Introduction
Lung cancer is one of main malignant tumors that
cause death in cancer patients, accounting for 10–13% of systemic
malignant tumors, with non-small cell lung cancer (NSCLC)
accounting for 80–85% of all lung cancers (1,2). Early
NSCLC has no obvious specific clinical manifestations, so it is
often ignored by patients who then miss the best treatment period,
leading to high prognostic mortality of NSCLC (3). According to Gettinger et al
(4), approximately 78% of NSCLC
patients are in the middle or advanced stage when diagnosed, losing
the chance of surgical cure. Planchard et al (5) reported that NSCLC patients have a
5-year survival rate of only 41.8%. At present, chemotherapy is the
main non-surgical medical treatment for NSCLC. The clinical
treatment of NSCLC chemotherapy is mainly to prolong the survival
time of patients and improve their quality of life (6). Due to its high incidence and mortality,
NSCLC is a hot research topic in the clinic.
The human 8-hydroxyguanine glycosidase 1 (hOGG1)
gene is a DNA oxidative damage repair gene (7). It can recognize and specifically excise
the oxidative damage product of DNA, 8-hydroxydeoxyguanine,
repairing damaged DNA (8). The hOGG1
gene has a C/G polymorphism in the 1245th base of exon 7, making
the 326th codon produce serine (S) or cysteine (C), resulting in
S→C amino acid replacement (9).
Related literature has reported (10,11) that
the mutation of base C to G will lead to a decrease in the activity
of hOGG1 enzyme, making the body susceptible to diseases such as
tumors. Studies have reported (12)
that the hOGG1 Ser326Cys polymorphism increases the susceptibility
of high-risk prostates. However, currently, there are less related
literature on the hOGG1 Ser326Cys (rs1052133) gene polymorphism and
the prognosis in NSCLC patients. The aim of this study was to
investigate the mRNA expression level and gene polymorphism of
hOGG1 in NSCLC patients and analyze the prognostic
significance.
Patients and methods
Patient data
A total of 182 NSCLC patients surgically treated in
Xiang Yang No. 1 People's Hospital, Hubei University of Medicine
(Xiangyang, China) from January 2008 to January 2012 were the NSCLC
group, including 104 males and 78 females, aged 26–71 years, with
an average age of 52.57±2.75 years. Two hundred healthy individuals
in Xiang Yang No. 1 People's Hospital, Hubei University of Medicine
were the control group, including 111 males and 89 females, aged
25–67 years, with an average age of 54.16±3.04 years. All the
patients had complete clinical and follow-up data.
Electrocardiogram, blood routine and liver and kidney function of
patients were normal. Patients with communication impairment or
cognitive dysfunction were excluded. All the patients and their
families signed an informed consent form and cooperated with
medical staff to complete relevant medical treatment. It was found
that there was no significant difference in terms of sex, age,
smoking, drinking and chronic diseases between the two groups
(P>0.05), indicating that the experimental results are accurate
and credible. Details of subject basic data are shown in Table I. This study was approved by Xiang
Yang No. 1 People's Hospital, Hubei University of Medicine.
 | Table I.Patient basic data of NSCLC group and
control group [n (%)]. |
Table I.
Patient basic data of NSCLC group and
control group [n (%)].
| Factors | NSCLC group
(n=182) | Control group
(n=200) | χ2
value | P-value |
|---|
| Sex |
|
| 0.105 | 0.747 |
| Male | 104 (57.14) | 111 (55.50) |
|
|
|
Female | 78
(42.86) | 89
(44.50) |
|
|
| Age (years) |
|
| 2.318 | 0.128 |
|
<40 | 85
(46.70) | 119 (59.50) |
|
|
| ≥40 | 97
(53.30) | 81
(40.50) |
|
|
| Smoking |
|
| 0.384 | 0.536 |
| Yes | 98
(53.85) | 114 (57.00) |
|
|
| No | 84
(46.15) | 86
(43.00) |
|
|
| Drinking |
|
| 0.025 | 0.876 |
| Yes | 75
(41.21) | 84
(42.00) |
|
|
| No | 107 (58.79) | 116 (58.00) |
|
|
| Chronic diseases |
|
| 0.153 | 0.696 |
| Yes | 81
(44.51) | 93
(46.50) |
|
|
| No | 101 (55.49) | 107 (53.50) |
|
|
| Pathological
diagnosis typing |
|
| – | – |
| Squamous
cell carcinoma | 93
(51.10) | – |
|
|
|
Adenocarcinoma | 54
(29.67) | – |
|
|
| Large
cell carcinoma | 35
(19.23) | – |
|
|
Extraction of total DNA and RNA
Fasting venous blood (5 ml) was collected from
patients in NSCLC group and control group into EDTA anticoagulant
tubes. The DNA in the blood was extracted strictly according to the
instructions of the animal tissue/cell genomic DNA extraction kit
(Shanghai Jingke Chemical Technology Co., Ltd., item no.: D1700).
Cancer tissues and adjacent tissues surgically resected from
patients in NSCLC group were added to the total RNA extraction
reagent (Shanghai Kanglang Biotechnology Co., Ltd., item no.:
KL101-151) to extract the RNA in tissues. The UV-9000S ultraviolet
spectrophotometer (Jingfeile Instrument Co., Ltd.) was used to
measure the quality and concentration of DNA and RNA at wavelengths
of 260 and 280 nm. At OD260/OD280 value of
approximately 1.8 the extracted DNA and RNA purity was considered
satisfactory, when necessary, re-extraction and identification were
performed.
RT-qPCR detection of hOGG1
expression
RT-PCR was used to detect the hOGG1 expression, and
TRIzol kit (Wuhan Purity Biotechnology Co., Ltd., item no.:
CD-13433-ML) to extract whole blood total RNA in strict accordance
with the instructions. After the detection of nucleic acid
concentration, the instructions of miRNA reverse transcription kit
(Beijing Baiaolaibo Technology Co., Ltd., item no.: ALH266-PTO) was
used for reverse transcription to generate cDNA. qPCR was
performed, with β-actin as an internal reference, according to the
PCR kit (Beijing Zhijiefangyuan Technology Co., Ltd., item no.:
RR047A) instructions. qPCR reaction conditions: Pre-denaturation at
95°C for 3 min, and then 95°C for 30 sec, 58°C for 30 sec, 72°C for
30 sec, for a total of 35 cycles, and extension at 72°C for 10 min.
After the reaction was completed, the temperature was raised to
95°C, then reduced to 60°C; then the temperature was gradually
heated to 95°C in 20 min, then maintained for 15 sec. ABI 7900 PCR
instrument was used for qPCR detection. The primer sequence of
hOGG1 was designed and synthesized by Shanghai Jima Pharmaceutical
Technology Co., Ltd., with β-actin as a reaction internal
reference. Each group of samples was repeated 3 times.
2−∆Cq method was used to analyze the expression level of
hOGG1 in the specimen (13). Primer
sequences are shown in Table
II.
 | Table II.hOGG1 primer and internal reference
sequences. |
Table II.
hOGG1 primer and internal reference
sequences.
| Primer sequence | Upstream primer | Downstream
primer | Length (bp) |
|---|
| hOGG1 |
5′-GGAAGGTGCTTGGGGAAT-3′ |
5′-ACTGTCACTAGTCTCACCAG-3′ | 200 |
| β-actin |
5′-ATCATGTTTGAGACCTTCAACA-3′ |
5′-CATCTCTTGCTCGAAGTCCA-3′ | 348 |
Genotyping
Taqman probe method was used for hOGG1 rs1052133
locus genotyping. The probe was designed and synthesized by
Shanghai Jima Pharmaceutical Technology Co., Ltd., with probe
sequences of 5′-FAM-CCAATGCCGCCATG-MGB-3′ and
5′-HEX-CGCCAATCCCGCCA-MGB-3′. A total of 5 µl Mix buffer, 0.5 µl
upstream primer, 0.5 µl downstream primer, probe sequence (0.25 µl
each) and 10 ng DNA (Invitrogen; Thermo Fisher Scientific, Inc.;
cat. no. 15279011) were added, and deionized water was added to
make a total volume of 10 µl. The American Bole PCR amplification
instrument (Zhejiang Tuopuyunnong Technology Co., Ltd.) was used
for amplification. PCR reaction conditions: pre-denaturation at
94°C for 3 min, at 94°C for 30 sec; annealing at 61°C for 30 sec,
at 72°C for 30 sec, for a total of 30 cycles, and then extension at
72°C for 10 min. Taq DNA polymerase was purchased from
GeneCopoeia.
Statistical analysis
SPSS 19.5 (Beijing Sichuangweida Information
Technology Co., Ltd.) software statistical package was used for
processing and analyzing data. Measurement data are expressed as
mean ± standard deviation, and analyzed by t-test. Enumeration data
were analyzed by Chi-square test. Survival data were analyzed using
Kaplan-Meier survival curve and log rank test. ROC curve was used
for the value of hOGG1 expression in the survival of patients with
NSCLC. The genotype distribution was analyzed by the
goodness-of-fit Chi-square test to determine whether it was
consistent with the Hardy-Weinberg equilibrium. P<0.05 was
considered to indicate a statistically significant difference.
Results
Comparison of subject general clinical
data between NSCLC group and control group
To achieve accurate and credible experimental
results, individuals were compared between two groups in terms of
sex, age, smoking, drinking and chronic diseases, with no
significant difference (P>0.05), which proves that the two
groups of individuals are comparable (Table I).
Comparison of hOGG1 expression between
cancer tissues and adjacent tissues of NSCLC patients
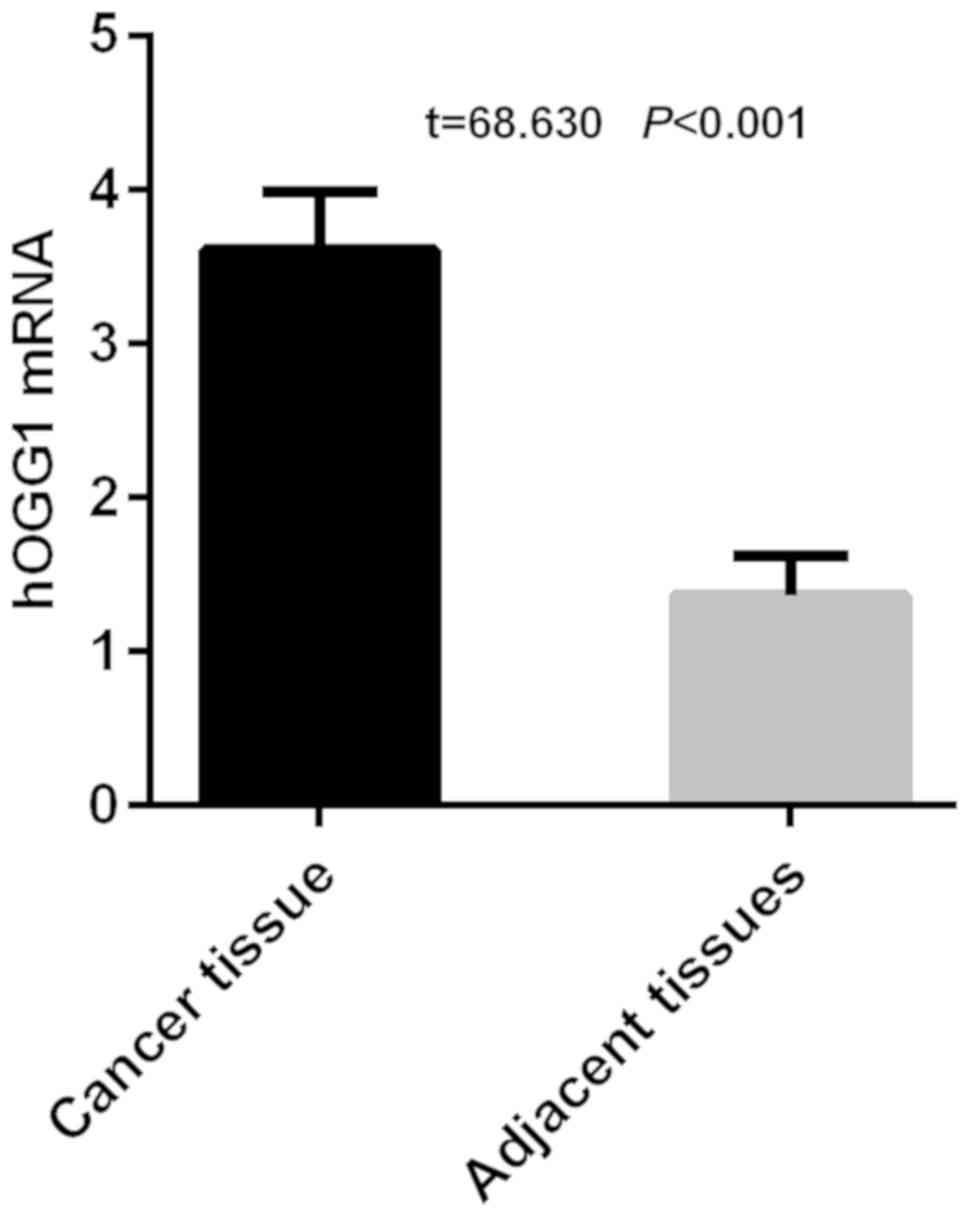
The results of RT-qPCR detection of hOGG1 showed
that the expression level of hOGG1 was 3.61±0.38 in cancer tissues
and 1.37±0.25 in adjacent tissues in NSCLC group of patients.
Expression of hOGG1 was significantly higher in cancer tissues than
that in adjacent tissues in NSCLC group of patients, with a
statistically significant difference (t=68.630, P<0.001)
(Fig. 1).
Hardy-Weinberg equilibrium test of
hOGG1 genotype distribution frequency and distribution of genotypes
in each group
The genotypes of hOGG1 polymorphism locus rs1052133
in NSCLC group and control group included S/S, S/C and C/C
genotypes, with the genotype distribution frequencies of NSCLC
group (χ2=0.732, P=0.411) and control group
(χ2=0.642, P=0.354) consistent with the Hardy-Weinberg
equilibrium. In NSCLC group of patients, there were 6 cases of S/S
type, 65 cases of S/C type and 111 cases of C/C type. In control
group of patients, there were 15 cases of S/S type, 90 cases of S/C
type and 95 cases of C/C type. Compared to S/S and S/C genotypes,
the C/C gene was found to be more common in NSCLC group than in
control group (P<0.05, OR=2.2, 95%, CI=1.27–4.52). Compared with
allele S, it was found that allele C was more common in NSCLC group
than in control group (P<0.05) (Table III).
 | Table III.Distribution of polymorphism locus
gene and genotypes in each group. |
Table III.
Distribution of polymorphism locus
gene and genotypes in each group.
| Polymorphism
locus | NSCLC group
(n=182) | Control group
(n=200) | χ2
value | P-value |
|---|
| rs1052133 |
|
|
|
|
|
Genotype |
|
|
|
|
|
S/S | 6
(3.30) | 15 (7.50) |
3.241 | 0.072a |
|
S/C | 65
(35.71) | 90 (45.00) |
5.638 | 0.018b |
|
C/C | 111 (60.99) | 95 (47.50) | 23.631 |
<0.001c |
|
Allele |
|
|
7.793 | 0.005 |
|
S | 77
(21.15) | 120 (30.00) |
|
|
|
C | 287 (78.85) | 280 (70.00) |
|
|
Expression and gene polymorphism of
hOGG1 and prognosis of NSCLC patients
Patients were followed up for 3 years by telephone,
review or letter, to the last date of follow up. i.e., December
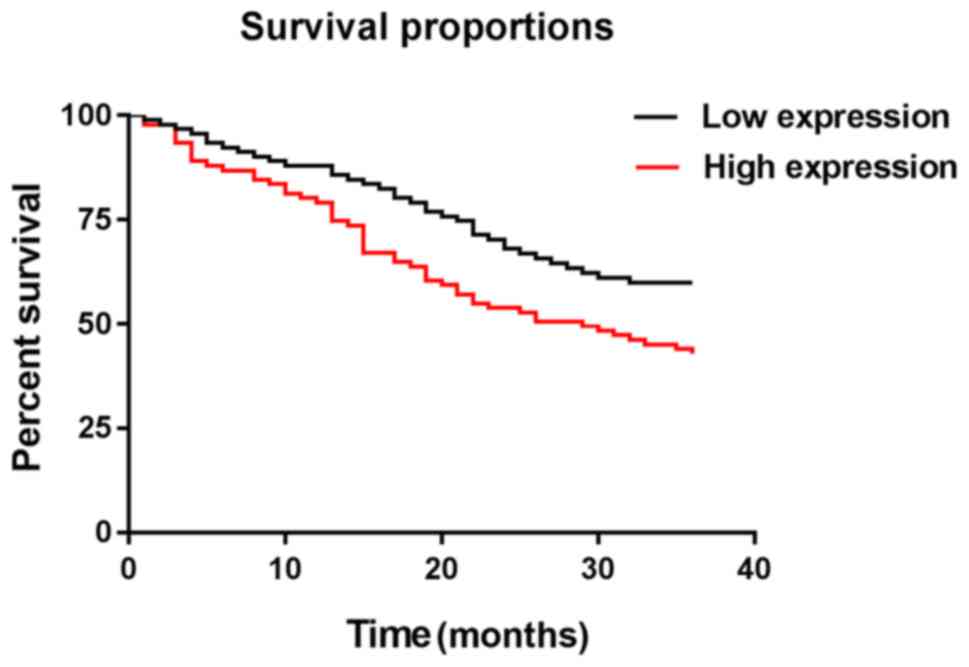
2017 or if patients died. The median value of the hOGG1 expression
level in detection results as the boundary, NSCLC patients were
divided into hOGG1 high expression group (≥3.61) with 91 cases and
hOGG1 low expression group (<3.61) with 91 cases. The 1-, 2- and
3-year survival rates of patients in hOGG1 low expression group
were 87.91, 70.33, and 63.74%, respectively. Those of patients in
hOGG1 high expression group were 79.12, 56.04, and 43.96%,
respectively. Those of patients in hOGG1 low expression group were
significantly higher than those in hOGG1 high expression group
(P=0.007). The number of S/S+S/C was small, so they were merged
into one group. Those of patients with hOGG1 rs1052133 of S/S+S/C
genotypes were 83.10, 52.11, and 45.07%, respectively, and those of
patients with C/C genotype were 80.18, 48.65, and 36.04%,
respectively, with no significant difference (P=0.365) (Figs. 2 and 3).
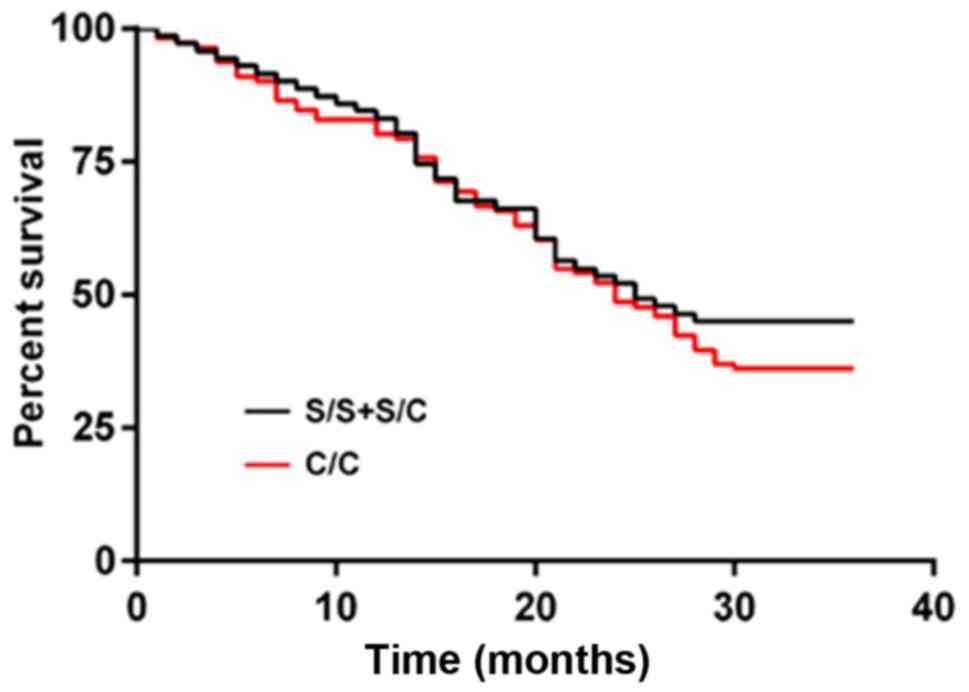 | Figure 3.hOGG1 gene polymorphism and prognosis
of NSCLC patients. The 1-, 2- and 3-year survival rates of patients
with hOGG1 rs1052133 of S/S+S/C genotypes were 83.10, 52.11, and
45.07%, respectively, and those of patients with C/C genotype were
80.18, 48.65, and 36.04%, respectively, with no significant
difference (P=0.365). hOGG1, human 8-hydroxyguanine glycosidase 1;
NSCLC, non-small cell lung cancer. |
Predictive value of hOGG1 expression
in survival of patients with NSCLC
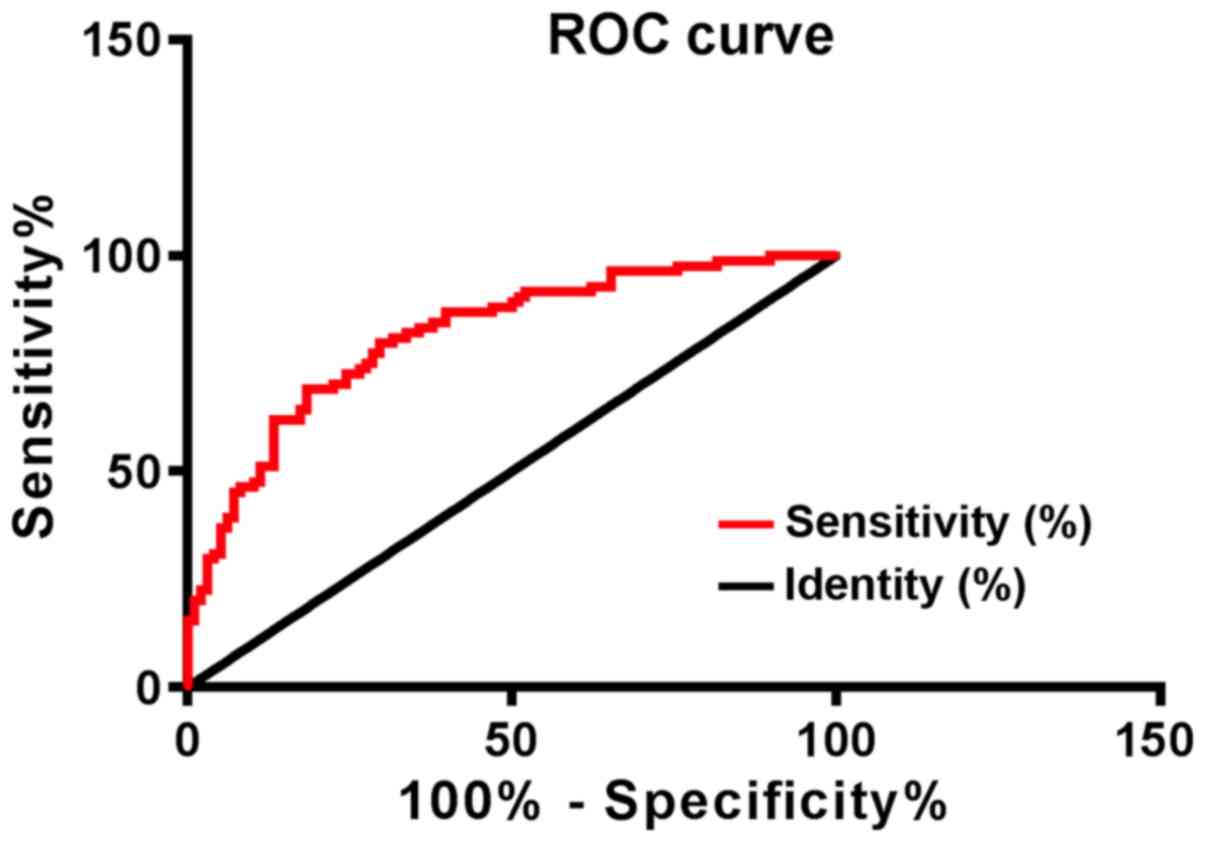
At 3 years, 98 patients with NSCLC survived, and 84
patients had died. Patients were seperated into the survival group
and the death group according to the survival. The hOGG1 expression
in the survival group was 3.32±0.34, and the hOGG1 expression in
the death group was 3.81±0.39. The sensitivity, specificity and AUC
of hOGG1 for survival prediction were 83.33%, 64.29%, and 0.816,
respectively (Fig. 4).
Discussion
Substantial data has shown that hOGG1 polymorphism
is closely related to a variety of malignant tumors, such as
bladder cancer, breast cancer and rectal cancer (14,15).
Therefore, the hOGG1 gene polymorphism and the prognosis were
analyzed. First, the Hardy-Weinberg equilibrium analysis was
performed on the hOGG1 genotype distribution frequency in NSCLC
group and control group of patients. The results showed that there
was no difference in the gene frequency between them. The genotypes
of hOGG1 polymorphism locus rs1052133 in NSCLC group and control
group included S/S, S/C, and C/C genotypes. In NSCLC group of
patients, there were 6 cases of S/S type, 65 cases of S/C type and
111 cases of C/C type. In control group of patients, there were 15
cases of S/S type, 90 cases of S/C type and 95 cases of C/C type.
Compared to S/S and S/C genotypes, the C/C gene was found to be
more common in NSCLC group than in control group. However, there
was no significant difference in the 1-, 2- and 3-year survival
rates of patients with hOGG1 rs1052133 of S/S+S/C genotypes and C/C
genotypes. The mutation of hOGG1 gene forms Ser326 and Cys326
results in low ability of hOGG1 to repair 8-hydroxy-deoxyguanine
(8-oxoG). Having high mutagenicity, 8-oxoG is closely related to
the occurrence and development of tumors, leading to an increased
risk of cancers (16). Studies have
reported (17) that when hOGG1
repairs DNA damage caused by 8-oxoG, it is found that the repair
ability of the C allele is approximately 7 times lower than that of
the S allele. According to reports by Wang et al (18), people in Southeast Asia who carry C/C
genotype have a significantly higher risk of developing liver
cancer than those who carry S/C or S/S genotypes. According to Xie
et al (19), the risk of
death in patients with nasopharyngeal carcinoma with hOGG1 mutant
genotype will be significantly higher than that of wild genotypes.
Their results are consistent with the results of this study, which
further support our experiments. It is indicated that rs1052133
locus can be used for NSCLC gene screening, but is not related to
survival prognosis.
A retrospective analysis of clinical data of 182
NSCLC patients and 200 healthy individuals was performed. The
results of RT-qPCR detection showed that the expression level of
hOGG1 was significantly higher in cancer tissues than that in
adjacent tissues in NSCLC group of patients, with a statistically
significant difference. The median value of the hOGG1 expression
level in detection results as the boundary, NSCLC patients were
divided into hOGG1 high expression group and hOGG1 low expression
group. The 1-, 2- and 3-year survival rates of patients in hOGG1
low expression group were significantly higher than those in hOGG1
high expression group. AUC of hOGG1 expression in the survival
prediction of patients with NSCLC was 0.816 which is a high
predictive value. Studies have reported that the expression of most
oncogenes and tumor suppressor genes can affect the proliferation
and differentiation of tumor cells, correlated with the occurrence
and development and prognosis of tumors (20). As a glycosidase that repairs
oxidative DNA damage in humans, hOGG1 is closely related to the
maintenance of the normal function of human genes and the
occurrence and development of cancers (21). hOGG1 gene polymorphism has been
reported to be important in a variety of digestive system cancers,
closely related to the prognosis of various cancers. hOGG1 gene can
inhibit the proliferation of lung cancer cells and the growth of
lung cancer tumors (22). According
to studies by Zhou et al (23), the hOGG1 expression in serum of
patients with acute leukemia depression is higher than that in
normal patients, which further supports our experimental
results.
There are still some shortcomings in this study. For
example, the specimen number selected for the study is small, due
to limited experimental conditions. In addition, there may be
differences in the hOGG1 gene polymorphism in NSCLC patients in
different regions. Moreover, individual differences after treatment
and intervention were not taken into consideration.
In summary, hOGG1 is highly expressed in NSCLC
tissues. Compared to S/S and S/C genotypes, the C/C gene was found
to be more common in NSCLC group than in control group. hOGG1 has a
high predictive value for patient survival. More studies on this
polymorphism locus will be carried out in subsequent studies to
determine whether this polymorphism can serve as a molecular marker
for the risk of NSCLC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JW drafted the manuscript. JW and PW conceived and
designed the study. PW performed RT-qPCR and genotyping. JW and PW
were equal contributors in extraction of total DNA and RNA. Both
authors read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by Xiang Yang No. 1 People's
Hospital, Hubei University of Medicine (Xiangyang, China). Signed
informed consents were obtained from the patients and/or
guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fehrenbacher L, Spira A, Ballinger M,
Kowanetz M, Vansteenkiste J, Mazieres J, Park K, Smith D,
Artal-Cortes A, Lewanski C, et al POPLAR Study Group, :
Atezolizumab versus docetaxel for patients with previously treated
non-small-cell lung cancer (POPLAR): A multicentre, open-label,
phase 2 randomised controlled trial. Lancet. 387:1837–1846. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Masters GA, Johnson DH and Temin S:
Systemic therapy for stage IV non-small-cell lung cancer: American
Society of Clinical Oncology Clinical Practice Guideline Update. J
Oncol Pract. 12:90–93. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jamal-Hanjani M, Wilson GA, McGranahan N,
Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R,
Rosenthal R, et al TRACERx Consortium, : Tracking the evolution of
non-small-cell lung cancer. N Engl J Med. 376:2109–2121. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gettinger S, Rizvi NA, Chow LQ, Borghaei
H, Brahmer J, Ready N, Gerber DE, Shepherd FA, Antonia S, Goldman
JW, et al: Nivolumab monotherapy for first-line treatment of
advanced non-small-cell lung cancer. J Clin Oncol. 34:2980–2987.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Planchard D, Yokoi T, McCleod MJ, Fischer
JR, Kim YC, Ballas M, Shi K and Soria JC: A phase III study of
durvalumab (MEDI4736) with or without tremelimumab for previously
treated patients with advanced NSCLC: Rationale and protocol design
of the ARCTIC study. Clin Lung Cancer. 17:232–236.e1. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Emdad L, Lebedeva IV, Su ZZ, Gupta P,
Sarkar D, Settleman J and Fisher PB: Combinatorial treatment of
non-small-cell lung cancers with gefitinib and Ad.mda-7 enhances
apoptosis-induction and reverses resistance to a single therapy. J
Cell Physiol. 210:549–559. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cravens SL and Stivers JT: Comparative
effects of ions, molecular crowding, and bulk DNA on the damage
search mechanisms of hOGG1 and hUNG. Biochemistry. 55:5230–5242.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kadıoğlu E, Taçoy G, Özçağlı E, Okyay K,
Akboğa MK, Çengel A and Şardaş S: The role of oxidative DNA damage
and GSTM1, GSTT1, and hOGG1 gene polymorphisms in coronary artery
disease risk. Anatol J Cardiol. 16:931–938. 2016.PubMed/NCBI
|
|
9
|
Alanazi M, Pathan AAK, Shaik JP, Alhadheq
A, Khan Z, Khan W, Al Naeem A and Parine NR: The hOGG1 Ser326Cys
gene polymorphism and breast cancer risk in Saudi population.
Pathol Oncol Res. 23:525–535. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sliwinska A, Kwiatkowski D, Czarny P, Toma
M, Wigner P, Drzewoski J, Fabianowska-Majewska K, Szemraj J, Maes
M, Galecki P, et al: The levels of 7,8-dihydrodeoxyguanosine
(8-oxoG) and 8-oxoguanine DNA glycosylase 1 (OGG1) - A potential
diagnostic biomarkers of Alzheimer's disease. J Neurol Sci.
368:155–159. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Z, Guan W, Li MX, Zhong ZY, Qian CY,
Yang XQ, Liao L, Li ZP and Wang D: Genetic polymorphism of DNA
base-excision repair genes (APE1, OGG1 and XRCC1) and their
correlation with risk of lung cancer in a Chinese population. Arch
Med Res. 42:226–234. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cao WJ, Lu JZ, Li C, Gao YJ, Lu KQ, Wang
HZ and Wang ZP: The hOGG1 Ser326Cys gene polymorphism and
susceptibility for bladder cancer: A meta-analysis. Int Braz J
Urol. 42:883–896. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sanjari Moghaddam A, Nazarzadeh M, Bidel
Z, Karamatinia A, Darvish H and Mosavi Jarrahi A: hOGG1 gene
polymorphism and breast cancer risk: A systematic review and
meta-analysis study. Breast J. 24:70–73. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang M and Mo R: Association of hOGG1
Ser326Cys polymorphism with colorectal cancer risk: An updated
meta-analysis including 5235 cases and 8438 controls. Tumour Biol.
35:12627–12633. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lukina MV, Kuznetsova AA, Kuznetsov NA and
Fedorova OS: The kinetic analysis of recognition of the damaged
nucleotides by mutant forms of the 8-oxoguanine DNA glycosylase
hOGG1. Russ J Bioorganic Chem. 43:1–12. 2017. View Article : Google Scholar
|
|
17
|
Kohno T, Shinmura K, Tosaka M, Tani M, Kim
SR, Sugimura H, Nohmi T, Kasai H and Yokota J: Genetic
polymorphisms and alternative splicing of the hOGG1 gene, that is
involved in the repair of 8-hydroxyguanine in damaged DNA.
Oncogene. 16:3219–3225. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang W, Dang S, Li Y, Sun M, Jia X, Wang R
and Liu J: hOGG1 Ser326Cys polymorphism and risk of hepatocellular
carcinoma among East Asians: A meta-analysis. PLoS One.
8:e601782013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xie Y, Wu Y, Zhou X, Yao M, Ning S and Wei
Z: Association of polymorphisms hOGGI rs1052133 and hMUTYH
rs3219472 with risk of nasopharyngeal carcinoma in a Chinese
population. Onco Targets Ther. 9:755–760. 2016.PubMed/NCBI
|
|
20
|
Tan H: On the protective effects of gene
SNPs against human cancer. EBioMedicine. 33:4–5. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Corella D, Ramírez-Sabio JB, Coltell O,
Ortega-Azorín C, Estruch R, Martínez-González MA, Salas-Salvadó J,
Sorlí JV, Castañer O, Arós F, et al: Effects of the Ser326Cys
polymorphism in the DNA repair OGG1 gene on cancer, cardiovascular
and all-cause mortality in the PREDIMED study: Modulation by diet.
J Acad Nutr Diet. 118:589–605. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Y, Gao X, Wei F, Zhang X, Yu J, Zhao
H, Sun Q, Yan F, Yan C, Li H, et al: The hOGG1 Ser326Cys
polymorphism contributes to digestive system cancer susceptibility:
Evidence from 48 case-control studies. Tumour Biol. 36:1029–1038.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou F, Zhang W, Wei Y, Zhou D, Su Z, Meng
X, Hui L and Tian W: The changes of oxidative stress and human
8-hydroxyguanine glycosylase1 gene expression in depressive
patients with acute leukemia. Leuk Res. 31:387–393. 2007.
View Article : Google Scholar : PubMed/NCBI
|