|
1
|
Lu C, Onn A and Vaporciyan A: 78: Cancer
of the lung. In: Holland-Frei Cancer Medicine. 8th edition.
People's Medical Publishing House. 2010.
|
|
2
|
Crinò L, Weder W, van Meerbeeck J and
Felip E; ESMO Guidelines Working Group, : Early stage and locally
advanced (non-metastatic) non-small-cell lung cancer: ESMO clinical
practice guidelines for diagnosis, treatment and follow-up. Ann
Oncol. 21 (Suppl 5):v103–v115. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yokoi K, Taniguchi T, Usami N, Kawaguchi
K, Fukui T and Ishiguro F: Surgical management of locally advanced
lung cancer. Gen Thorac Cardiovasc Surg. 62:522–530. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Biomarkers Definitions Working Group, :
Biomarkers and surrogate endpoints: Preferred definitions and
conceptual framework. Clin Pharmacol Ther. 69:89–95. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chan DS, Yang H, Kwan MH, Cheng Z, Lee P,
Bai L, Jiang Z, Wong C, Fong W, Leung C and Ma D: Biochimie
Structure-based optimization of FDA-approved drug methylene blue as
a c-myc G-quadruplex DNA stabilizer. Biochimie. 93:1055–1064. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ma DL, Lin S, Wang W, Yang C and Leung CH:
Luminescent chemosensors by using cyclometalated iridium(III)
complexes and their applications. Chem Sci. 8:878–889. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Miao X, Wang W, Kang T, Liu J, Shiu KK,
Leung CH and Ma DL: Ultrasensitive electrochemical detection of
miRNA-21 by using an iridium(III) complex as catalyst. Biosens
Bioelectron. 86:454–458. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tian S and Suárez-fariñas M:
Hierarchical-TGDR: Combining biological hierarchy with a
regularization method for multi-class classification of lung cancer
samples via high-throughput gene-expression data. Systems
Biomedicine. 1:93–102. 2013. View Article : Google Scholar
|
|
9
|
Ben-hamo R, Boue S, Martin F, Talikka M
and Efroni S: Classification of lung adenocarcinoma and squamous
cell carcinoma samples based on their gene expression profile in
the sbv IMPROVER diagnostic signature challenge. Systems
Biomedicine. 1:83–92. 2013. View Article : Google Scholar
|
|
10
|
Tian S: Classification and survival
prediction for early-stage lung adenocarcinoma and squamous cell
carcinoma patients. Oncol Lett. 14:5464–5470. 2017.PubMed/NCBI
|
|
11
|
Tarca AL, Lauria M, Unger M, Bilal E, Boue
S, Kumar Dey K, Hoeng J, Koeppl H, Martin F, Meyer P, et al:
Strengths and limitations of microarray-based phenotype prediction:
Lessons learned from the IMPROVER diagnostic signature challenge.
Bioinformatics. 29:2892–2899. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mramor M, Leban G, Demsar J and Zupan B:
Visualization-based cancer microarray data classification analysis.
Bioinformatics. 23:2147–2154. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang L, Wang L, Du B, Wang T, Tian P and
Tian S: Classification of non-small cell lung cancer using
significance analysis of microarray-gene set reduction algorithm.
Biomed Res Int. 2016:24916712016.PubMed/NCBI
|
|
14
|
Boutros PC, Lau SK, Pintilie M, Liu N,
Shepherd FA, Der SD, Tsao MS, Penn LZ and Jurisica I: Prognostic
gene signatures for non-small-cell lung cancer. Proc Natl Acad Sci
USA. 106:2824–2828. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu CQ, Ding K, Strumpf D, Weir BA,
Meyerson M, Pennell N, Thomas RK, Naoki K, Ladd-Acosta C, Liu N, et
al: Prognostic and predictive gene signature for adjuvant
chemotherapy in resected non-small-cell lung cancer. J Clin Oncol.
28:4417–4424. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Der SD, Sykes J, Pintilie M, Zhu CQ,
Strumpf D, Liu N, Jurisica I, Shepherd FA and Tsao MS: Validation
of a histology-independent prognostic gene including stage ia
patients. J Thorac Oncol. 9:59–64. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hira ZM and Gillies DF: A review of
feature selection and feature extraction methods applied on
microarray data. Adv Bioinformatics. 2015:1983632015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rahmatallah Y, Emmert-Streib F and Glazko
G: Gene set analysis approaches for RNA-seq data: performance
evaluation and application guideline. Brief Bioinform. 17:393–407.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hrdlickova R, Toloue M and Tian B: RNA-Seq
mthods for transcriptome analysis. Wiley Interdsicrip Rev RNA.
8:e13642017. View Article : Google Scholar
|
|
20
|
Law CW, Chen Y, Shi W and Smyth GK: Voom:
Precision weights unlock linear model analysis tools for RNA-seq
read counts. Genome Biol. 15:R292014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ramasamy A, Mondry A, Holmes CC and Altman
DG: Key issues in conducting a meta-analysis of gene expression
microarray datasets. PLoS Med. 5:e1842008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Blettner M, Sauerbrei W, Schlehofer B,
Scheuchenpflug T and Friedenreich C: Traditional reviews,
meta-analyses and pooled analyses in epidemiology. Int J Epidemiol.
28:1–9. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu J, Huang J and Ma S: Integrative
analysis of multiple cancer genomic datasets under the
heterogeneity model. Stat Med. 32:3509–3521. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Krzystanek M, Moldvay J, Szüts D, Szallasi
Z and Eklund AC: A robust prognostic gene expression signature for
early stage lung adenocarcinoma. Biomark Res. 4:42016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lu Y, Lemon W, Liu PY, Yi Y, Morrison C,
Yang P, Sun Z, Szoke J, Gerald WL, Watson M, et al: A gene
expression signature predicts survival of patients with stage I
non-small cell lung cancer. PLoS Med. 3:e4672006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Skrzypski M, Dziadziuszko R, Jassem E,
Szymanowska-Narloch A, Gulida G, Rzepko R, Biernat W, Taron M,
Jelitto-Górska M, Marjański T, et al: Main histologic types of
non-small-cell lung cancer differ in expression of
prognosis-related genes. Clin Lung Cancer. 14:666–673. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tian S, Wang C and An MW: Test on
existence of histology subtype-specific prognostic signatures among
early stage lung adenocarcinoma and squamous cell carcinoma
patients using a Cox-model based filter. Biol Direct. 10:152015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tian S: Identification of subtype-specific
prognostic genes for early-stage lung adenocarcinoma and squamous
cell carcinoma patients using an embedded feature selection
algorithm. PLoS One. 10:e01346302015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Safran M, Dalah I, Alexander J, Rosen N,
Stein TI, Shmoish M, Nativ N, Bahir I, Doniger T, Krug H, et al:
GeneCards Version 3: The human gene integrator. Database (Oxford).
2010:baq0202010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zeng XQ, Li GZ, Yang JY, Yang MQ and Wu
GF: Dimension reduction with redundant gene elimination for tumor
classification. BMC Bioinformatics. 9 (Suppl 6):S82008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bild AH, Yao G, Chang JT, Wang Q, Potti A,
Chasse D, Joshi M, Harpole D, Lancaster JM, Berchuck A, et al:
Oncogenic pathway signatures in human cancers as a guide to
targeted therapies. Nature. 439:353–357. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Botling J, Edlund K, Lohr M, Hellwig B,
Holmberg L, Lambe M, Berglund A, Ekman S, Bergqvist M, Pontén F, et
al: Biomarker discovery in non-small cell lung cancer: Integrating
gene expression profiling, meta-analysis and tissue microarray
validation. Clin Cancer Res. 19:194–204. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rousseaux S, Debernardi A, Jacquiau B,
Vitte A, Vesin A, Nagy-mignotte H, Moro-sibilot D, Brichon P,
Hainaut P, Laffaire J, et al: Ectopic activation of germline and
placental genes identifies aggressive metastasis-prone lung
cancers. Sci Transl Med. 5:186ra662013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
McCall MN, Bolstad BM and Irizarry RA:
Frozen robust multiarray analysis (fRMA). Biostatistics.
11:242–253. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Smyth GK: Limma: linear models for
microarray data. In: Bioinformatics and Computational Biology
Solutions Using R and Bioconductor. Gentleman R, Carey V, Dudoit S,
Irizarry R and Huber W: Springer; New York, NY: pp. 397–420.
2005
|
|
36
|
DerSimonian R and Laird NM: Meta-analysis
in clinical trials. Control Clin Trials. 7:177–188. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Choi JK, Yu U, Kim S and Yoo OJ: Combining
multiple microarray studies and modeling interstudy variation.
Bioinformatics. 19 (Suppl 1):i84–i90. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J Roy Statist Soc. 57:289–300. 1995.
|
|
39
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res 41
(Database Issue). D808–D815. 2013.
|
|
40
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ogata H, Goto S, Sato K, Fujibuchi W, Bono
H and Kanehisa M: KEGG: Kyoto encyclopedia of genes and genomes.
Nucleic Acids Res. 27:29–34. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Uno H, Cai T, Pencina MJ, D'Agostino RB
and Wei LJ: On the C-statistics for evaluating overall adequacy of
risk prediction procedures with censored survival data. Stat Med.
30:1105–1117. 2011.PubMed/NCBI
|
|
43
|
Laimighofer M, Krumsiek J, Buettner F and
Theis FJ: Unbiased prediction and feature selection in
high-dimensional survival regression. J Comput Biol. 23:279–290.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang W, Stabile LP, Keohavong P, Romkes
M, Grandis JR, Traynor AM and Siegfried JM: Mutation and
polymorphism in the EGFR-TK domain associated with lung cancer. J
Thorac Oncol. 1:635–647. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu J, Yang XY and Shi WJ: Identifying
differentially expressed genes and pathways in two types of
non-small cell lung cancer: Adenocarcinoma and squamous cell
carcinoma. Genet Mol Res. 13:95–102. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Choi CM, Jang SJ, Park SY, Choi YB, Jeong
JH, Kim DS, Kim HK, Park KS, Nam BH, Kim HR, et al:
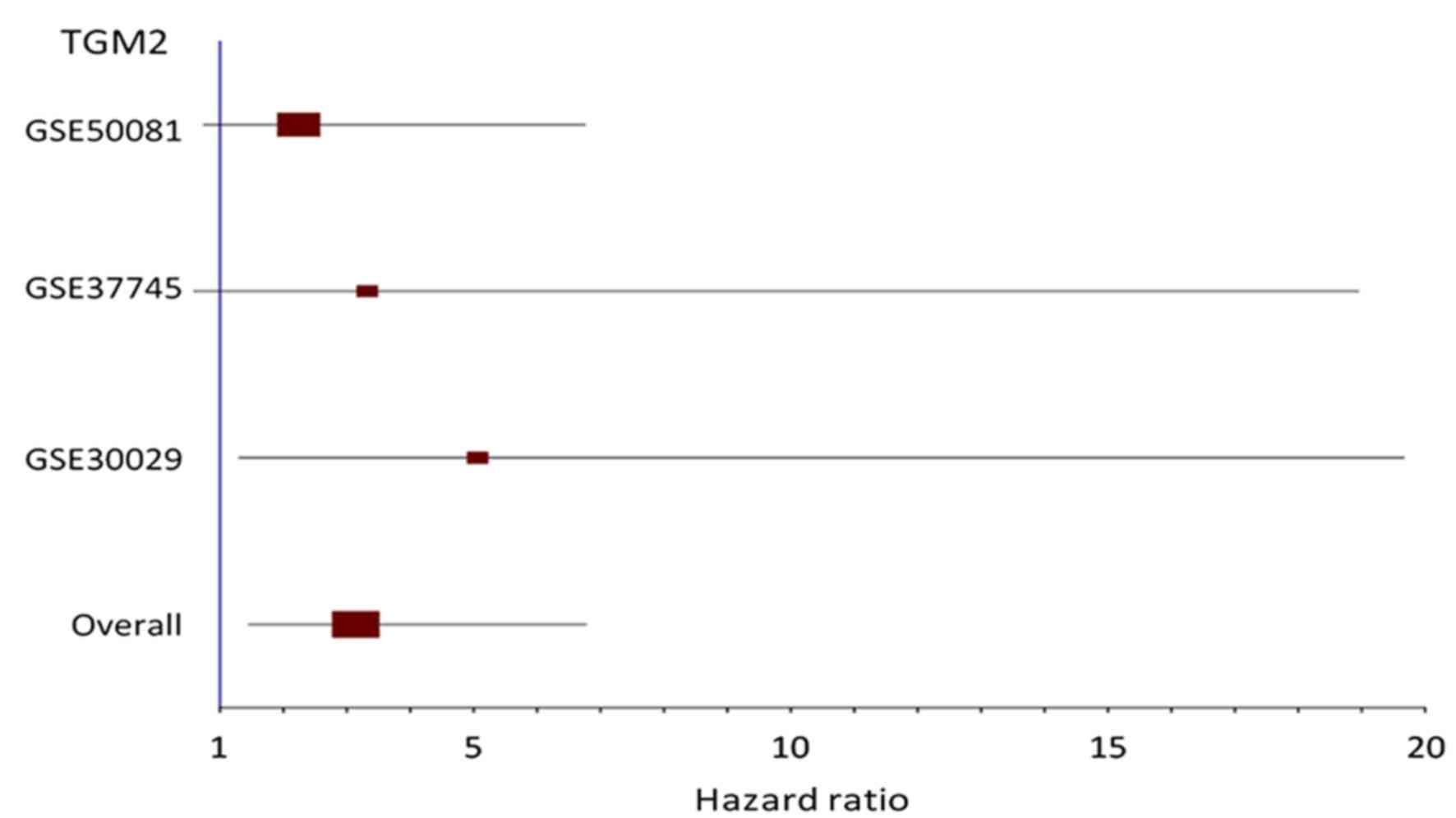
Transglutaminase 2 as an independent prognostic marker for survival
of patients with non-adenocarcinoma subtype of non-small cell lung
cancer. Mol Cancer. 10:1192011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ma S and Huang J: Regularized gene
selection in cancer microarray meta-analysis. BMC bioinformatics.
10:12009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zeng XQ and Li GZ: Supervised redundant
feature detection for tumor classification. BMC Med Genomics. 7
(Suppl 2):S52014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ge R, Zhou M, Luo Y, Meng Q, Mai G, Ma D,
Wang G and Zhou F: McTwo: A two-step feature selection algorithm
based on maximal information coefficient. BMC Bio. 1–14. 2016.
|
|
50
|
Gu JL, Lu Y, Liu C and Lu H: Multiclass
classification of sarcomas using pathway based feature selection
method. J Theor Biol. 362:3–8. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tian S: Identification of subtype-specific
prognostic signatures using Cox models with redundant gene
elimination. Oncol Lett. 15:8545–8555. 2018.PubMed/NCBI
|
|
52
|
Saeys Y, Inza I and Larrañaga P: A review
of feature selection techniques in bioinformatics. Bioinformatics.
23:2507–2517. 2007. View Article : Google Scholar : PubMed/NCBI
|