Introduction
Endometrial carcinoma (EC) occurs in the endometrium
and is one of the most common female gynecological tumors. It was
estimated that >63,230 new cases of EC and nearly 11,350 deaths
from EC occurred in 2018 in the USA (1). EC usually includes two types based on
its estrogen dependency (2).
Generally, the majority of ECs that occur in postmenopausal women
are type I, with an increased dependency on estrogen. The prognosis
of type I EC is good, and this type accounts for 70–80% of ECs
(3). By contrast, type II EC usually
has a poor prognosis. Surgery is the primary treatment for EC that
occurs in postmenopausal women. Despite successful surgical
intervention, the recurrence rate within 5 years for type II EC is
10–15%, with poor results and short survival time (4).
Similar to that of other tumors, the process of EC
initiation, development and metastasis involves genetic
aberrations, and changes in the tumor microenvironment (5). The literature has also reported that
the development and occurrence of EC are closely associated with a
variety of genes and cellular pathways (6,7).
However, there is currently little literature available regarding
the treatment of type II EC due to the complex nature of the
molecular mechanisms and the unfavorable biological behavior. Thus,
it is important to study and understand the cellular mechanisms by
which type II EC proliferates or initiates apoptosis when searching
for diagnostic tools and measures. The present study aimed to
identify the difference between type I and type II EC at the
genetic level, and to identify candidate biomarkers or therapeutic
targets for type II EC.
In life sciences, high-throughput sequencing
analysis of gene expression has become increasingly valued as a
promising tool for the early diagnosis and grading of cancer, the
prediction of prognosis and the discovery of new anticancer
targeted drugs. In the present study, by applying genomic
techniques, particularly large-scale genome sequencing analyses,
the genomic variation map of human EC was drawn and systematically
analyzed to find all the minor mutations of carcinogenic and tumor
suppressor genes, which can improve the ability to diagnose, treat
and prevent cancer. In the present analysis, 177 samples were
selected that contained 20,531 gene expression profiles from The
Cancer Genome Atlas (TCGA) database. Morpheus (https://software.broadinstitute.org/morpheus/) was
utilized to select differentially expressed genes (DEGs). Gene
Ontology (GO) (8) and the Kyoto
Encyclopedia of Genes and Genomes (KEGG) (9) pathway enrichment analyses were then
performed to investigate the biological processes, functions and
pathways associated with the selected DEGs. Next, the
protein-protein interaction (PPI) network was investigated via
Cytoscape v3.6.1(http://www.cytoscape.org/) to identify key genes
involved in type I and type II EC. In addition, the overall
survival (OS) of these the dominant genes was analyzed using the
Kaplan-Meier method with GraphPad Prism software version 6.0
(GraphPad Software, Inc.). Finally, the potential associations
between genes were analyzed using a visual image according to TCGA
database. New clues were identified to guide treatment, with
gene-targeted therapy also expected to be an effective treatment
for type II EC.
Materials and methods
Identification of DEGs from TCGA
The gene expression profiles of EC were collected
from the TCGA (10) database through
cBioPortal (https://www.cbioportal.org/). The Uterine Corpus
Endometrial Carcinoma (TCGA, Provisional) which is updated
regularly, was downloaded (11,12); it
contained the genetic information and clinical data of 547
patients. In total, 177 EC specimens, which included 119 subtype I
(endometrioid endometrial adenocarcinoma) and 58 subtype II (serous
endometrial adenocarcinoma) samples and corresponded to complete
clinical basic information, were included. Thus, the group of 177
cases was a subset of the 547 cohort. The clinical data of the
patients, including the demographic features, International
Federation of Gynecology and Obstetrics (FIGO) stage (13), and histological type according to the
World Health Organization 2014 classification (14), as well as performance status, were
retrospectively analyzed. OS and progression-free survival (PFS)
were assessed using Kaplan-Meier analyses for the 119 patients with
type I disease and the 58 patients with type II disease. Next, the
gene expression profile data of type I and type II patients were
downloaded from TCGA database and saved in a file, which was
uploaded to the Morpheus online tool, which is a versatile matrix
visualization and analysis software. A series of processing
selection operations were performed, including ‘Annotate
Selection’, ‘Maker Selection’ and ‘T-test’. Finally, the results
were presented in the form of a heat map and downloaded.
Differential genes with P-values <0.05 were screened by heat
maps using Morpheus online tools.
Analysis of the DEGs with GO and KEGG
online tools
The Database for Annotation, Visualization and
Integrated Discovery (DAVID; http://david.ncifcrf.gov/) is a bioinformatics
database that integrates biological data and analysis tools to
deliver systematic, comprehensive bio-functional annotation
information for large-scale gene or protein lists. KEGG (https://www.genome.jp/) is a database that integrates
information on genomic, chemical and systemic functions to reveal
the genetic and chemical blueprints of life processes (15). In addition, KEGG has powerful
capabilities that use graphics to introduce numerous metabolic
pathways and associations between pathways. The DAVID online
software analyzed the potential pathways of the core genes with
P-values <0.05.
PPI network construction and module
analysis
The Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING; http://string-db.org/) is a tool that studies genetic
interactions and can assess PPI (16). Therefore, the STRING database
(version 10.5) in Cytoscape (version 3.6.1) (17) was used to identify and map potential
associations between the DEGs. The cut-off criteria were a
confidence score ≥0.4 and a maximum number of iterations equal to
0. Modules in the PPI network were extracted from Cytoscape using
the MCODE, with the following criteria: Degree cut-off, 2; node
score cut-off, 0.2; k-core, 2; and maximum depth, 100. On this
basis, only modules with a score >5 were selected for further
function and pathway enrichment analyses. The DAVID tool aided the
analysis of the pathways between genes within these modules. In
addition, the potential associations between the 13 core genes were
assessed using KEGG and GO analysis. These genes with confidence
scores between >0.4 and <5 maximum number of interactions
were assigned to STRING.
Potential molecular mechanisms of the
top 13 aberrantly expressed genes in EC
cBioPortal (https://www.cbioportal.org/) is a database with
integrated genetic data, including DNA mutations, methylations,
gene amplifications, homozygous deletions, protein alterations and
phosphorous abundance. The present study contains results from 10
published tumor studies and >20 results from TCGA. Similarly,
the Uterine Corpus Endometrial Carcinoma (TCGA, Provisional) was
selected for further analysis in the cBioPortal database.
Therefore, possible genetic alterations of the top 13 aberrantly
expressed genes in EC were summarized using the visual cBioPortal
v1.11.3 analysis tool, and the clinical value following genetic
alterations were further analyzed.
Statistical analysis
All statistical analyses were performed using SPSS
software (version 22.0; IBM Corp.). Comparisons of the demographics
and general characteristics of the EC sample groups were performed
using the χ2 and Fisher's exact tests. A paired
Student's t-test was used to analyse quantitative data. The
survival rate was estimated using the Kaplan-Meier method and the
comparison between the type I and type II groups was calculated
using the log-rank test. Adjusted P-value was used to find genuine
tendencies towards co-occurring genes with very low error rates
using the Benjamini-Hochberg false discovery rate (FDR) correction
procedure. P<0.05 was considered to indicate a statistically
significant result.
Results
Identification of the DEGs
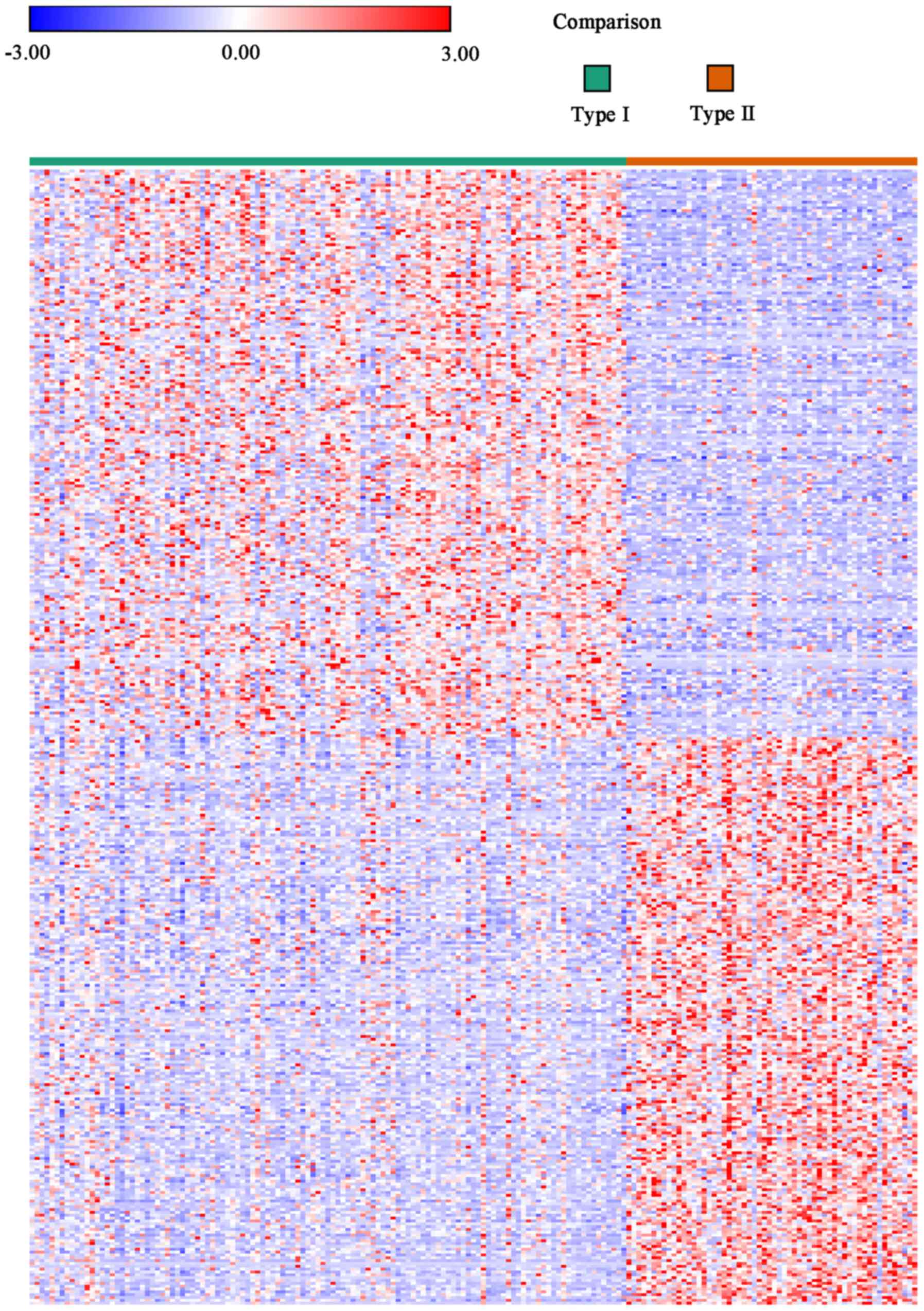
The heat maps from the Morpheus online tool were
used to compare the data of the type I samples with that of the
type II samples, and DEGs were determined using the threshold of
P<0.05. In total, 9,671 genes were screened as DEGs from the
heat maps. Among them, 5,962 genes were identified as being
upregulated, whereas 3,709 were downregulated in type I samples
when compared with type II samples. The expression heat maps
revealing the top 200 genes in the matched 9,671 DEGs are presented
in Fig. 1, and detailed information
is also presented in Table SI.
Demographics and clinical
characteristics of patients with type I and type II EC
The patients were subdivided into type I and type II
groups according to their histopathological type following surgery.
In the present study, 119 cases of pathologically confirmed
endometrioid adenocarcinoma were selected as the type I group and
58 cases of serous carcinomas were selected as the type II group.
There were significant differences in menopausal stage,
histological subtype, FIGO stage and vital status. However, there
were no significant differences in age, body mass index or
ethnicity between the aforementioned two groups. The demographics
and clinical characteristics of these two groups are summarized in
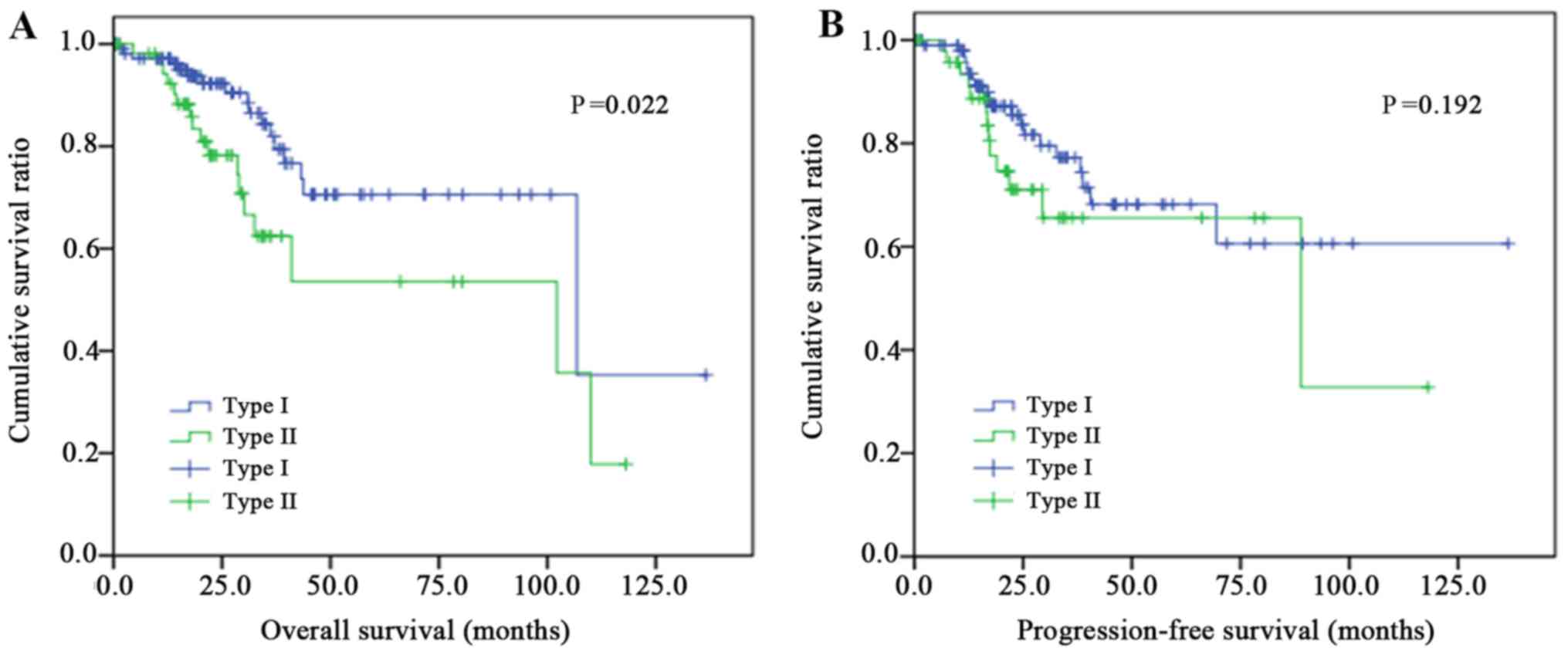
Table I. The Kaplan-Meier analysis
curves of OS of the two groups are presented in Fig. 2A. A significant statistical
difference was observed in the OS between the two groups (P=0.022),
with the type I group experiencing increased survival times,
whereas no significant difference was revealed in the PFS time
between the two groups, as presented in Fig. 2B (P=0.192).
 | Table I.Comparison of the demographics and
general characteristics of endometrial carcinoma sample groups. |
Table I.
Comparison of the demographics and
general characteristics of endometrial carcinoma sample groups.
|
Characteristics | Type I | Type II | P-value |
|---|
| Total patients,
n | 119 | 58 |
|
| Age range,
years | 33–88 | 51–90 | 0.068 |
| BMI | 35.29±19.48 | 30.02±7.90 | 0.065 |
| Menopausal, n
(%) |
|
| 0.019 |
|
Pre- | 14 (11.76) | 0 (0.00) |
|
|
Post- | 96 (80.67) | 51 (87.93) |
|
| Data
not available | 9 (7.56) | 7 (12.07) |
|
| Ethnicity, n
(%) |
|
| 0.466 |
|
Caucasian | 58 (48.74) | 34 (58.62) |
|
|
African-American | 43 (36.13) | 17 (29.31) |
|
|
Othera | 18 (15.13) | 7 (12.07) |
|
| Histological
subtype, n (%) |
|
| 0.001 |
| Grade
1 | 13 (10.92) | 1 (1.72) |
|
| Grade
2 | 19 (15.97) | 1 (1.72) |
|
| Grade
3 | 87 (73.11) | 56 (96.55) |
|
| FIGO stage, n
(%) |
|
| <0.001 |
| I | 75 (63.03) | 24 (41.38) |
|
| II | 19 (15.97) | 4 (6.90) |
|
|
III | 22 (18.49) | 23 (39.66) |
|
| IV | 3 (2.52) | 7 (12.07) |
|
| Vital status, n
(%) |
|
| 0.022 |
|
Alive | 102 (85.71) | 41 (70.69) |
|
|
Dead | 17 (14.29) | 17 (29.31) |
|
GO function and KEGG pathway
enrichment analyses
All DEGs were uploaded to the functional annotation
tool DAVID Bioinformatics Resources (version 6.7), to identify the
over-represented GO categories and KEGG pathways. From the GO
analysis, the top 5 biological process (BP), cell component and
molecular function results are presented in Table II according to their P-values in
reverse order.
 | Table II.GO analysis of differentially
expressed genes associated with patients with different types of
endometrial carcinoma. |
Table II.
GO analysis of differentially
expressed genes associated with patients with different types of
endometrial carcinoma.
| Category | Term | Count | % | P-value |
|---|
| Upregulated |
|
GOTERM_BP_FAT | GO:0043065~positive
regulation of apoptosis | 12 | 0.8 |
6.9×10−4 |
|
GOTERM_BP_FAT | GO:0043068~positive
regulation of programmed cell death | 12 | 0.8 |
7.3×10−4 |
|
GOTERM_BP_FAT | GO:0010942~positive
regulation of cell death | 12 | 0.8 |
7.6×10−4 |
|
GOTERM_BP_FAT |
GO:0008629~induction of apoptosis by
intracellular signals | 5 | 0.3 |
9.3×10−4 |
|
GOTERM_BP_FAT | GO:0006631~fatty
acid metabolic process | 8 | 0.5 |
1.1×10−3 |
|
GOTERM_CC_FAT | GO:0009898~internal
side of plasma membrane | 7 | 0.4 |
4.1×10−2 |
|
GOTERM_MF_FAT | GO:0017137~Rab
GTPase binding | 3 | 0.2 |
2.5×10−2 |
|
GOTERM_MF_FAT | GO:0008565~protein
transporter activity | 4 | 0.3 |
3.4×10−2 |
|
GOTERM_MF_FAT | GO:0003705~RNA
polymerase II transcription factor activity, enhancer binding | 3 | 0.2 |
4.0×10−2 |
|
GOTERM_MF_FAT | GO:0019207~kinase
regulator activity | 4 | 0.3 |
4.0×10−2 |
|
GOTERM_MF_FAT |
GO:0003857~3-hydroxyacyl-CoA dehydrogenase
activity | 2 | 0.1 |
4.8×10−2 |
| Downregulated |
|
GOTERM_BP_FAT |
GO:0006351~transcription,
DNA-dependent | 7 | 0.6 |
1.1×10−2 |
|
GOTERM_BP_FAT | GO:0032774~RNA
biosynthetic process | 7 | 0.6 |
1.2×10−2 |
|
GOTERM_BP_FAT |
GO:0006915~apoptosis | 10 | 0.8 |
1.5×10−2 |
|
GOTERM_BP_FAT |
GO:0012501~programmed cell death | 10 | 0.8 |
1.6×10−2 |
|
GOTERM_BP_FAT |
GO:0008637~apoptotic mitochondrial
changes | 3 | 0.2 |
1.7×10−2 |
|
GOTERM_CC_FAT |
GO:0005654~nucleoplasm | 16 | 1.3 |
4.9×10−4 |
|
GOTERM_CC_FAT |
GO:0070013~intracellular organelle
lumen | 24 | 1.9 |
7.2×10−4 |
|
GOTERM_CC_FAT | GO:0031981~nuclear
lumen | 21 | 1.7 |
7.9×10−4 |
|
GOTERM_CC_FAT |
GO:0044451~nucleoplasm part | 12 | 1.0 |
8.9×10−4 |
|
GOTERM_CC_FAT |
GO:0043233~organelle lumen | 24 | 1.9 |
9.9×10−4 |
|
GOTERM_MF_FAT |
GO:0000166~nucleotide binding | 27 | 2.2 |
2.2×10−3 |
|
GOTERM_MF_FAT | GO:0005524~ATP
binding | 19 | 1.5 |
7.3×10−3 |
|
GOTERM_MF_FAT | GO:0032559~adenyl
ribonucleotide binding | 19 | 1.5 |
8.4×10−3 |
|
GOTERM_MF_FAT | GO:0030554~adenyl
nucleotide binding | 19 | 1.5 |
1.4×10−2 |
|
GOTERM_MF_FAT | GO:0032555~purine
ribonucleotide binding | 21 | 1.7 |
1.5×10−2 |
The results of the GO analysis revealed that the
upregulated DEGs were primarily enriched in the BP for ‘positive
regulation of apoptosis’, ‘positive regulation of programmed cell
death’ and ‘positive regulation of cell death’, as well as
‘induction of apoptosis by intracellular signals’ and ‘fatty acid
metabolic process’. The downregulated DEGs focused on
‘transcription, DNA-dependent pathways’, ‘RNA biosynthetic
process’, ‘apoptosis’, ‘programmed cell death’ and ‘apoptotic
mitochondrial changes’. It was revealed that the upregulated DEGs
were significantly enriched in the ‘p53 signaling pathway’ and
‘lysine degradation’ processes, while the downregulated DEGs were
enriched in ‘pathways in cancer’, ‘tight junction’ regulation, the
‘cell cycle’, the ‘Wnt signaling pathway’, ‘chronic myeloid
leukemia’ development and ‘small-cell lung cancer’ development
according to the results of the KEGG pathway enrichment analysis
(Table III).
 | Table III.Kyoto Encyclopedia of Genes and
Genomes pathway analysis of differentially expressed genes
associated with patients with different types of endometrial
carcinoma. |
Table III.
Kyoto Encyclopedia of Genes and
Genomes pathway analysis of differentially expressed genes
associated with patients with different types of endometrial
carcinoma.
| Term | Count | % | P-value | Genes |
|---|
| Upregulated |
|
hsa04115: p53 signaling
pathway | 5 | 0.3 |
5.6×10−3 | CDKN1A, TNFRSF10B,
BBC3, SESN1, TP73 |
|
hsa00310: Lysine
degradation | 3 | 0.2 |
3.4×10−2 | AADAT, DOT1L,
HADH |
| Downregulated |
|
hsa05200: Pathways in
cancer | 10 | 0.8 |
2.7×10−4 | DVL3, ACVR1B, E2F3,
CDKN2A, CDKN2B, LAMA5, PAX8, TFG, BCL2L1, WNT7A |
|
hsa04530: Tight junction | 4 | 0.3 |
7.8×10−3 | PARD6B, CLDN6,
PRKCI, TJAP1 |
|
hsa04110: Cell cycle | 5 | 0.4 |
1.0×10−2 | E2F3, CDKN2A,
CDKN2B, TFDP2, TTK |
|
hsa04310: Wnt signaling
pathway | 4 | 0.3 |
1.1×10−2 | SENP2, DVL3,
TBL1XR1, WNT7A |
|
hsa05220: Chronic myeloid
leukemia | 4 | 0.3 |
1.4×10−2 | ACVR1B, E2F3,
CDKN2A, BCL2L1 |
|
hsa05222: Small cell lung
cancer | 4 | 0.3 |
2.3×10−2 | E2F3, CDKN2B,
LAMA5, BCL2L1 |
Screening for core genes and modules
via the PPI network
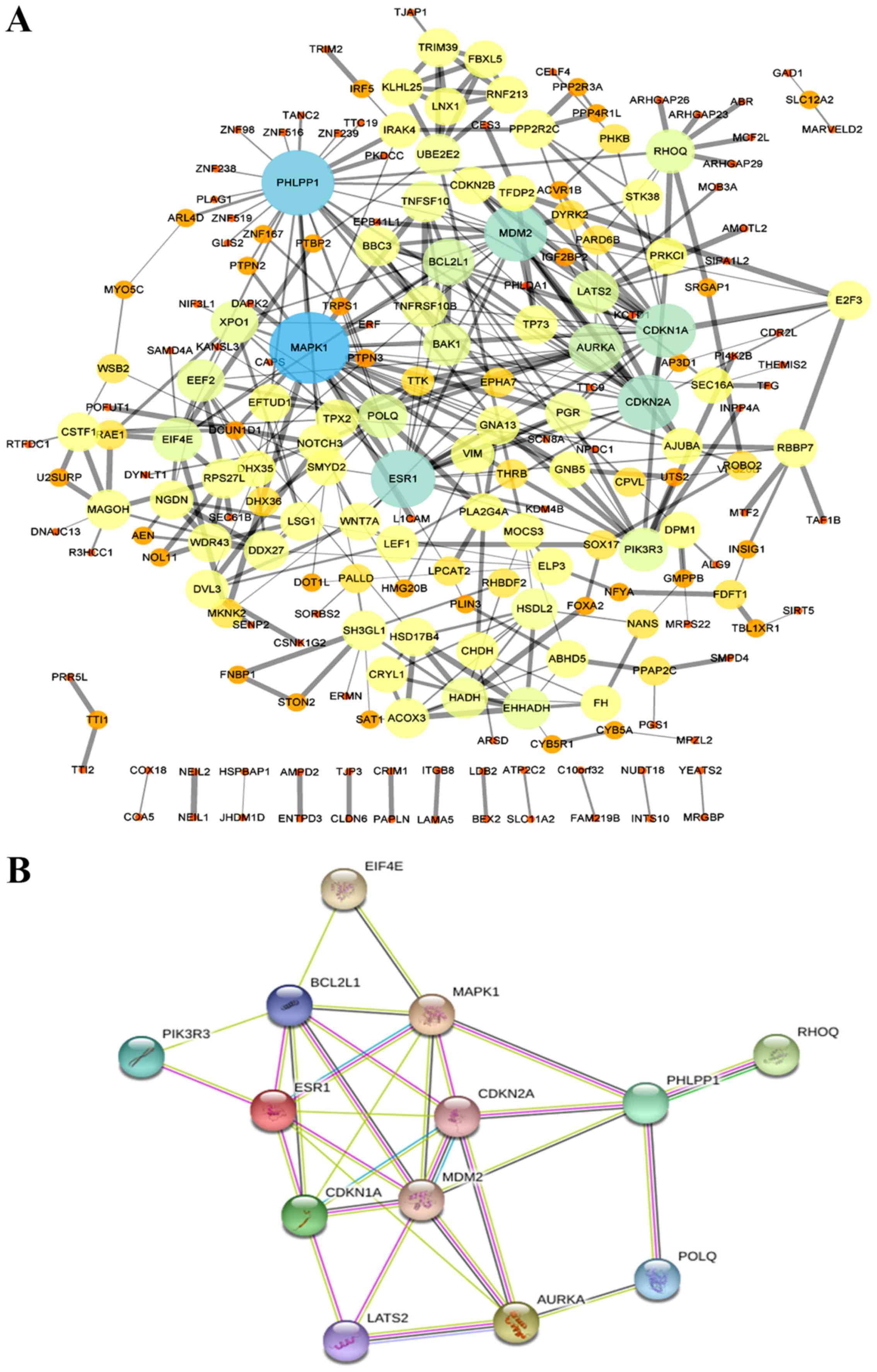
An MCODE plug-in was used in Cytoscape to build a
PPI network with 212 nodes and 380 edges (Fig. 3A). Using the Network Analyzer tool,
the top 13 hub DEGs with the highest node degrees were obtained
(Table IV). As presented in
Table IV, the hub genes were MAPK1,
PHLPP1, ESR1, MDM2, CDKN2A, CDKN1A, AURKA, BCL2L1, POLQ, PIK3R3,
RHOQ, EIF4E and LATS2. A PPI network of the top 13 central genes
with the highest connectivity was established through the STRING
protein analysis (Fig. 3B).
 | Table IV.Top 13 hub genes of the
protein-protein interaction network with higher degrees of
connectivity. |
Table IV.
Top 13 hub genes of the
protein-protein interaction network with higher degrees of
connectivity.
| Hub gene | Gene name | Node degree |
|---|
| MAPK1 | Mitogen-activated
protein kinase 1 | 32 |
| PHLPP1 | Ph domain and
leucine-rich repeat protein phosphatase 1 | 27 |
| ESR1 | Estrogen receptor
1 | 21 |
| MDM2 | Mdm2
proto-oncogene | 20 |
| CDKN2A | Cyclin-dependent
kinase inhibitor 2a (Melanoma, p16, Inhibits Cdk4) | 19 |
| CDKN1A | Cyclin-dependent
kinase inhibitor 1a | 18 |
| AURKA | Aurora kinase
a | 14 |
| BCL2L1 | Bcl-2-like 1 | 12 |
| POLQ | Dna polymerase
τ | 10 |
| PIK3R3 |
Phosphoinositide-3-kinase regulatory
subunit 3 | 10 |
| RHOQ | Ras homolog family
member q | 10 |
| EIF4E | Eukaryotic
translation initiation factor 4e | 10 |
| LATS2 | Large tumor
suppressor kinase 2 | 10 |
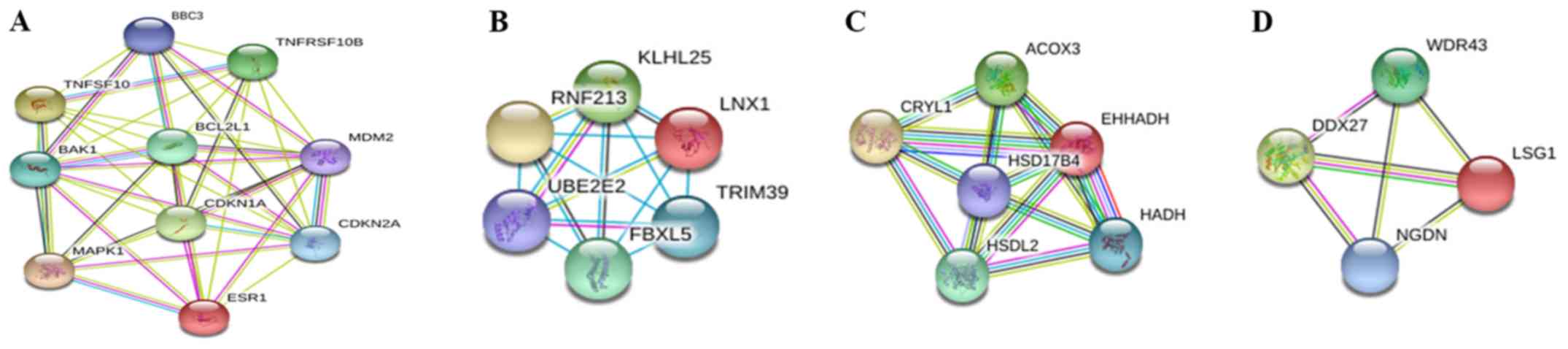
In addition, the MCODE plug-in extracted an
important module with a score >4, and then performed function
and pathway enrichment analyses (Fig.
4A). This module consisted of 10 nodes and 41 edges, which were
primarily involved in the ‘positive regulation of apoptosis’,
‘positive regulation of programmed cell death’ and ‘positive
regulation of cell death’ (Table V).
Three other modules were also revealed (Fig. 4B-D), whose further function and
pathway enrichment analysis data were not available in the GO and
KEGG databases.
 | Table V.GO and Kyoto Encyclopedia of Genes
and Genomes enrichment analyses of genes in the significant module,
with only the top 3 BPs presented. |
Table V.
GO and Kyoto Encyclopedia of Genes
and Genomes enrichment analyses of genes in the significant module,
with only the top 3 BPs presented.
| Category | Term | P-value | Genes |
|---|
|
GOTERM_BP_FAT | GO:0043065~positive
regulation of apoptosis |
1.1×10−9 | BAK1, MAPK1,
CDKN1A, TNFSF10, CDKN2A, TNFRSF10B, BBC3, BCL2L1 |
|
GOTERM_BP_FAT | GO:0043068~positive
regulation of programmed cell death |
1.1×10−9 | BAK1, MAPK1,
CDKN1A, TNFSF10, CDKN2A, TNFRSF10B, BBC3, BCL2L1 |
|
GOTERM_BP_FAT | GO:0010942~positive
regulation of cell death |
1.2×10−9 | BAK1, MAPK1,
CDKN1A, TNFSF10, CDKN2A, TNFRSF10B, BBC3, BCL2L1 |
| KEGG_PATHWAY | hsa04115: p53
signaling pathway |
9.9×10−7 | CDKN1A, CDKN2A,
TNFRSF10B, BBC3, MDM2 |
| KEGG_PATHWAY | hsa05220: Chronic
myeloid leukemia |
1.5×10−6 | MAPK1, CDKN1A,
CDKN2A, MDM2, BCL2L1 |
| KEGG_PATHWAY | hsa05219: Bladder
cancer |
1.8×10−5 | MAPK1, CDKN1A,
CDKN2A, MDM2, |
Top 13 expression genes in EC
The OncoPrint module in cBioPortal, an online tool,
was used to analyze genetic alterations, and showed that 25%
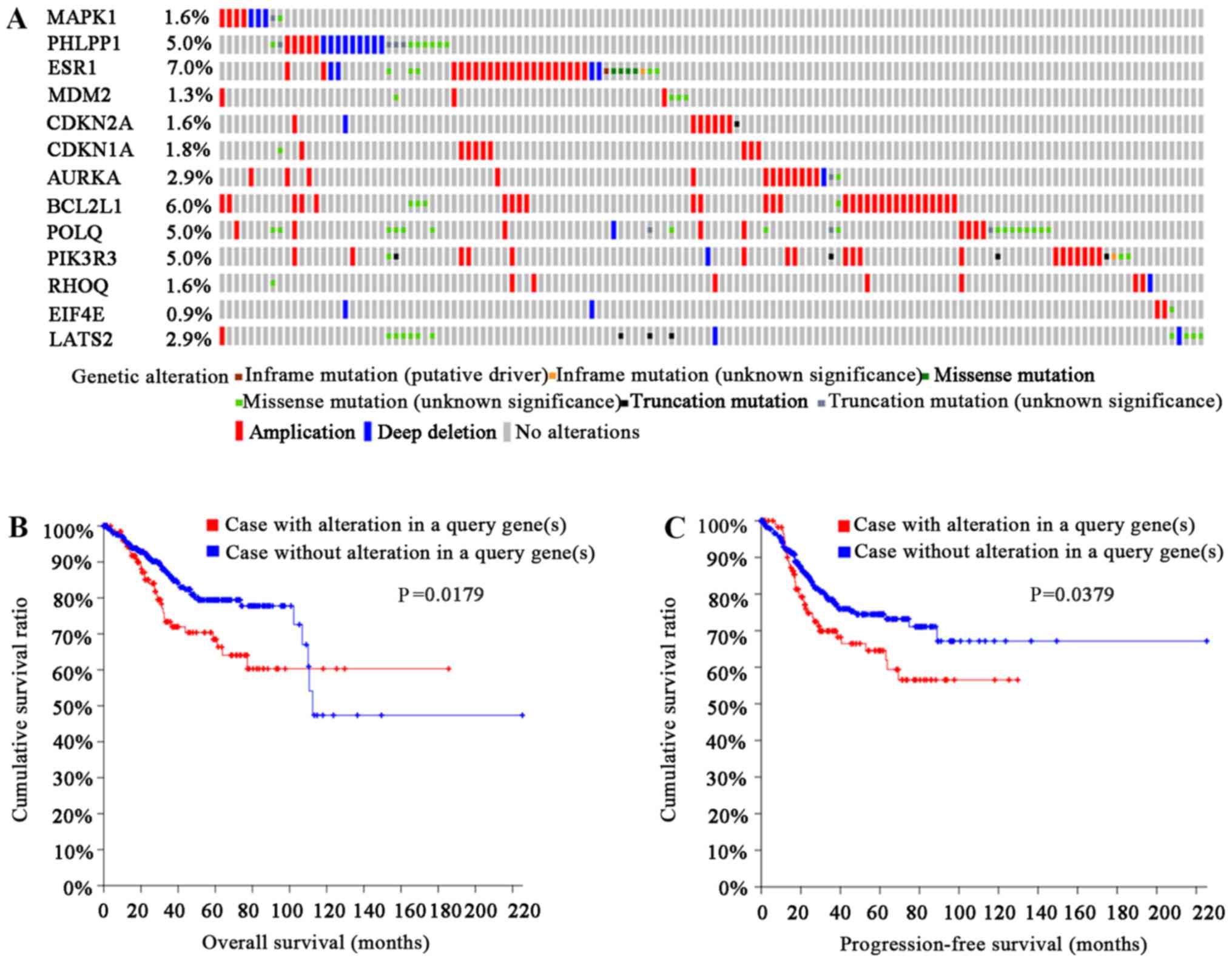
(136/547) of the cases had undergone genetic changes (Fig. 5A). The types of genetic alterations
included amplification, deep deletion and mRNA upregulation.
Notably, patients with genetic alterations had shorter OS and PFS
times than those without alterations (log rank, P=0.0179; Fig. 5B; and log rank, P=0.0379; Fig. 5C, respectively). Furthermore, as
presented in Table VI, there were 6
pairs of genes identified via cBioPortal with significant
tendencies towards co-occurrence. These genes were PHLPP1 and
LATS2, POLQ and LATS2, PHLPP1 and POLQ, ESR1 and CDKN1A, PHLPP1 and
PIK3R3, and POLQ and ESR1.
 | Table VI.Results of mutual exclusivity and
co-occurrence analysis by cBioPortal. |
Table VI.
Results of mutual exclusivity and
co-occurrence analysis by cBioPortal.
| Gene A | Gene B | P-value | Adjusted
P-value | Association |
|---|
| PHLPP1 | LATS2 | <0.001 | 0.002 | Tendency towards
co-occurrence |
| POLQ | LATS2 | <0.001 | 0.007 | Tendency towards
co-occurrence |
| PHLPP1 | POLQ | <0.001 | 0.014 | Tendency towards
co-occurrence |
| ESR1 | CDKN1A | <0.001 | 0.014 | Tendency towards
co-occurrence |
| PHLPP1 | PIK3R3 | <0.001 | 0.030 | Tendency towards
co-occurrence |
| POLQ | ESR1 | <0.001 | 0.046 | Tendency towards
co-occurrence |
| MDM2 | LATS2 | <0.001 | 0.052 | Tendency towards
co-occurrence |
| AURKA |
BCL2L1 | 0.002 | 0.142 | Tendency towards
co-occurrence |
| ESR1 | LATS2 | 0.002 | 0.186 | Tendency towards
co-occurrence |
| PHLPP1 | BCL2L1 | 0.003 | 0.214 | Tendency towards
co-occurrence |
Discussion
EC is a common malignant neoplasm of the female
reproductive tract (1). The
histological type of disease, determined postoperatively, has a
crucial impact on the survival and prognosis of patients with EC.
Pathologically determined serous carcinoma usually leads to a poor
prognosis with metastasis and more difficult surgery. Furthermore,
the 5-year survival rate only reaches 25–30% (18). Consequently, a comprehensive
understanding of the molecular mechanism behind the occurrence and
development of EC is of importance for management and therapy, as
is the development of novel targeted therapies to improve OS time.
With the widespread use of next-generation sequencing technologies
and microarrays, the expression levels of genes in the human genome
are openly included in public databases. Therefore, the molecular
mechanisms of different pathological types of EC and microarray
data can be investigated in order to facilitate the genetic
research of EC.
In the present study, it was observed through GO
analysis that upregulated DEGs were primarily enriched in the
‘positive regulation of apoptosis’, ‘positive regulation of
programmed cell death’, ‘positive regulation of cell death’,
‘induction of apoptosis by intracellular signals’ and ‘fatty acid
metabolic process’, and that the downregulated DEGs were primarily
involved in ‘transcription, DNA-dependent pathways’, ‘RNA
biosynthetic process’, ‘apoptosis’, ‘programmed cell death’ and
‘apoptotic mitochondrial changes’. These results are compatible
with the suggestion that the main causes of tumor development and
metastasis are defective functions in the regulation of apoptosis
and cell death regulators (19). The
metabolic patterns, such as that of glycolysis, oxidative
phosphorylation, amino acid metabolism, fatty acid metabolism and
nucleic acid metabolism, are significantly altered during cell
carcinogenesis. Among these processes, fatty acid metabolism is
essential for energy storage, membrane proliferation and the
synthesis of signaling molecules (20). Thus, studying the mechanism of de
novo fatty acid synthesis and its association with the
development and progression of tumors may be a new idea for
improving tumor diagnosis, prevention and treatment. A previous
study revealed that the risk of EC is significantly reduced by the
consumption of ω-3 polyunsaturated fatty acids (PUFAs) in high-fat
diets. However, the underlying molecular mechanism of action of
PUFAs in EC is not well understood (21). Furthermore, the latest research has
also demonstrated that ω-6 is strongly associated with the risk of
breast cancer (22,23).
In addition, upregulated DEGs are primarily involved
in the ‘p53 signaling pathway’ and the ‘lysine degradation’
processes in the KEGG pathway enrichment analysis. Previous studies
have revealed that markers of the p53 pathway improved the
stratification of EC and can provide novel insights into the effect
of this pathway in the morphological classification of high-risk EC
(24,25). Furthermore, the phenomenon that the
p53 signaling pathway is disturbed in EC has been widely noted. In
addition, lysine-specific demethylase 1 (LSD1) plays a vital role
in the regulation of chromatin and can affect the occurrence and
development of many types of malignant tumor by regulating the
proliferation, invasion and metastasis of tumor cells. Therefore,
the special role of LSD1 allows for it to become a new antitumor
target (26). The downregulated DEGs
were enriched in ‘pathways in cancer’, ‘tight junction’ regulation,
the ‘cell cycle’, the ‘Wnt signaling pathway’, ‘chronic myeloid
leukemia’ development’ and ‘small-cell lung cancer’ development. As
reported in the literature, alteration of pathways such as the
Janus kinase/signal transducer and activator of transcription
proteins signaling pathway, the Wnt signaling pathway and the
phosphoinositide 3-kinase (PI3K)/protein kinase B (Akt)/mammalian
target of rapamycin pathway were verified in a number of different
types of cancer (27,28). For example, stabilizing DVL3
expression can activate the Wnt/β-catenin signaling pathway in
hepatocellular carcinoma (29), and
E2F3 expression, as a potent transcriptional inducer of cell-cycle
progression, can promote non-small cell lung cancer progression
through the microRNA-377-3p-E2F3 pathway (30). These altered DEGs may be co-expressed
in different types of cancer and participate in tumorigenesis, such
as that of chronic myeloid leukemia and small cell lung cancer,
according to KEGG pathway analysis. In the multi-stage development
of cancer, the imbalance of the equilibrium steady state of the
activity of signaling pathways responsible for cell cycle
regulation and division will lead to the inhibition of apoptosis
and the enhancement of cell proliferation (31). Tight junctions are protein structures
that control the transport of water, ions and macromolecules across
cell layers (32). Previous studies
have demonstrated that low levels of tight junction plaque
molecules such as zonula occludens-1 and multi-PDZ domain protein-1
in breast cancer are associated with a poor prognosis (33,34). The
Wnt signaling pathway is a highly conserved and complicated
network, in which the abnormal activation of the canonical
Wnt/β-catenin pathway can lead to the anomalous expression of
tumor-associated genes and impact EC progression. Chen et al
(35) reported that β-catenin and
c-myc were activated due to the upregulation of Wnt10b expression,
which ultimately promoted the proliferation of Ishikawa cells and
inhibited cell apoptosis. Inhibiting the canonical Wnt/β-catenin
pathway or interfering with the regulation of its upstream signals
may be a target for anticancer therapy (36–38).
According to the results of the PPI network with
DEGs in the present study, the 13 top hub genes are as follows:
MAPK1, PHLPP1, ESR1, MDM2, CDKN2A, CDKN1A, AURKA, BCL2L1, POLQ,
PIK3R3, RHOQ, EIF4E and LATS2.
The MAPK1 gene, which encodes a member of the MAPK
family, is involved in cell proliferation, differentiation and
apoptosis. In addition, MAPKs are also known to be extracellular
signal-regulated kinases that act as an integration point for
biochemical signals. MAPK1 is overexpressed in various solid
tumors, such as EC, non-small cell lung cancer, prostate cancer and
colorectal cancer (39,40). Wang et al (41) demonstrated that insulin receptors are
activated by visfatin, which in turn activates the PI3K/Akt and
MAPK/ERK signaling pathways. This process is thought to be one of
the molecular mechanisms by which visfatin promotes the progression
of EC.
PHLPP1 is encoded by a member of the
serine/threonine phosphatase family. The function of encoded
proteins is to act as tumor inhibitors in a number of different
types of cancer, and their possible molecular mechanism is to
regulate apoptosis by dephosphorylation and inactivate the
serine/threonine kinase Akt. Thus, PHLPP1 has an effect on
mediating the activation of the PI3K/AKT signaling pathway
(42). It has been reported that
significant downregulation or loss of PHLPP1 is closely associated
with metastasis in a variety of different types of tumor, such as
colorectal cancer, breast cancer and gastric cancer (43,44).
This finding coincides with the results from the present study, in
that PHLPP1 was more significantly downregulated in type II EC than
in type I EC. PHLPP1 phosphatases have a clinically relevant role
in the pathogenesis of insulin resistance-associated diseases,
including EC (45). To the best of
our knowledge, there has been no study to date regarding the role
of PHLPP1 in EC.
The third hub gene, ESR1, encodes an estrogen
receptor and has been noted in several pathological processes of
breast cancer and endometrial cancer (46). ESR1 is an estrogen-dependent
transcription factor discovered by Jensen in 1971 (47). ESR1 is also a member of the nuclear
receptor superfamily, which can bind to estrogen, transfer to the
nucleus of breast cancer cells and participate in the regulation of
gene transcription signaling pathways, thus promoting the
proliferation of breast cancer cells (48). The ESR1 gene has been a focus of
breast cancer studies for quite some time, but it is also
clinically relevant in endometrial, ovarian and other types of
cancer.
The MDM2 gene is an oncogene closely associated with
the function of p53. MDM2 has been studied in regard to tumors such
as lung cancer and uterine sarcoma (49), but its mechanism in the occurrence
and development of EC has rarely been reported. The MDM2 protein is
overexpressed in both primary and metastatic EC, which also
indicates that MDM2 protein expression is closely associated with
the development of EC (50). In
addition, Wu and Luo (51) revealed
that the MDM2 rs2279744 polymorphism was significantly associated
with EC risk in the Chinese Han population.
CDKN1A is a gene that encodes a potent inhibitor of
cyclin. The encoded protein CDKN1A regulates the progression of the
cell cycle and focuses on the G1 phase of cell division,
as it directly and indirectly binds to and inhibits the activity of
complexes with cyclin-dependent kinase 2 or cyclin-dependent kinase
4 (52). The diseases associated
with CDKN1A include bladder cancer (53) and pancreatic inflammation (54). The loss of CDKN2A has been
demonstrated to be an important event in various types of tumor.
Although no clinical trials have been conducted on targeted
therapies as of yet, the impact of CDKN1A on the prognosis and
chemotherapy resistance in pancreatic and breast cancer have been
studied (55).
A protein that seems to mediate microtubule
formation and/or stability at the spindle end during chromosome
segregation is encoded by the AURKA gene, which is essentially a
protein kinase capable of regulating the self-reporting cycle.
Recent evidence indicates that the overexpression of AURKA is
associated with high tumor grade, severe histological type and
paclitaxel sensitivity in EC (56).
Furthermore, previous studies have demonstrated that patients with
EC who are resistant to conventional surgical treatment and
chemotherapy have short survival times and poor prognosis (57). However, the combination of an AURKA
inhibitor and paclitaxel can be effective in these patients with
EC, so it is speculated that AURKA may be a promising target in EC
therapy (58).
Another hub gene, BCL2L1 is a member of the BCL-2
protein family and forms hetero- or homodimers to act as
anti-apoptotic or pro-apoptotic regulators that are involved in
programmed cell death or apoptosis activities (59). Studies have revealed that both lung
cancer and pancreatic cancer are associated with BCL2L1. The
associated pathways of BCL2L1 include apoptosis regulation, signal
transduction and chronic myeloid leukemia progression (60).
PIK3R3 serves as a second messenger in growth
signaling pathways; it encodes the regulatory subunit PI3K that can
activate (phosphorylate) tyrosine protein kinases via binding to
the SH2 domain (61). A previous
study (62) revealed that the mRNA
expression and the expression of PIK3R3 was markedly increased in
ovarian cancer samples compared with that in normal ovarian
samples, and the difference was statistically significant. Cellular
experiments further demonstrated that the proliferation, migration
and invasion of the ovarian cancer SKOV3 cells were regulated by
the interaction between HOTAIR and PIK3R3 (63). However, given the limited research
available, it is not possible to speculate on the association
between PIK3R3 and EC.
Furthermore, the small GTPases that are encoded by
RHOQ belong to the ρ family. GTPases alternate between inactive GDP
and the active GTP binding state, and act as a molecular switch in
the signal transduction cascade. The associated pathways include
ERK signaling and Akt signaling. In diffuse subtypes of gastric
cancer, the overexpression of the RHOQ gene is correlated with a
poor prognosis (64).
One of the proteins that make up the eukaryotic
translation initiation factor 4F complex is encoded by the EIF4E
gene, which can recognize mRNAs through the structure of the
7-methylguanosine cap at the 5′ tail. The overexpression of the
EIF4E gene and high expression levels of its proteins were noted in
the EC specimens, and were associated with FIGO stage, histological
grade and degree of lymphatic metastasis (65). The positive expression rates of eIF4E
were not only associated with the EC stages as determined by FIGO
stage but were also associated with tumor cell differentiation
degree and lymphatic metastasis (66). As noted, EIF4E may serve a crucial
role in the pathogenesis of EC and become a potential trigger in
carcinogenesis.
LATS2 is a tumor suppressor that encodes the
corresponding protein kinase, namely, serine/threonine protein
kinase. LATS2 mediates the mutation of normal cells into tumor
cells by regulating the cell cycle, proliferation and apoptosis of
tumor cells. LATS2 not only interacts with p53 in a positive
feedback loop to participate in the response to cytoskeletal
injury, but LATS2 is also regarded as a co-inhibitory factor with
androgen. By limiting proliferation and promoting apoptosis,
negative regulators of YAP1 involved in the regulation of the Hippo
signaling pathway play a crucial role in organ size control and
tumor inhibition (67). Zhao et
al (68) showed that prostatic
invasion and metastasis were notably downregulated by LATS2
expression in the LAST2-Yap1 signaling pathway. Notably, in a
previous study, LATS2 was confirmed as the target of MIR31 in EC
cells, as LATS2 was downregulated among MIR31-overexpressing cells
(69). These studies were consistent
with the results from the present study, showing that LATS2 was
significantly downregulated in type II EC.
Currently, the biological functions of CDKN2A,
BCL2L1, POLQ and RHOQ in EC are unknown.
Module analysis of the PPI network revealed that the
progression and histological types of EC were positively associated
with the regulation of apoptosis and programmed cell death.
Regarding histogenesis of these type II tumors, the concept of
resting/atrophic endometrial transformation into endometrial
intraepithelial carcinoma has been proposed and increasing evidence
has demonstrated that p53 features are associated with the dynamic
transformation of endometrial carcinoma development (70). Serous carcinoma typically displays
significant overexpression of p53 and p16 (71). There was a large difference in the
functions of the DEGs between type I and type II endometrial cancer
in the present study, which suggested that there were also great
differences in the biological characteristics of these two types of
endometrial cancer.
In conclusion, by comparing the type I and type II
EC samples, a total of 9,671 DEGs, including 5,962 upregulated and
3,709 downregulated DEGs, were identified. The 13 top hub genes,
MAPK1, PHLPP1, ESR1, MDM2, CDKN2A, CDKN1A, AURKA, BCL2L1, POLQ,
PIK3R3, RHOQ, EIF4E and LATS2, were identified from the PPI
network. The key pathways and genes associated with the
histological types of EC, endometrioid adenocarcinoma and serous
adenocarcinoma, were identified. Based on the enrichment analysis
of DEGs, they were primarily associated with the positive
regulation of apoptosis, programmed cell death and cell death. The
altered DEGs (MAPK1, MDM2, AURKA, EIF4E and LATS2) may play roles
in the tumor differentiation of EC. The present study highlights
the novel molecular features of serous adenocarcinoma. These
identified key genes may help to identify the potential molecular
mechanism underlying tumorigenesis, and additionally serve as
candidate biomarkers and potential targeted therapy key points for
type II EC. However, the present study had a limitation, as the
findings were not validated in clinical samples. More in-depth
experimental studies are necessary to sufficiently confirm the role
of these DEGs in these two different types of EC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was funded by the Natural Science
Foundation of China (grant nos. 81772790 and 81572568), the
Postgraduate Innovation Fund of 13th Five-year comprehensive
investment, Tianjin Medical University (grant no. YJSCX201812) and
the Natural Science Foundation of Tianjin (grant no.
15JCYBJC25900).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KZ drafted this manuscript. KZ and HL conceived and
designed the study. YY, KL and YZ collected the data and performed
the analyses. YW and FX helped to design the study and revised the
manuscript. All authors approved the final version of
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bokhman JV: Two pathogenetic types of
endometrial carcinoma. Gynecol Oncol. 15:10–17. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Koh WJ, Abu-Rustum NR, Bean S, Bradley K,
Campos SM, Cho KR, Chon HS, Chu C, Cohn D, Crispens MA, et al:
Uterine neoplasms, version 1.2018, NCCN clinical practice
guidelines in oncology. J Natl Compr Canc Netw. 16:170–199. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Buhtoiarova TN, Brenner CA and Singh M:
Endometrial carcinoma: Role of current and emerging biomarkers in
resolving persistent clinical dilemmas. Am J Clin Pathol. 145:8–21.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Felix AS, Yang HP, Bell DW and Sherman ME:
Epidemiology of endometrial carcinoma: Etiologic importance of
hormonal and metabolic influences. Adv Exp Med Biol. 943:3–46.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mirakhor Samani S, Ezazi Bojnordi T,
Zarghampour M, Merat S and Fouladi DF: Expression of p53, Bcl-2 and
Bax in endometrial carcinoma, endometrial hyperplasia and normal
endometrium: A histopathological study. J Obstet Gynaecol.
38:999–1004. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen HX, Xu XX, Tan BZ, Zhang Z and Zhou
XD: MicroRNA-29b inhibits angiogenesis by targeting VEGFA through
the MAPK/ERK and PI3K/Akt signaling pathways in endometrial
carcinoma. Cell Physiol Biochem. 41:933–946. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Du J, Yuan Z, Ma Z, Song J, Xie X and Chen
Y: KEGG-PATH: Kyoto encyclopedia of genes and genomes-based pathway
analysis using a path analysis model. Mol Biosyst. 10:2441–2447.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cancer Genome Atlas Research Network, ;
Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA,
Ellrott K, Shmulevich I, Sander C and Stuart JM: The Cancer Genome
Atlas pan-cancer analysis project. Nat Genet. 45:1113–1120. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guan J, Xie L, Luo X, Yang B, Zhang H, Zhu
Q and Chen X: The prognostic significance of estrogen and
progesterone receptors in grade I and II endometrioid endometrial
adenocarcinoma: Hormone receptors in risk stratification. J Gynecol
Oncol. 30:e132019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tan W, Zhong Z, Carney RP, Men Y, Li J,
Pan T and Wang Y: Deciphering the metabolic role of AMPK in cancer
multi-drug resistance. Semin Cancer Biol. 56:56–71. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nadaraja S, Jørgensen TL, Matzen LE and
Herrstedt J: Impact of age, comorbidity, and FIGO stage on
treatment choice and mortality in older danish patients with
gynecological cancer: A retrospective register-based cohort study.
Drugs Real World Outcomes. 5:225–235. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lax SF: New features in the 2014 WHO
classification of uterine neoplasms. Pathologe. 37:500–511.
2016.(In German). View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kanehisa M, Goto S, Furumichi M, Tanabe M
and Hirakawa M: KEGG for representation and analysis of molecular
networks involving diseases and drugs. Nucleic Acids Res.
38:D355–D360. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ono K, Demchak B and Ideker T: Cytoscape
tools for the web age: D3.js and Cytoscape.js exporters. F1000Res.
3:1432014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Monterossi G, Ghezzi F, Vizza E, Zannoni
GF, Uccella S, Corrado G, Restaino S, Quagliozzi L, Casarin J,
Dinoi G, et al: Minimally invasive approach in type II endometrial
cancer: Is it wise and safe? J Minim Invasive Gynecol. 24:438–445.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu Y, Nan F, Lu K and Wang Y, Liu Y, Wei
S, Wu R and Wang Y: Identification of key genes in endometrioid
endometrial adenocarcinoma via TCGA database. Cancer Biomark.
21:11–21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Corbet C, Pinto A, Martherus R, Santiago
DJJ, Polet F and Feron O: Acidosis drives the reprogramming of
fatty acid metabolism in cancer cells through Changes in
mitochondrial and histone acetylation. Cell Metab. 24:311–323.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pan J, Cheng L, Bi X, Zhang X, Liu S, Bai
X, Li F and Zhao AZ: Elevation of omega-3 polyunsaturated fatty
acids attenuates PTEN-deficiency induced endometrial cancer
development through regulation of COX-2 and PGE2 production. Sci
Rep. 5:149582015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
de Lorgeril M and Salen P: New insights
into the health effects of dietary saturated and omega-6 and
omega-3 polyunsaturated fatty acids. BMC Med. 10:502012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zanoaga O, Jurj A, Raduly L,
Cojocneanu-Petric R, Fuentes-Mattei E, Wu O, Braicu C, Gherman CD
and Berindan-Neagoe I: Implications of dietary omega-3 and omega-6
polyunsaturated fatty acids in breast cancer. Exp Ther Med.
15:1167–1176. 2018.PubMed/NCBI
|
|
24
|
Edmondson RJ, Crosbie EJ, Nickkho-Amiry M,
Kaufmann A, Stelloo E, Nijman HW, Leary A, Auguste A, Mileshkin L,
Pollock P, et al: Markers of the p53 pathway further refine
molecular profiling in high-risk endometrial cancer: A TransPORTEC
initiative. Gynecol Oncol. 146:327–333. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nout RA, Bosse T, Creutzberg CL,
Jürgenliemk-Schulz IM, Jobsen JJ, Lutgens LC, van der Steen-Banasik
EM, van Eijk R, Ter Haar NT and Smit VT: Improved risk assessment
of endometrial cancer by combined analysis of MSI, PI3K-AKT,
Wnt/β-catenin and P53 pathway activation. Gynecol Oncol.
126:466–473. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Theisen ER, Gajiwala S, Bearss J, Sorna V,
Sharma S and Janat-Amsbury M: Reversible inhibition of lysine
specific demethylase 1 is a novel anti-tumor strategy for poorly
differentiated endometrial carcinoma. BMC Cancer. 14:7522014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gao X, Sun J, Huang C, Hu X, Jiang N and
Lu C: RNAi-mediated silencing of NOX4 inhibited the invasion of
gastric cancer cells through JAK2/STAT3 signaling. Am J Transl Res.
9:4440–4449. 2017.PubMed/NCBI
|
|
28
|
Shen Y, Bian R, Li Y, Gao Y, Liu Y, Xu Y,
Song X and Zhang Y: Liensinine induces gallbladder cancer apoptosis
and G2/M arrest by inhibiting ZFX-induced PI3K/AKT pathway. Acta
Biochim Biophys Sin (Shanghai). 607–614. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lo RC, Leung CO, Chan KK, Ho DW, Wong CM,
Lee TK and Ng IO: Cripto-1 contributes to stemness in
hepatocellular carcinoma by stabilizing Dishevelled-3 and
activating Wnt/beta-catenin pathway. Cell Death Differ.
25:1426–1441. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang J, Li Y, Dong M and Wu D: Long
non-coding RNA NEAT1 regulates E2F3 expression by competitively
binding to miR-377 in non-small cell lung cancer. Oncol Lett.
14:4983–4988. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu T, Jia W, An Q, Cao X and Xiao G:
Bioinformatic analysis of GLI1 and related signaling pathways in
chemosensitivity of gastric cancer. Med Sci Monit. 24:1847–1855.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang L, Feng T and Spicer LJ: The role of
tight junction proteins in ovarian follicular development and
ovarian cancer. Reproduction. 155:R183–R198. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Martin TA, Watkins G, Mansel RE and Jiang
WG: Loss of tight junction plaque molecules in breast cancer
tissues is associated with a poor prognosis in patients with breast
cancer. Eur J Cancer. 40:2717–2725. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Youakim A and Ahdieh M: Interferon-gamma
decreases barrier function in T84 cells by reducing ZO-1 levels and
disrupting apical actin. Am J Physiol. 276:G1279–G1288.
1999.PubMed/NCBI
|
|
35
|
Chen H, Wang Y and Xue F: Expression and
the clinical significance of Wnt10a and Wnt10b in endometrial
cancer are associated with the Wnt/β-catenin pathway. Oncol Rep.
29:507–514. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Han X, Cao Y, Wang K and Zhu G: HMGA1
facilitates tumor progression through regulating Wnt/beta-catenin
pathway in endometrial cancer. Biomed Pharmacother. 82:312–318.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang ZM, Wan XH, Sang GY, Zhao JD, Zhu QY
and Wang DM: miR-15a-5p suppresses endometrial cancer cell growth
via Wnt/β-catenin signaling pathway by inhibiting WNT3A. Eur Rev
Med Pharmacol Sci. 21:4810–4818. 2017.PubMed/NCBI
|
|
38
|
Wang T, Wang M, Fang S, Wang Q, Fang R and
Chen J: Fibulin-4 is associated with prognosis of endometrial
cancer patients and inhibits cancer cell invasion and metastasis
via Wnt/β-catenin signaling pathway. Oncotarget. 8:18991–19012.
2017.PubMed/NCBI
|
|
39
|
Chen QG, Zhou W, Han T, Du SQ, Li ZH,
Zhang Z, Shan GY and Kong CZ: MiR-378 suppresses prostate cancer
cell growth through downregulation of MAPK1 in vitro and in vivo.
Tumour Biol. 37:2095–2103. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wei WT, Nian XX, Wang SY, Jiao HL, Wang
YX, Xiao ZY, Yang RW, Ding YQ, Ye YP and Liao WT: miR-422a inhibits
cell proliferation in colorectal cancer by targeting AKT1 and
MAPK1. Cancer Cell Int. 17:912017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang Y, Gao C, Zhang Y, Gao J, Teng F,
Tian W, Yang W, Yan Y and Xue F: Visfatin stimulates endometrial
cancer cell proliferation via activation of PI3K/Akt and
MAPK/ERK1/2 signalling pathways. Gynecol Oncol. 143:168–178. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kanan Y, Matsumoto H, Song H, Sokolov M,
Anderson RE and Rajala RV: Serine/threonine kinase akt activation
regulates the activity of retinal serine/threonine phosphatases,
PHLPP and PHLPPL. J Neurochem. 113:477–488. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li X, Stevens PD, Liu J, Yang H, Wang W,
Wang C, Zeng Z, Schmidt MD, Yang M, Lee EY and Gao T: PHLPP is a
negative regulator of RAF1, which reduces colorectal cancer cell
motility and prevents tumor progression in mice. Gastroenterology.
146:1301–1312.e1-e10. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Huang YS, Chang CC, Lee SS, Jou YS and
Shih HM: Xist reduction in breast cancer upregulates AKT
phosphorylation via HDAC3-mediated repression of PHLPP1 expression.
Oncotarget. 7:43256–43266. 2016.PubMed/NCBI
|
|
45
|
Hribal ML, Mancuso E, Spiga R, Mannino GC,
Fiorentino TV, Andreozzi F and Sesti G: PHLPP phosphatases as a
therapeutic target in insulin resistance-related diseases. Expert
Opin Ther Targets. 20:663–675. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Toderow V, Rahmeh M, Hofmann S, Kirn V,
Mahner S, Jeschke U and von Schönfeldt V: Promotor analysis of ESR1
in endometrial cancer cell lines, endometrial and endometriotic
tissue. Arch Gynecol Obstet. 296:269–276. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jensen EV, Block GE, Smith S, Kyser K and
DeSombre ER: Estrogen receptors and breast cancer response to
adrenalectomy. Natl Cancer Inst Monogr. 34:55–70. 1971.PubMed/NCBI
|
|
48
|
Zhang CZ, Wang SX, Zhang Y, Chen JP and
Liang XM: In vitro estrogenic activities of Chinese medicinal
plants traditionally used for the management of menopausal
symptoms. J Ethnopharmacol. 98:295–300. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hall KL, Teneriello MG, Taylor RR, Lemon
S, Ebina M, Linnoila RI, Norris JH, Park RC and Birrer MJ: Analysis
of Ki-ras, p53, and MDM2 genes in uterine leiomyomas and
leiomyosarcomas. Gynecol Oncol. 65:330–335. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jeczen R, Skomra D, Cybulski M,
Schneider-Stock R, Szewczuk W, Roessner A, Rechberger T and Semczuk
A: P53/MDM2 overexpression in metastatic endometrial cancer:
Correlation with clinicopathological features and patient outcome.
Clin Exp Metastasis. 24:503–511. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wu JP and Luo X: The association between
murine double minute 2 (MDM2) rs2279744 and endometrial cancer risk
in a Chinese Han population. Cell Mol Biol (Noisy-le-grand).
63:128–130. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Dutto I, Tillhon M, Cazzalini O, Stivala
LA and Prosperi E: Biology of the cell cycle inhibitor p21(CDKN1A):
Molecular mechanisms and relevance in chemical toxicology. Arch
Toxicol. 89:155–178. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhao J, Shi L, Zeng S, Ma C, Xu W, Zhang
Z, Liu Q, Zhang P, Sun Y and Xu C: Importin-11 overexpression
promotes the migration, invasion, and progression of bladder cancer
associated with the deregulation of CDKN1A and THBS1. Urol Oncol.
36:11.e1–311.e13. 2018. View Article : Google Scholar
|
|
54
|
Seleznik GM, Reding T, Peter L, Gupta A,
Steiner SG, Sonda S, Verbeke CS, Dejardin E, Khatkov I, Segerer S,
et al: Development of autoimmune pancreatitis is independent of
CDKN1A/p21-mediated pancreatic inflammation. Gut. 67:1663–1673.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hayashi H, Kohno T, Ueno H, Hiraoka N,
Kondo S, Saito M, Shimada Y, Ichikawa H, Kato M, Shibata T, et al:
Utility of assessing the number of mutated KRAS, CDKN2A, TP53, and
SMAD4 genes using a targeted deep sequencing assay as a prognostic
biomarker for pancreatic cancer. Pancreas. 46:335–340. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Umene K, Banno K, Kisu I, Yanokura M,
Nogami Y, Tsuji K, Masuda K, Ueki A, Kobayashi Y, Yamagami W, et
al: Aurora kinase inhibitors: Potential molecular-targeted drugs
for gynecologic malignant tumors. Biomed Rep. 1:335–340. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tanaka Y, Ueda Y, Nakagawa S, Matsuzaki S,
Kobayashi E, Shiki Y, Nishio Y, Takemura M, Yamamoto T, Sawada K,
et al: A phase I/II study of GLIF combination chemotherapy for
taxane/platinum-refractory/resistant endometrial cancer (GOGO-EM2).
Cancer Chemother Pharmacol. 82:585–592. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Umene K, Yanokura M, Banno K, Irie H,
Adachi M, Iida M, Nakamura K, Nogami Y, Masuda K, Kobayashi Y, et
al: Aurora kinase A has a significant role as a therapeutic target
and clinical biomarker in endometrial cancer. Int J Oncol.
46:1498–1506. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Inoue-Yamauchi A, Jeng PS, Kim K, Chen HC,
Han S, Ganesan YT, Ishizawa K, Jebiwott S, Dong Y, Pietanza MC, et
al: Targeting the differential addiction to anti-apoptotic BCL-2
family for cancer therapy. Nat Commun. 8:160782017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Lucas CM, Milani M, Butterworth M, Carmell
N, Scott LJ, Clark RE, Cohen GM and Varadarajan S: High CIP2A
levels correlate with an antiapoptotic phenotype that can be
overcome by targeting BCL-XL in chronic myeloid leukemia. Leukemia.
30:1273–1281. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Sato K, Suzuki T, Yamaguchi Y, Kitade Y,
Nagase T and Ueda H: PLEKHG2/FLJ00018, a Rho family-specific
guanine nucleotide exchange factor, is tyrosine phosphorylated via
the EphB2/cSrc signaling pathway. Cell Signal. 26:691–696. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhang L, Huang J, Yang N, Greshock J,
Liang S, Hasegawa K, Giannakakis A, Poulos N, O'Brien-Jenkins A,
Katsaros D, et al: Integrative genomic analysis of
phosphatidylinositol 3′-kinase family identifies PIK3R3 as a
potential therapeutic target in epithelial ovarian cancer. Clin
Cancer Res. 13:5314–5321. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Dong L and Hui L: HOTAIR promotes
proliferation, migration, and invasion of ovarian cancer SKOV3
cells through regulating PIK3R3. Med Sci Monit. 22:325–331. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhang C, Min L, Liu J, Tian W, Han Y, Qu L
and Shou C: Integrated analysis identified an intestinal-like and a
diffuse-like gene sets that predict gastric cancer outcome. Tumour
Biol. Nov 17–2016.(Epub ahead of print). View Article : Google Scholar
|
|
65
|
Choi CH, Lee JS, Kim SR, Lee YY, Kim CJ,
Lee JW, Kim TJ, Lee JH, Kim BG and Bae DS: Direct inhibition of
eIF4E reduced cell growth in endometrial adenocarcinoma. J Cancer
Res Clin Oncol. 137:463–469. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Shi ZM, Liu YN, Fu B, Shen YF and Li LM:
Expression profile of eukaryotic translation initiation factor and
matrix metalloproteinase 9 in endometrial cancer tissue. J Biol
Regul Homeost Agents. 31:1053–1059. 2017.PubMed/NCBI
|
|
67
|
Guo C, Liang C, Yang J, Hu H, Fan B and
Liu X: LATS2 inhibits cell proliferation and metastasis through the
Hippo signaling pathway in glioma. Oncol Rep. 41:2753–2761.
2019.PubMed/NCBI
|
|
68
|
Zhao B, Li L, Wang L, Wang CY, Yu J and
Guan KL: Cell detachment activates the Hippo pathway via
cytoskeleton reorganization to induce anoikis. Genes Dev. 26:54–68.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Mitamura T, Watari H, Wang L, Kanno H,
Kitagawa M, Hassan MK, Kimura T, Tanino M, Nishihara H, Tanaka S
and Sakuragi N: microRNA 31 functions as an endometrial cancer
oncogene by suppressing Hippo tumor suppressor pathway. Mol Cancer.
13:972014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yasuda M: Immunohistochemical
characterization of endometrial carcinomas: Endometrioid, serous
and clear cell adenocarcinomas in association with genetic
analysis. J Obstet Gynaecol Res. 40:2167–2176. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Alkushi A, Köbel M, Kalloger SE and Gilks
CB: High-grade endometrial carcinoma: Serous and grade 3
endometrioid carcinomas have different immunophenotypes and
outcomes. Int J Gynecol Pathol. 29:343–350. 2010. View Article : Google Scholar : PubMed/NCBI
|