To explore the genetic alterations in head and neck
tumors, identify new high-specificity and high-sensitivity tumor
markers, and identify potentially effective therapeutic targets,
in silico methods were used to study HNC. In the present
study, GSE58911 was downloaded and analyzed from the Gene
Expression Omnibus (GEO) database to obtain differentially
expressed genes (DEGs) between HNC tissues and non-cancerous
tissues. Subsequently, Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathway analysis, Gene Ontology (GO) enrichment analysis,
and protein-protein interaction (PPI) network analysis was
performed to characterize the molecular mechanisms underlying
carcinogenesis and progression of HNC. A total of 648
differentially expressed genes and 26 hub genes were identified,
which may be potential targets for clinical diagnosis and therapy
of HNC.
A comprehensive set of functional annotation tools
were provided by the Database for Annotation, Visualization, and
Integrated Discovery (DAVID; http://david.ncifcrf.gov) (version 6.8). DAVID is an
online biological information database for investigators to
understand biological significance underlying a large number of
genes (11). KEGG (http://www.kegg.jp), an integrated database resource,
is used for the biological interpretation of genome sequences and
other high-throughput data (12).
The GO (www.geneontology.org) project is a
major bioinformatics tool and represents the most comprehensive
resource currently available for computable knowledge regarding the
functions of genes and gene products (13). Enrichment analysis from GO and KEGG
pathways for differentially expressed genes was obtained using
DAVID. P<0.05 was considered to indicate a statistically
significant difference.
A PPI network of DEGs was constructed using the
Search Tool for the Retrieval of Interacting Genes (STRING) online
database (version 10.5; http://string-db.org) (14). Through the STRING database, DEGs with
a combined score ≥0.4 were chosen to construct a PPI network which
could be visualized using Cytoscape software (version 3.4.0;
www.cytoscape.org) (15). The functional modules of the PPI
network were then identified using the Molecular Complex Detection
(MCODE) (version 1.4.2) plug-in of Cytoscape (16). The criteria for selection were as
follows: Max depth, 100; degree cut-off, 2; k-score, 2 and node
score cut-off, 0.2.
Hub genes were selected using Cytoscape software. A
network of hub genes and their co-expressed genes was analyzed
using the cBioPortal for Cancer Genomics (http://www.cbioportal.org) (17,18),
which allows for visualization, analysis, and download of
large-scale cancer genomics data sets. Hierarchical clusters of hub
genes were constructed using the next generation University of
California Santa Cruz (UCSC) Cancer Browser: UCSC Xena (http://xena.ucsc.edu) (19). The sample source ‘The Cancer Genome
Atlas Head-Neck Squamous Cell Carcinoma (HNSC)’ was selected for
these 26 hub genetic analyses and 604 samples were selected for
analysis. The overall survival and disease-free survival rate
analyses of hub genes was performed by constructing Kaplan-Meier
curves using the cBioPortal online platform (statistical analysis
performed is a log-rank test). Furthermore, the relationship
between expression patterns, tumor grades, and HPV infection status
was analyzed using Oncomine (https://www.oncomine.org) (20–29).
After the standardization of the microarray results,
648 differentially expressed genes were identified between HNC
tissues and normal tissues. The results from the GSE58911 dataset
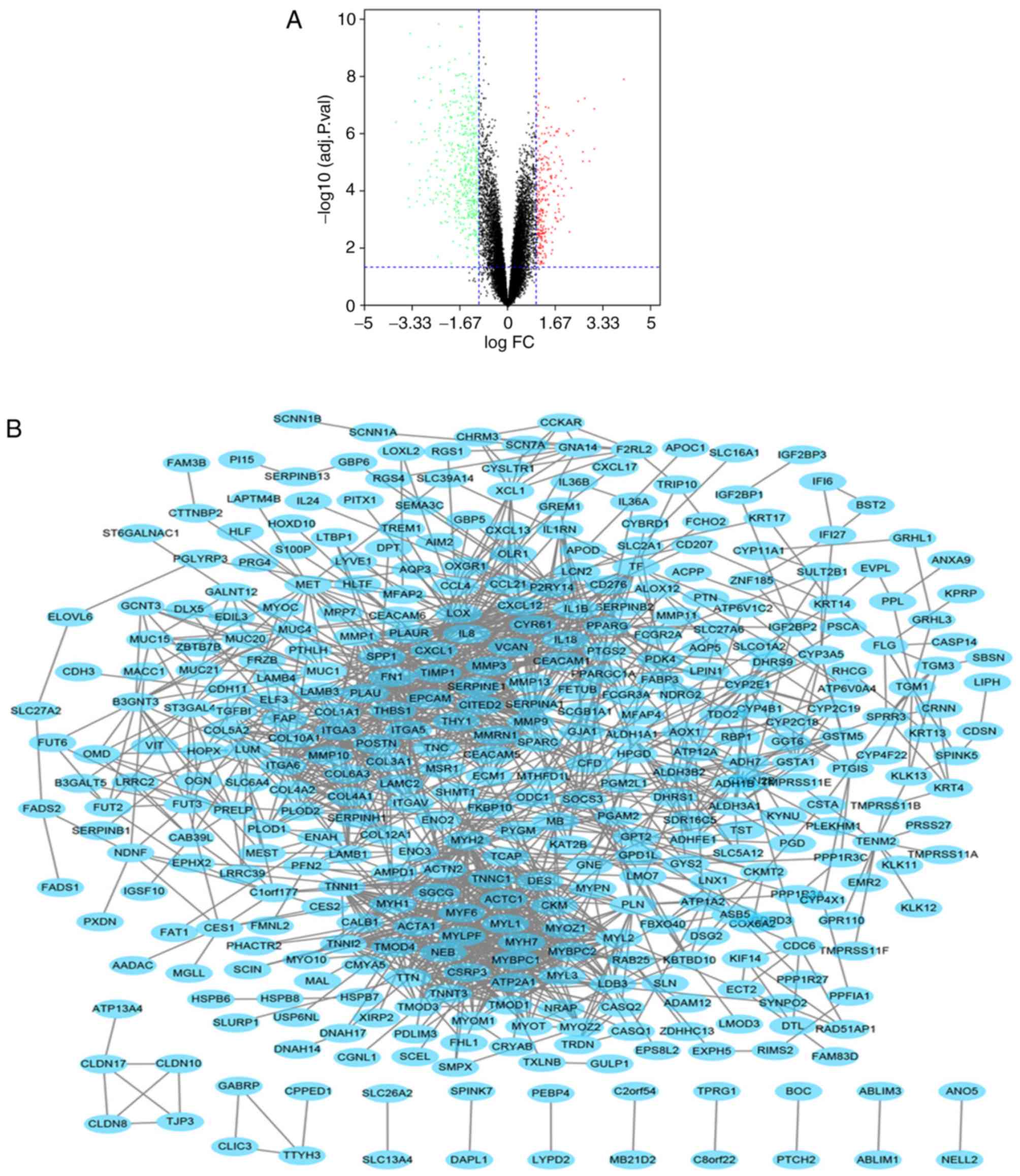
are represented as a volcano plot (Fig.
1A). The PPI network of DEGs was constructed (Fig. 1B). There were 554 nodes and 1574
edges in the PPI network, and the average node score was 5.68
(Fig. 1B).
To analyze the biological classification of DEGs, GO
and KEGG pathway enrichment analyses were performed using DAVID
(Table I). The results of GO
analysis showed that changes in biological processes of DEGs were
mainly ‘enriched in muscle system process’, ‘extracellular matrix
organization’, ‘muscle contraction’, ‘extracellular structure
organization’, and ‘muscle filament sliding’. Molecular function
DEGs included ‘actin binding’, ‘structural constituent of muscle’,
‘cytoskeletal protein binding’, ‘structural molecule activity’ and
‘actinin binding’. Cell component DEGs included ‘extracellular
region part’, ‘contractile fiber’, ‘extracellular region’,
‘sarcomere’, and ‘myofibril’. KEGG pathway analysis showed that the
DEGs were mainly enriched in ‘extracellular matrix (ECM)-receptor
interaction’, ‘focal adhesion’, ‘amoebiasis’, ‘drug
metabolism-cytochrome P450’, ‘chemical carcinogenesis’, ‘dilated
cardiomyopathy’, ‘small cell lung cancer’, ‘hypertrophic
cardiomyopathy’, and ‘retinol metabolism’.
Using the MCODE plug-in of Cytoscape, 26 genes were
identified as hub genes. The results of GO and KEGG pathway
analyses indicated that the hub genes were mainly enriched in
‘extracellular matrix organization’, ‘collagen catabolic process’,
‘extracellular structure organization’, ‘multicellular organism
catabolic process’, ‘collagen metabolic process’, ‘serine-type
endopeptidase activity’, ‘extracellular matrix’, ‘proteinaceous
extracellular matrix’, ‘extracellular space’, ‘extracellular region
part’, ‘extracellular region’, and ‘complement and coagulation
cascades’ (Table II). The
abbreviations, official full names, and synonyms for these hub
genes are shown in Table III. A
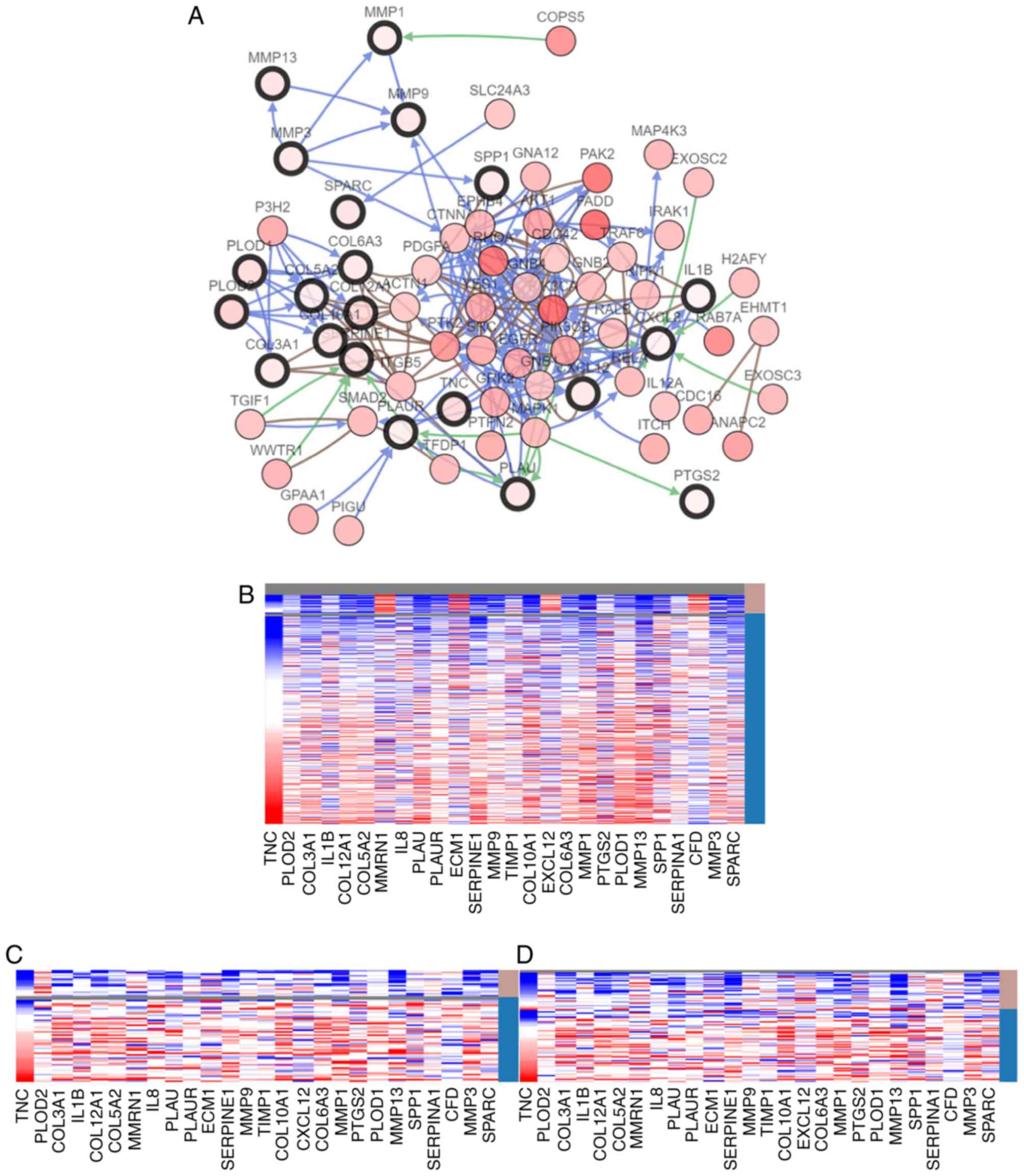
network of the hub genes and their co-expressed genes was analyzed
using cBioPortal for Cancer Genomics (Fig. 2A). Hierarchical clustering revealed
that the expression of hub genes could differentiate the HNC
samples from normal samples (Fig.
2B). From figure 2B, it can be
seen that 22 of the 26 hub genes were highly expressed in head and
neck tumors compared with normal tissues, whereas expression of
four genes (MMRN1/ECM1/EXCL12/CFD) was relatively high in the
normal tissues. Furthermore, hierarchical clustering showed that
HPV infection status determined by fluorescent in situ
hybridization (FISH) testing (Fig.
2C) and P16 testing (Fig. 2D)
was negatively associated with expression of the gene, although the
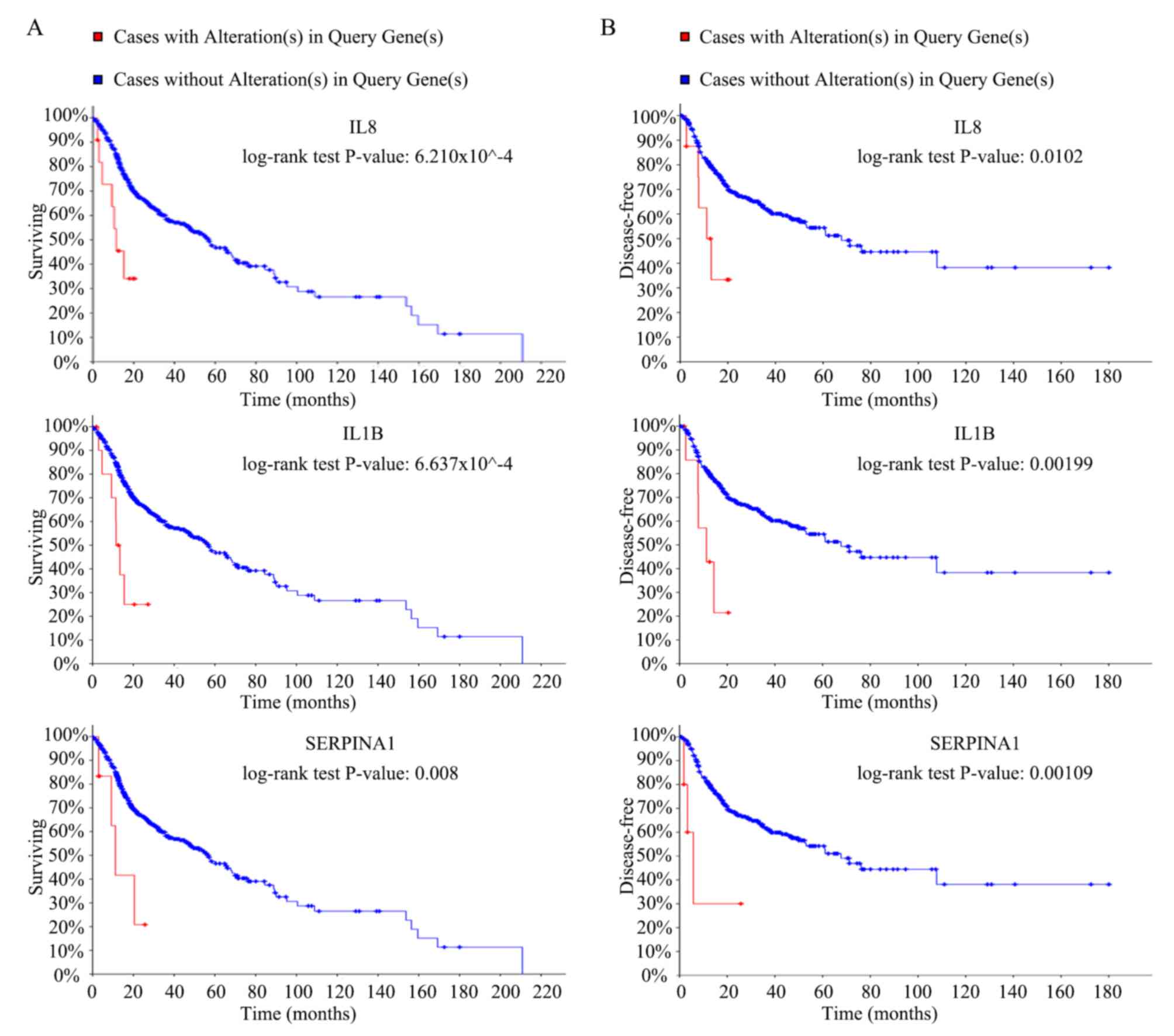
mechanisms remains unknown. Overall survival rate analysis of the
hub genes was performed using Kaplan-Meier curves in the cBioPortal
online platform. Patients with HNC and high expression of
interleukin (IL)8, IL1B and serpin family A member 1 (SERPINA1) had
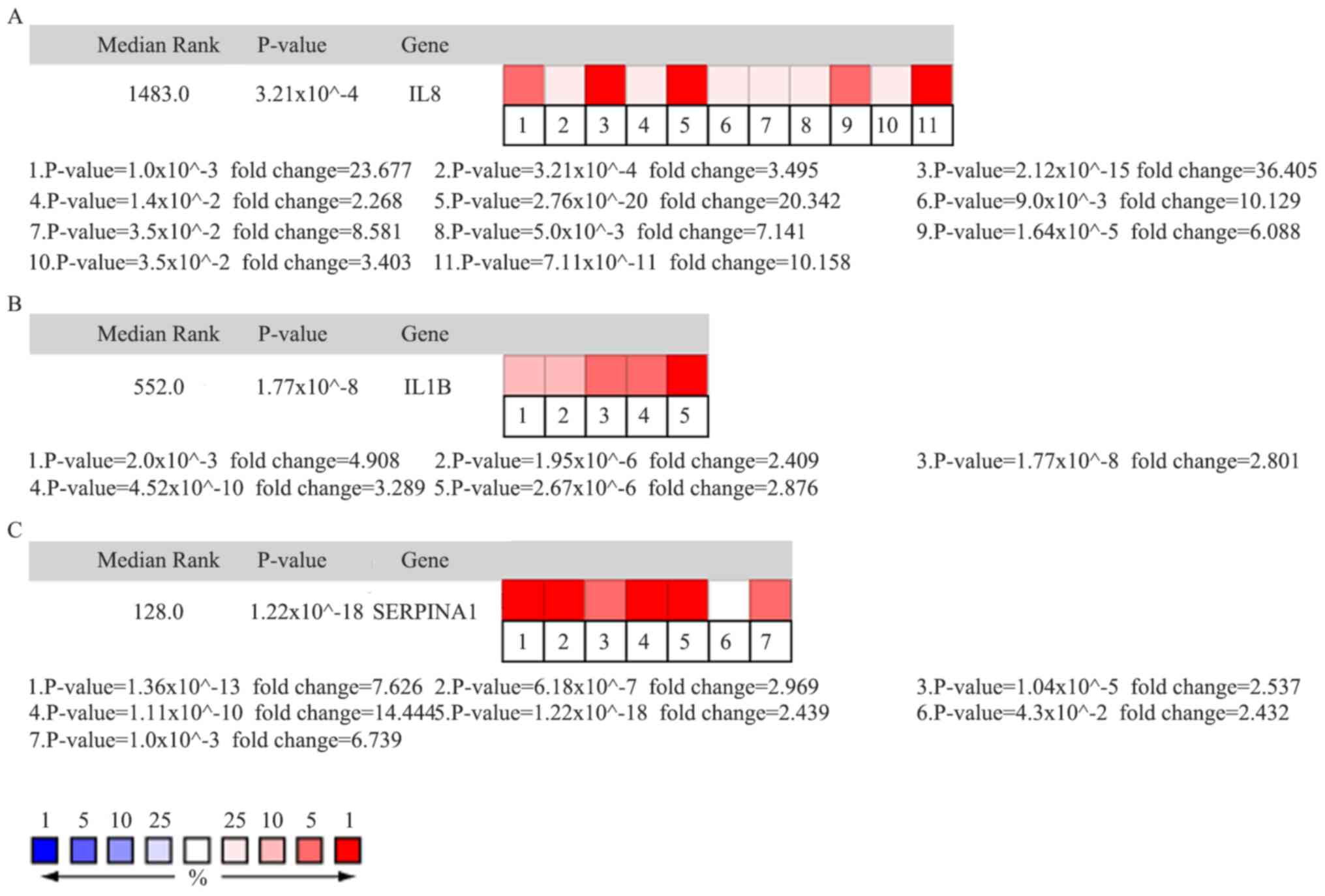
worse overall survival and worse disease-free survival (Fig. 3A and B). Oncomine analysis of cancer
vs. normal tissues indicated that IL8, IL1B, and SERPINA1 were
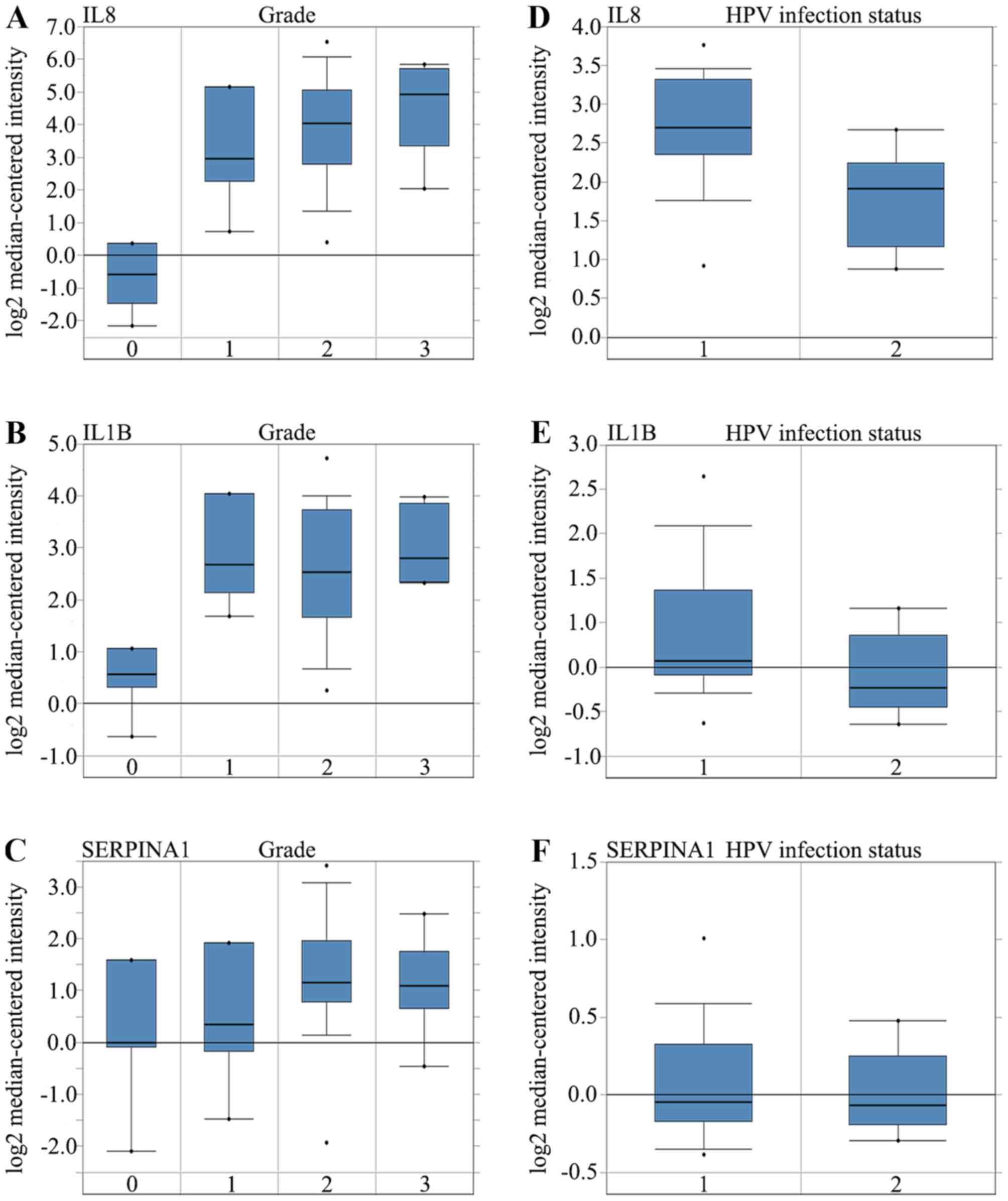
over-expressed in HNC in the different datasets (Fig. 4A, B and C). Higher mRNA expression
levels of IL8 was associated with tumor grade (P=0.001). However,
the mRNA expression levels of IL1B and SERPINA1 were not associated
with tumor grade (P>0.05). Higher mRNA expression levels of IL8
(P=6.30×10−9) and IL1B (P=3.48×10−6) were
associated with HPV infection status. The mRNA expression levels of
SERPINA1 however, were not associated with HPV infection status
(Fig. 5A-F).
HNC is the sixth most common cancer worldwide and is
associated with severe disease- and treatment-related morbidity,
with a 5-year survival rate of <60% (1,2). The
survival rate has not improved across more than two decades due to
lack of early detection (30,31).
There are two primary causes of HNC: Tobacco and alcohol use, and
human papillomavirus infection (32). Previous studies have shown that HPV
infection plays a role in the pathogenesis of head and neck tumors
(33–35). The oncomine analysis of cancer vs.
normal tissue for IL8, IL1B and SERPINA1 demonstrated that the
expression was compared with the normal tissues (Fig. 4). mRNA expressions of IL8, IL1B and
SERPINA1 were higher in the HPV-negative group compared with that
in the HPV-positive group (Fig.
5D-F). These seemingly contradictory results are
understandable. There are many studies suggesting that HPV-positive
head and neck tumors were associated with improved disease-free and
overall survival (32,36). According to the present study, IL8,
IL1B and SERPINA1 are highly expressed in the HNC, and the results
in Fig. 3 indicate that the overall
survival rate and disease-free survival rate of patients with high
expression of these 3 genes are worse. Therefore, it is
understandable that IL8, IL1B and SERPINA1 have higher expressions
in HPV-negative tumor patients. However, the underlying molecular
mechanisms of HNC remain unclear. Abnormal expression of
transglutaminase 3, regenerating islet-derived protein 3, keratin
8, and phosphatase and tensin homolog is associated with HNC
(37–40). In addition, mutations within tumor
protein p53, notch receptor 1,
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α,
X-ray repair cross complementing 1 and epidermal growth factor
receptor have been reported to be involved in HNC (41–44).
Patients with HNC that do not detect the cancer early have no
effective treatments available except receiving palliative care,
which leads to poor prognosis and quality of life, and a high rate
of mortality (30). Therefore, there
is an urgent need to identify potential target biomarkers that can
be used to efficiently diagnose and treat HNC. Bioinformatics
technology allows us to explore genetic differences between HNC and
normal tissues, which can be used to identify potential biomarkers.
Then, effective genes can be selected through screening and
experimental validation for early diagnosis, clinical prognosis,
and treatment of HNC.
In the present study, the dataset GSE58911 was
analyzed to obtain differentially expressed genes between HNC and
non-cancerous tissues. A total of 648 DEGs were identified. GO and
KEGG enrichment analyses were performed to explore interactions
among these genes and they were mainly enriched in ‘extracellular
matrix organization’, ‘actin binding’, ‘extracellular region’,
‘ECM-receptor interaction’, ‘drug metabolism-cytochrome P450’, and
‘chemical carcinogenesis’. Previous studies reported that
‘extracellular matrix organization’, ‘actin binding’, and
‘ECM-receptor interaction’ play important roles in the
carcinogenesis, progression, and metastasis of tumors (45–48). In
addition, previous data indicated that focal adhesion, drug
metabolism-cytochrome P450, and chemical carcinogenesis are
involved in radio- and chemotherapy (49–52).
Thus, the findings from the present study are consistent with
results from previous studies. GO enrichment analysis indicated
that changes in hub genes were mainly enriched in ‘extracellular
matrix organization’, ‘collagen catabolic process’, ‘serine-type
endopeptidase activity’, ‘extracellular matrix’, and ‘proteinaceous
extracellular matrix’, while changes according to KEGG pathway
analysis were mainly enriched in ‘complement and coagulation
cascades’.
A total of 26 DEGs were selected as hub genes, among
which survival rates and disease-free survival rates between
patients with head and neck tumors and patients without tumors were
significantly associated with the expression of IL8, IL1B, and
SERPINA1. IL8 is a chemotactic factor that attracts neutrophils,
basophils, and T-cells, but not monocytes (53) and can be released by several cell
types in response to inflammatory stimuli (53). Higher IL8 expression was observed in
HNSCC tissue (54,55). Furthermore, IL8 stimulated the
proliferation of HNSC cells (55,56). In
addition, a previous study showed that the tumor microenvironment
plays a vital role in HNC initiation, progression, and metastasis
(57). Tumor-associated macrophages
can promote cancer initiation and progression by releasing
cytokines and may facilitate papillary thyroid carcinoma (PTC) cell
metastasis through IL8 and its paracrine interaction with C-X-C
chemokine receptor CXCR1 and CXCR2 (58). Thus, IL8 may be a potential
therapeutic target.
IL1B is a potent pro-inflammatory cytokine.
Initially discovered as the major endogenous pyrogen, IL1B induces
prostaglandin synthesis, neutrophil influx and activation, cytokine
production, T cell and B cell activation, antibody production,
collagen production, and fibroblast proliferation (59). A recent study of IL1B has shown that
it plays a major role in tumor chemotherapy resistance. Anakinra
can block the IL-1 pathway and overcome erlotinib resistance in
HNSCC, which may represent a novel strategy to overcome EGFR
inhibitor resistance, allowing for more effective treatment of
patients with HNSCC (60).
Furthermore, high expression of inflammatory cytokines (IL8, IL1B)
and shorter progression-free survival are significantly associated.
The expression level of inflammatory cytokines may help to identify
which patients with recurrent and/or metastatic squamous cell
carcinoma of the head and neck are likely to benefit from
dacomitinib (61).
SERPINA1, an inhibitor of serine proteases,
irreversibly inhibits trypsin, chymotrypsin, and plasminogen
activator (62). Its primary target
is elastase, but it also has a moderate affinity for plasmin and
thrombin (62). A recent study
showed a higher abundance of SERPINA1 candidate biomarkers in the
saliva of patients with oral squamous cell carcinoma (OSCC),
demonstrating that SERPINA1 is related to OSCC development
(63). Moreover, SERPINA1 may be
related to PTC by responding to steroid hormone stimuli and
regulating the epithelial-to-mesenchymal transition (64). Based on these associations, SERPINA1
may be an effective mRNA marker of PTC (65). Oncomine analysis indicated that
higher mRNA levels of IL8, IL1B, and SERPINA1 were associated with
tumor grade and HPV infection status, indicating vital roles of
IL8, IL1B, and SERPINA1 in the carcinogenesis or progression of
HNC.
In addition to IL1B, lL8 and SERPINA1, which were
associated with the survival rate of patients with head and neck
cancer, other relevant hub genes that were identified in the
present study are discussed.
Tenascin C (TNC), a gene associated with tumor
metastatic potential, was upregulated in the OSCC cell line
LNMTca8113 (66). In addition,
vascular density and higher tumor stage were associated with
differences in immuno-expression of stromal TNC, demonstrating its
role in the tumorigenesis of juvenile nasopharyngeal angiofibroma
(67).
A previous study showed that microRNA-29a/b could
regulate the expression of collagen type III alpha 1 chain to
enhance migration and invasion ability of nasopharyngeal carcinoma
cells (68). The markers, the
combination of collagen type V alpha 1 chain (COL5A1) and
hemoglobin subunit beta and COL5A1 itself can better predict the
treatment response in patients with oral tongue squamous cell
carcinoma (69).
Poor disease-free survival and increased progression
or relapse risk were associated with high plasminogen activator,
urokinase (PLAU) expression. Moreover, circulating PLAU levels were
significantly higher in the plasma of patients with HNSCC compared
with that in healthy individuals (70).
Extracellular matrix protein 1 (ECM1) levels
gradually increased from benign laryngeal lesions to precancerous
to malignant lesions, and ECM1 was expressed at lower levels in
laryngeal carcinomas without metastasis (71,72).
These results demonstrated that ECM1 facilitated development and
metastasis of laryngeal carcinoma.
Overexpression of SERPINE1 promotes tumor migration
and invasion and plays an important role in metastasis and poor
prognosis of HNSCC (73). In
addition, many researchers regard SERPINE1 as a prognostic marker
based on its ability to stratify patients with HNSCC according to
their recurrence risk (74).
Matrix metalloproteinases (MMPs) are a family of
proteolytic enzymes that promote invasion and metastasis of various
cancers due to their ability to degrade components of the
extracellular matrix. MMP1, MMP3, MMP9, and MMP13 are predictors of
poor clinical outcomes in patients with HNC (75–78).
Furthermore, specific tissue inhibitors of matrix
metalloproteinases (TIMPs) can regulate MMP activity. In addition
to HNC, the majority of tumors are associated with alterations in
MMPs and TIMPs. Imbalance between matrix metalloproteinases and
their inhibitors contributes greatly to the progression and
prognosis of HNC (76,79).
Compared with normal oral mucosa, secreted
phosphoprotein 1 was expressed at significantly higher levels in
OSCC (80). According to a previous
study, secreted protein acidic and cysteine rich had significant
prognostic value, especially in the stroma surrounding OSCC
(81). A literature search revealed
that the interaction between HNC and the hub genes
procollagen-lysine 2-oxoglutarate 5-dioxygenase (PLOD)-2, collagen
type XII alpha 1 chain, multimerin 1 (MMRN1), plasminogen activator
urokinase receptor, collagen type X α 1 chain, collagen type VI α 3
chain, prostaglandin-endoperoxide synthase 2, PLOD1, and complement
factor D (CFD) has not been widely reported.
There were several limitations associated with the
present study. First, only one series (GSE58911) downloaded and
used from the GEO database. The number of tumor and normal samples
in this series were both 15. This sample size was insufficient.
Second, genes were analyzed that may be related to the
carcinogenesis or progression of head and neck tumors from the
results of the bioinformatics analyses. The functions of these
genes have not been verified in vitro and in vivo. In
the present study, the expression levels of IL8 (C-X-C motif
chemokine ligand 8), IL1B and SERPINA1 in tumor tissues and normal
tissues of patients with head and neck tumors were not verified
further. In addition, phenotypic function was also not verified in
head and neck tumor cell lines. In future studies this will be
investigated. Third, the number of hub genes (modes with bold black
circles) in Fig. 2A is 21, and there
are 5 hub genes (MMRN1, ECM1, TIMP metallopeptidase inhibitor 1,
SERPINA1 and CFD) that do not appear in the network map. In
particular, SERPINA1 is among one of the identified three genes
following further analysis of the data. This could be due to the
following reasons: i) These 5 genes may not be closely related to
other genes, and have other roles and mechanisms in the occurrence
and development of tumors, so they were excluded from the network
map; ii) the 26 hub genes were obtained by analyzing 648 DEGs using
the Cytoscape plug-in, MCODE. cBioPortal for Cancer Genomics was
subsequently used to analyze these 26 hub genes to obtain a network
map of hub genes and their co-expressed genes. The computer
algorithms that each database performed for analysis may differ,
and may also cause differences; iii) in addition, 3 hub genes (IL8,
IL1B and SERPINA1) were selected for more in-depth analysis as
cBioPortal for Cancer Genomics was used to analyze overall survival
and disease-free survival rate for the 26 hub genes. Changes in the
expression of IL8, IL1B and SERPINA1 in patients with head and neck
tumors were associated with overall survival and disease-free
survival rate, and were statistically significant; iv), a holistic
analysis of 26 hub genes from 628 DEGs was performed in an attempt
to obtain an inductive result (Fig.
2). In addition, 2 detection methods (FISH and P16 tests) of
HPV infection were used to obtain more accurate results for further
analysis. However, the profiles of IL8, IL1B and SERPINA1 in
Fig. 2C and D are not consistent.
This may be due to different detection methods or detection of HPV
subtypes. This may require a more accurate method to detect the
infection status of HPV for a more accurate analysis; and v), there
are differences in the monitoring of overall survival and
disease-free survival of patients between the gene alterations of
IL8, IL1B and SERPINA1 (Fig. 3).
Monitoring was performed for over 180 months (over 15 years) when
there were no alteration(s) in these 3 genes while alterations in
these genes was monitored for only 20 months. This may be due to
the following reasons: i) These samples were obtained at different
time points over a 10-year period and as such the samples may have
degraded; ii) some patients cannot be contacted during follow-up or
have died due to illness. An open source, free database was used
therefore information pertaining to when the samples were collected
and whether it was in different decades. In this regard, the lack
of data beyond 20 months may be considered a limitation associated
with the present study. However, using this database for analysis
is reliable and credible. This database is used in many articles on
bioinformatics analysis (82–84).
These limitations will be addressed in future studies. Despite
these limitations, research in the present study is important as it
elucidated molecular mechanisms underlying development of HNC, and
also provides potential target genes for clinical diagnosis and
targeted therapy. In addition, this provides direction for future
studies of HNC.
In conclusion, the present study identified DEGs
that may be involved in the carcinogenesis or progression of HNC. A
total of 648 DEGs and 26 hub genes were identified and may have
potential as target biomarkers for HNC. Further studies are needed
to elucidate the biological functions of these genes in HNC.
Not applicable.
This study was supported by the National Natural
Science Foundation of China (grant nos. 81372880 and 81670910) and
the Guidance fund of the Renmin Hospital of Wuhan University (grant
no. RMYD2018Z12).
All data are fully available without
restriction.
FC and ZT conceived and designed the study. FC, AZ,
FL, SW, SC and ZT processed the data. FC wrote the paper. FC, AZ,
FL, SW, SC, and ZT reviewed and edited the manuscript. All authors
read and approved the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bray F, Ferlay J, Laversanne M, Brewster
DH, Gombe MC, Kohler B, Piñeros M, Steliarova-Foucher E,
Swaminathan R, Antoni S, et al: cancer incidence in five
continents: Inclusion criteria, highlights from volume X and the
global status of cancer registration. Int J Cancer. 137:2060–2071.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Thibodeau BJ, Geddes TJ, Fortier LE, Ahmed
S, Pruetz BL, Wobb J, Chen P, Wilson GD and Akervall JA: Gene
expression characterization of HPV positive head and neck cancer to
predict response to chemoradiation. Head Neck Pathol. 9:345–353.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Min SK, Lee SK, Park JS, Lee J, Paeng JY,
Lee SI, Lee HJ, Kim Y, Pae HO, Lee SK and Kim EC: Endoplasmic
reticulum stress is involved in hydrogen peroxide induced apoptosis
in immortalized and malignant human oral keratinocytes. J Oral
Pathol Med. 37:490–498. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Skvortsov S, Dudás J, Eichberger P,
Witsch-Baumgartner M, Loeffler-Ragg J, Pritz C, Schartinger VH,
Maier H, Hall J, Debbage P, et al: Rac1 as a potential therapeutic
target for chemo-radioresistant head and neck squamous cell
carcinomas (HNSCC). Br J Cancer. 110:2677–2687. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Y, Chen C, Wang X, Jin F, Liu Y, Liu
H, Li T and Fu J: Lower DSC1 expression is related to the poor
differentiation and prognosis of head and neck squamous cell
carcinoma (HNSCC). J Cancer Res Clin Oncol. 142:2461–2468. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res 41 (Database Issue).
D991–D995. 2013.
|
|
10
|
Lobert S, Graichen ME, Hamilton RD, Pitman
KT, Garrett MR, Hicks C and Koganti T: Prognostic biomarkers for
HNSCC using quantitative real-time PCR and microarray analysis:
β-tubulin isotypes and the p53 interactome. Cytoskeleton (Hoboken).
71:628–637. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kanehisa M, Sato Y, Kawashima M, Furumichi
M and Tanabe M: KEGG as a reference resource for gene and protein
annotation. Nucleic Acids Res. 44(D1): D457–D462. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pinoli P, Chicco D and Masseroli M:
Computational algorithms to predict gene ontology annotations. BMC
Bioinformatics. 16 (Suppl 6):S42015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res 43
(Database issue). D447–D452. 2015. View Article : Google Scholar
|
|
15
|
Su G, Morris JH, Demchak B and Bader GD:
Biological network exploration with Cytoscape 3. Curr Protoc
Bioinformatics. 47:8.13.1–24. 2014. View Article : Google Scholar
|
|
16
|
Bandettini WP, Kellman P, Mancini C,
Booker OJ, Vasu S, Leung SW, Wilson JR, Shanbhag SM, Chen MY and
Arai AE: Multi contrast delayed enhancement (MCODE) improves
detection of subendocardial myocardial infarction by late
gadolinium enhancement cardiovascular magnetic resonance: A
clinical validation study. J Cardiovasc Magn Reson. 14:832012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rosenbloom KR, Armstrong J, Barber GP,
Casper J, Clawson H, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo
L, Haeussler M, et al: The UCSC Genome Browser database: 2015
update. Nucleic Acids Res 43 (Database Issue). D670–D681. 2015.
View Article : Google Scholar
|
|
20
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cromer A, Carles A, Millon R, Ganguli G,
Chalmel F, Lemaire F, Young J, Dembélé D, Thibault C, Muller D, et
al: Identification of genes associated with tumorigenesis and
metastatic potential of hypopharyngeal cancer by microarray
analysis. Oncogene. 23:2484–2498. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Estilo CL, O-charoenrat P, Talbot S, Socci
ND, Carlson DL, Ghossein R, Williams T, Yonekawa Y, Ramanathan Y,
Boyle JO, et al: Oral tongue cancer gene expression profiling:
Identification of novel potential prognosticators by
oligonucleotide microarray analysis. BMC Cancer. 9:112009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ginos MA, Page GP, Michalowicz BS, Patel
KJ, Volker SE, Pambuccian SE, Ondrey FG, Adams GL and Gaffney PM:
Identification of a gene expression signature associated with
recurrent disease in squamous cell carcinoma of the head and neck.
Cancer Res. 64:55–63. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He H, Jazdzewski K, Li W, Liyanarachchi S,
Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, et al:
The role of microRNA genes in papillary thyroid carcinoma. Proc
Natl Acad Sci USA. 102:19075–19080. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Peng CH, Liao CT, Peng SC, Chen YJ, Cheng
AJ, Juang JL, Tsai CY, Chen TC, Chuang YJ, Tang CY, et al: A novel
molecular signature identified by systems genetics approach
predicts prognosis in oral squamous cell carcinoma. PLoS One.
6:e234522011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pyeon D, Newton MA, Lambert PF, den Boon
JA, Sengupta S, Marsit CJ, Woodworth CD, Connor JP, Haugen TH,
Smith EM, et al: Fundamental differences in cell cycle deregulation
in human papillomavirus-positive and human papillomavirus-negative
head/neck and cervical cancers. Cancer Res. 67:4605–4619. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ye H, Yu T, Temam S, Ziober BL, Wang J,
Schwartz JL, Mao L, Wong DT and Zhou X: Transcriptomic dissection
of tongue squamous cell carcinoma. BMC Genomics. 9:692008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Giordano TJ, Au AY, Kuick R, Thomas DG,
Rhodes DR, Wilhelm KJ Jr, Vinco M, Misek DE, Sanders D, Zhu Z, et
al: Delineation, functional validation, and bioinformatic
evaluation of gene expression in thyroid follicular carcinomas with
the PAX8-PPARG translocation. Clin Cancer Res. 12:1983–1993. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vasko V, Espinosa AV, Scouten W, He H,
Auer H, Liyanarachchi S, Larin A, Savchenko V, Francis GL, de la
Chapelle A, et al: Gene expression and functional evidence of
epithelial-to-mesenchymal transition in papillary thyroid carcinoma
invasion. Proc Natl Acad Sci USA. 104:2803–2808. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Forastiere A, Koch W, Trotti A and
Sidransky D: Head and neck cancer. N Engl J Med. 345:1890–900.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bozec A, Ilie M, Dassonville O, Long E,
Poissonnet G, Santini J, Chamorey E, Ettaiche M, Chauviere D,
Peyrade F, et al: Significance of circulating tumor cell detection
using the Cell search system in patients with locally advanced head
and neck squamous cell carcinoma. Eur Arch Otorhinolaryngol.
270:2745–2749. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rettig EM and D'Souza G: Epidemiology of
head and neck cancer. Surg Oncol Clin N Am. 24:379–396. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Marullo R, Werner E, Zhang H, Chen GZ,
Shin DM and Doetsch PW: HPV16 E6 and E7 proteins induce a chronic
oxidative stress response via NOX2 that causes genomic instability
and increased susceptibility to DNA damage in head and neck cancer
cells. Carcinogenesis. 36:1397–1406. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Boscolo-Rizzo P, Zorzi M, Del Mistro A, Da
Mosto MC, Tirelli G, Buzzoni C, Rugge M, Polesel J and Guzzinati S;
AIRTUM Working Group, : The evolution of the epidemiological
landscape of head and neck cancer in Italy: Is there evidence for
an increase in the incidence of potentially HPV-related carcinomas?
PLoS One. 13:e01926212018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cheraghlou S, Torabi SJ, Husain ZA,
Otremba MD, Osborn HA, Mehra S, Yarbrough WG, Burtness BA and
Judson BL: HPV status in unknown primary head and neck cancer:
Prognosis and treatment outcomes. Laryngoscope. 129:684–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kreimer AR, Clifford GM, Boyle P and
Franceschi S: Human papillomavirus types in head and neck squamous
cell carcinomas worldwide: A systematic review. Cancer Epidemiol
Biomarkers Prev. 14:467–475. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu X, Cao W, Wang X, Zhang J, Lv Z, Qin X,
Wu Y and Chen W: TGM3, a candidate tumor suppressor gene,
contributes to human head and neck cancer. Mol Cancer. 12:1512013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Masui T, Ota I, Itaya-Hironaka A, Takeda
M, Kasai T, Yamauchi A, Sakuramoto-Tsuchida S, Mikami S, Yane K,
Takasawa S and Hosoi H: Expression of REG III and prognosis in head
and neck cancer. Oncol Rep. 30:573–578. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Andratschke M, Hagedorn H and Nerlich A:
Expression of the epithelial cell adhesion molecule and cytokeratin
8 in head and neck squamous cell cancer: A comparative study.
Anticancer Res. 35:3953–3960. 2015.PubMed/NCBI
|
|
40
|
Squarize CH, Castilho RM, Abrahao AC,
Molinolo A, Lingen MW and Gutkind JS: PTEN deficiency contributes
to the development and progression of head and neck cancer.
Neoplasia. 15:461–471. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gross AM, Orosco RK, Shen JP, Egloff AM,
Carter H, Hofree M, Choueiri M, Coffey CS, Lippman SM, Hayes DN, et
al: Multi-tiered genomic analysis of head and neck cancer ties TP53
mutation to 3p loss. Nat Genet. 46:939–943. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Psyrri A, Seiwert TY and Jimeno A:
Molecular pathways in head and neck cancer: EGFR, PI3K, and more.
Am Soc Clin Oncol Educ Book. 246–255. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mahjabeen I, Baig RM, Masood N, Sabir M,
Inayat U, Malik FA and Kayani MA: Genetic variations in XRCC1 gene
in sporadic head and neck cancer (HNC) patients. Pathol Oncol Res.
19:183–188. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Boeckx C, Weyn C, Vanden Bempt I,
Deschoolmeester V, Wouters A, Specenier P, Van Laer C, Van den
Weyngaert D, Kockx M, Vermorken JB, et al: Mutation analysis of
genes in the EGFR pathway in Head and Neck cancer patients:
Implications for anti-EGFR treatment response. BMC Res Notes.
7:3372014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gilkes DM, Semenza GL and Wirtz D: Hypoxia
and the extracellular matrix: Drivers of tumour metastasis. Nat Rev
Cancer. 14:430–439. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Malik R, Lelkes PI and Cukierman E:
Biomechanical and biochemical remodeling of stromal extracellular
matrix in cancer. Trends Biotechnol. 33:230–236. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Trulsson M, Yu H, Gisselsson L, Chao Y,
Urbano A, Aits S, Mossberg AK and Svanborg C: HAMLET binding to
α-actinin facilitates tumor cell detachment. PLoS One.
6:e171792011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang HJ, Tao J, Sheng L, Hu X, Rong RM,
Xu M and Zhu TY: Twist2 promotes kidney cancer cell proliferation
and invasion by regulating ITGA6 and CD44 expression in the
ECM-receptor interaction pathway. Onco Targets Ther. 9:1801–1812.
2016.PubMed/NCBI
|
|
49
|
Eke I and Cordes N: Focal adhesion
signaling and therapy resistance in cancer. Semin Cancer Biol.
31:65–75. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Blackstone BN, Li R, Ackerman WT, Ghadiali
SN, Powell HM and Kniss DA: Myoferlin depletion elevates focal
adhesion kinase and paxillin phosphorylation and enhances
cell-matrix adhesion in breast cancer cells. Am J Physiol Cell
Physiol. 308:C642–C649. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Johnson AL, Edson KZ, Totah RA and Rettie
AE: Cytochrome P450 ω-hydroxylases in inflammation and cancer. Adv
Pharmacol. 74:223–262. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ravegnini G, Sammarini G, Hrelia P and
Angelini S: Key genetic and epigenetic mechanisms in chemical
carcinogenesis. Toxicol Sci. 148:2–13. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Van Damme J, Rampart M, Conings R, Decock
B, Van Osselaer N, Willems J and Billiau A: The
neutrophil-activating proteins interleukin 8 and
beta-thromboglobulin: In vitro and in vivo comparison of
NH2-terminally processed forms. Eur J Immunol. 20:2113–2118. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lo MC, Yip TC, Ngan KC, Cheng WW, Law CK,
Chan PS, Chan KC, Wong CK, Wong RN, Lo KW, et al: Role of
MIF/CXCL8/CXCR2 signaling in the growth of nasopharyngeal carcinoma
tumor spheres. Cancer Lett. 335:81–92. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chan LP, Wang LF, Chiang FY, Lee KW, Kuo
PL and Liang CH: IL-8 promotes HNSCC progression on
CXCR1/2-meidated NOD1/RIP2 signaling pathway. Oncotarget.
7:61820–61831. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Christofakis EP, Miyazaki H, Rubink DS and
Yeudall WA: Roles of CXCL8 in squamous cell carcinoma proliferation
and migration. Oral Oncol. 44:920–926. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Curry JM, Sprandio J, Cognetti D,
Luginbuhl A, Bar-ad V, Pribitkin E and Tuluc M: Tumor
microenvironment in head and neck squamous cell carcinoma. Semin
Oncol. 41:217–234. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Fang W, Ye L, Shen L, Cai J, Huang F, Wei
Q, Fei X, Chen X, Guan H, Wang W, et al: Tumor-associated
macrophages promote the metastatic potential of thyroid papillary
cancer by releasing CXCL8. Carcinogenesis. 35:1780–1787. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Tominaga K, Yoshimoto T, Torigoe K,
Kurimoto M, Matsui K, Hada T, Okamura H and Nakanishi K: IL-12
synergizes with IL-18 or IL-1beta for IFN-gamma production from
human T cells. Int Immunol. 12:151–160. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Stanam A, Gibson-Corley KN, Love-Homan L,
Ihejirika N and Simons AL: Interleukin-1 blockade overcomes
erlotinib resistance in head and neck squamous cell carcinoma.
Oncotarget. 7:76087–76100. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Kim HS, Kwon HJ, Jung I, Yun MR, Ahn MJ,
Kang BW, Sun JM, Kim SB, Yoon DH, Park KU, et al: Phase II clinical
and exploratory biomarker study of dacomitinib in patients with
recurrent and/or metastatic squamous cell carcinoma of head and
neck. Clin Cancer Res. 21:544–552. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kwon CH, Park HJ, Choi JH, Lee JR, Kim HK,
Jo HJ, Kim HS, Oh N, Song GA and Park DY: Snail and serpinA1
promote tumor progression and predict prognosis in colorectal
cancer. Oncotarget. 6:20312–20326. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Kawahara R, Bollinger JG, Rivera C,
Ribeiro AC, Brandão TB, Paes Leme AF and MacCoss MJ: A targeted
proteomic strategy for the measurement of oral cancer candidate
biomarkers in human saliva. Proteomics. 16:159–173. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Qu T, Li YP, Li XH and Chen Y:
Identification of potential biomarkers and drugs for papillary
thyroid cancer based on gene expression profile analysis. Mol Med
Rep. 14:5041–5048. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Vierlinger K, Mansfeld MH, Koperek O,
Nöhammer C, Kaserer K and Leisch F: Identification of SERPINA1 as
single marker for papillary thyroid carcinoma through microarray
meta analysis and quantification of its discriminatory power in
independent validation. BMC Med Genomics. 4:302011. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Fialka F, Gruber RM, Hitt R, Opitz L,
Brunner E, Schliephake H and Kramer FJ: CPA6, FMO2, LGI1, SIAT1 and
TNC are differentially expressed in early- and late-stage oral
squamous cell carcinoma-a pilot study. Oral Oncol. 44:941–948.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Renkonen S, Heikkilä P, Haglund C, Mäkitie
AA and Hagström J: Tenascin-C, GLUT-1, and syndecan-2 expression in
juvenile nasopharyngeal angiofibroma: Correlations to vessel
density and tumor stage. Head Neck. 35:1036–1042. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Qiu F, Sun R, Deng N, Guo T, Cao Y, Yu Y,
Wang X, Zou B, Zhang S, Jing T, et al: miR-29a/b enhances cell
migration and invasion in nasopharyngeal carcinoma progression by
regulating SPARC and COL3A1 gene expression. PLoS One.
10:e01209692015. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Suresh A, Vannan M, Kumaran D, Gümüs ZH,
Sivadas P, Murugaian EE, Kekatpure V, Iyer S, Thangaraj K and
Kuriakose MA: Resistance/response molecular signature for oral
tongue squamous cell carcinoma. Dis Markers. 32:51–64. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Sepiashvili L, Hui A, Ignatchenko V, Shi
W, Su S, Xu W, Huang SH, O'Sullivan B, Waldron J, Irish JC, et al:
Potentially novel candidate biomarkers for head and neck squamous
cell carcinoma identified using an integrated cell line-based
discovery strategy. Mol Cell Proteomics. 11:1404–1415. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Gu M, Guan J, Zhao L, Ni K, Li X and Han
Z: Correlation of ECM1 expression level with the pathogenesis and
metastasis of laryngeal carcinoma. Int J Clin Exp Pathol.
6:1132–1137. 2013.PubMed/NCBI
|
|
72
|
Meng XY, Liu J, Lv F, Liu MQ and Wan JM:
Study on the correlation between extracellular matrix protein-1 and
the growth, metastasis and angiogenesis of laryngeal carcinoma.
Asian Pac J Cancer Prev. 16:2313–2316. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Pavón MA, Arroyo-Solera I, Céspedes MV,
Casanova I, León X and Mangues R: uPA/uPAR and SERPINE1 in head and
neck cancer: Role in tumor resistance, metastasis, prognosis and
therapy. Oncotarget. 7:57351–57366. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Pavón MA, Arroyo-Solera I, Téllez-Gabriel
M, León X, Virós D, López M, Gallardo A, Céspedes MV, Casanova I,
López-Pousa A, et al: Enhanced cell migration and apoptosis
resistance may underlie the association between high SERPINE1
expression and poor outcome in head and neck carcinoma patients.
Oncotarget. 6:29016–29033. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Van Tubergen EA, Banerjee R, Liu M, Vander
Broek R, Light E, Kuo S, Feinberg SE, Willis AL, Wolf G, Carey T,
et al: Inactivation or loss of TTP promotes invasion in head and
neck cancer via transcript stabilization and secretion of MMP9,
MMP2, and IL-6. Clin Cancer Res. 19:1169–1179. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Pietruszewska W, Bojanowska-Poźniak K and
Kobos J: Matrix metalloproteinases MMP1, MMP2, MMP9 and their
tissue inhibitors TIMP1, TIMP2, TIMP3 in head and neck cancer: An
immunohistochemical study. Otolaryngol Pol. 70:32–43. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Zhang C, Li C, Zhu M, Zhang Q, Xie Z, Niu
G, Song X, Jin L, Li G and Zheng H: Meta-analysis of MMP2, MMP3,
and MMP9 promoter polymorphisms and head and neck cancer risk. PLoS
One. 8:e620232013. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Vincent-Chong VK, Salahshourifar I,
Karen-Ng LP, Siow MY, Kallarakkal TG, Ramanathan A, Yang YH, Khor
GH, Rahman ZA, Ismail SM, et al: Overexpression of MMP13 is
associated with clinical outcomes and poor prognosis in oral
squamous cell carcinoma. ScientificWorldJournal. 2014:8975232014.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Pradhan-Palikhe P, Vesterinen T, Tarkkanen
J, Leivo I, Sorsa T, Salo T and Mattila PS: Plasma level of tissue
inhibitor of matrix metalloproteinase-1 but not that of matrix
metalloproteinase-8 predicts survival in head and neck squamous
cell cancer. Oral Oncol. 46:514–518. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Huang CF, Yu GT, Wang WM, Liu B and Sun
ZJ: Prognostic and predictive values of SPP1, PAI and caveolin-1 in
patients with oral squamous cell carcinoma. Int J Clin Exp Pathol.
7:6032–6039. 2014.PubMed/NCBI
|
|
81
|
Aquino G, Sabatino R, Cantile M, Aversa C,
Ionna F, Botti G, La Mantia E, Collina F, Malzone G, Pannone G, et
al: Expression analysis of SPARC/osteonectin in oral squamous cell
carcinoma patients: From saliva to surgical specimen. Biomed Res
Int. 2013:7364382013. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Li L, Lei Q, Zhang S, Kong L and Qin B:
Screening and identification of key biomarkers in hepatocellular
carcinoma: Evidence from bioinformatic analysis. Oncol Rep.
38:2607–2618. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Saha SK, Jeong Y, Cho S and Cho SG:
Systematic expression alteration analysis of master reprogramming
factor OCT4 and its three pseudogenes in human cancer and their
prognostic outcomes. Sci Rep. 8:148062018. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Yang Y, Dong X, Xie B, Ding N, Chen J, Li
Y, Zhang Q, Qu H and Fang X: Databases and web tools for cancer
genomics study. Genomics Proteomics Bioinformatics. 13:46–50. 2015.
View Article : Google Scholar : PubMed/NCBI
|