCL-2
family for cancer therapy. Nat Commun. 8:160782017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Lucas CM, Milani M, Butterworth M, Carmell
N, Scott LJ, Clark RE, Cohen GM and Varadarajan S: High CIP2A
levels correlate with an antiapoptotic phenotype that can be
overcome by targeting BCL-XL in chronic myeloid leukemia. Leukemia.
30:1273–1281. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Sato K, Suzuki T, Yamaguchi Y, Kitade Y,
Nagase T and Ueda H: PLEKHG2/FLJ00018, a Rho family-specific
guanine nucleotide exchange factor, is tyrosine phosphorylated via
the EphB2/cSrc signaling pathway. Cell Signal. 26:691–696. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhang L, Huang J, Yang N, Greshock J,
Liang S, Hasegawa K, Giannakakis A, Poulos N, O'Brien-Jenkins A,
Katsaros D, et al: Integrative genomic analysis of
phosphatidylinositol 3′-kinase family identifies PIK3R3 as a
potential therapeutic target in epithelial ovarian cancer. Clin
Cancer Res. 13:5314–5321. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Dong L and Hui L: HOTAIR promotes
proliferation, migration, and invasion of ovarian cancer SKOV3
cells through regulating PIK3R3. Med Sci Monit. 22:325–331. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhang C, Min L, Liu J, Tian W, Han Y, Qu L
and Shou C: Integrated analysis identified an intestinal-like and a
diffuse-like gene sets that predict gastric cancer outcome. Tumour
Biol. Nov 17–2016.(Epub ahead of print). View Article : Google Scholar
|
|
65
|
Choi CH, Lee JS, Kim SR, Lee YY, Kim CJ,
Lee JW, Kim TJ, Lee JH, Kim BG and Bae DS: Direct inhibition of
eIF4E reduced cell growth in endometrial adenocarcinoma. J Cancer
Res Clin Oncol. 137:463–469. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Shi ZM, Liu YN, Fu B, Shen YF and Li LM:
Expression profile of eukaryotic translation initiation factor and
matrix metalloproteinase 9 in endometrial cancer tissue. J Biol
Regul Homeost Agents. 31:1053–1059. 2017.PubMed/NCBI
|
|
67
|
Guo C, Liang C, Yang J, Hu H, Fan B and
Liu X: LATS2 inhibits cell proliferation and metastasis through the
Hippo signaling pathway in glioma. Oncol Rep. 41:2753–2761.
2019.PubMed/NCBI
|
|
68
|
Zhao B, Li L, Wang L, Wang CY, Yu J and
Guan KL: Cell detachment activates the Hippo pathway via
cytoskeleton reorganization to induce anoikis. Genes Dev. 26:54–68.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Mitamura T, Watari H, Wang L, Kanno H,
Kitagawa M, Hassan MK, Kimura T, Tanino M, Nishihara H, Tanaka S
and Sakuragi N: microRNA 31 functions as an endometrial cancer
oncogene by suppressing Hippo tumor suppressor pathway. Mol Cancer.
13:972014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yasuda M: Immunohistochemical
characterization of endometrial carcinomas: Endometrioid, serous
and clear cell adenocarcinomas in association with genetic
analysis. J Obstet Gynaecol Res. 40:2167–2176. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Alkushi A, Köbel M, Kalloger SE and Gilks
CB: High-grade endometrial carcinoma: Serous and grade 3
endometrioid carcinomas have different immunophenotypes and
outcomes. Int J Gynecol Pathol. 29:343–350. 2010. View Article : Google Scholar : PubMed/NCBI
|