Introduction
Breast cancer is the most widespread cancer and the
second-leading cause of cancer-associated mortality among females
worldwide (data from Cancer Statistics 2018), despite the
availability of effective chemotherapy agents (1,2).
Cytotoxic and molecular targeted anti-cancer therapies remain the
major treatment options for breast cancer (3). The greatest challenges in the
management of patients with breast cancer include the determination
of prognosis and the identification of appropriate adjuvant
systemic therapies (4). Therefore,
there is a requirement for effective prognostic biomarkers for
patients with refractory breast cancer.
WD-repeat domain (WDR)-containing proteins are
characterized by a common sequence repeat of tryptophan and
aspartic acid pairs are generally found at the end of their
40-residue-long amino acid sequences (5). WDRs, including DR13, DCAF4L2, WD48,
BOP1 and CIRH1A, serve as platforms for the assembly of protein
complexes or mediators of transient interplay between other
proteins (6).
WD domain repeat 34 (WDR34), a member of the WDR
superfamily, encodes a highly conserved protein consisting of 536
amino acids, with human and mouse WDR34 sharing 83% homology. WDR34
acts as a mitogen-activated protein kinase kinase kinase
7-associated inhibitor of the interleukin-1 receptor/Toll-like
receptor (TLR)3/TLR4-induced nuclear factor-κB activation pathway
(7,8). Studies have revealed that WDR34
mutations may result in short-rib polydactyly syndrome type III or
severe asphyxiating thoracic dysplasia (8,9). A study
by Wu et al (10) indicated
that WDR34-mutant mice succumb in mid-gestation and exhibit open
brain and polydactyly phenotypes. WDR34 downregulation was
identified in oral squamous cell carcinoma tissues compared with
normal control tissues (11).
Research from cDNA microarrays demonstrated that WDR34 had a
6.8-fold difference in expression between cases with and without
recurrence in patients with bladder cancer (12). Although numerous studies have
suggested that WDR34 expression is associated with the progression
of cancer, the role of WDR34 expression in the tumorigenesis and
prognosis of breast cancer remains unknown.
In the present study, a bioinformatics analysis
based on a number of public clinical databases was performed in
order to investigate WDR34 expression in breast cancer and normal
tissues. The aim of the present study was to identify a possible
biomarker suitable for the prognostic prediction of patients with
breast cancer.
Materials and methods
Oncomine analysis
The WDR34 mRNA expression level was analyzed in
breast cancer and matched normal tissues based on the The Oncomine
Platform (www.oncomine.org), which consists of 715
datasets and 86,733 samples. The analysis was conducted using the
following filters: i) Gene, WDR34; ii) differential analysis,
cancer vs. normal analysis; iii) cancer type, breast cancer; and
iv) data type, mRNA. In the current study, all statistical methods
and statistical values were obtained directly from the
corresponding database. The threshold for statistical significance
was set as P<0.01; fold change >2; and gene rank, top
10%.
The present study involved a meta-analysis using a
random permutation method and further illustrated WDR34 gene
expression in different breast cancer datasets from Oncomine
(13).
Breast Cancer Gene-Expression Miner
(bc-GenExMiner)
In the present study, the expression of WDR34 mRNA
in different subtypes of breast cancer and the correlation between
genes or identified clusters of correlated co-expressed genes were
analyzed using bc-GenExMiner (version 4.1;
bcgenex.centregauducheau.fr). bc-GenExMiner includes 36 annotated
genomic datasets and 5,861 patients with breast cancer (14,15).
Cancer Cell Line Encyclopedia (CCLE)
analysis
WDR34 mRNA expression in breast cancer cell lines
was analyzed using the CCLE database (portals.broadinstitute.org/ccle/home), which provides
public access to genomic data, analysis and visualization for 947
human cancer cell lines.
Kaplan-Meier survival curve
analysis
The prognostic value of WDR34 mRNA expression in
breast cancer was assessed according to overall survival (OS) and
relapse-free survival (RFS) using Kaplan-Meier plotter (kmplot.com/analysis) up to June 30 2018 (16). Log-rank P-values and hazard ratios
(HRs) with 95% confidence intervals were determined on the
webpage.
The Cancer Genome Atlas (TCGA) data
and cBioPortal
The invasive breast carcinoma dataset (TCGA,
Provisional), consisting of 1,105 samples with pathology reports,
was selected for further analysis of WDR34 expression using
cBioPortal (www.cbioportal.org) (17,18). The
selected genomic profiles included putative copy-number alterations
from GISTIC_2.0 (http://portals.broadinstitute.org/cgi-bin/cancer/publications/pub_paper.cgi?mode=view&paper_id=216&p=t),
and the selected patient/case sets included tumor samples with RNA
data (RNA Seq V2). Disease-free survival (DFS) or OS results were
derived from cBioPortal using the OS Kaplan-Meier estimate. The
proportion and distribution of samples with WDR34 alterations were
presented in the Oncoprint (http://www.canvasxpress.org/html/oncoprint-2.html),
which is a visual image used to show changes in different genomes,
including mutations, copy number changes and mRNA expression. All
statistical methods and statistical results in this study came from
the corresponding online database.
Results
mRNA expression profiles of WDR34 in
different tumor types
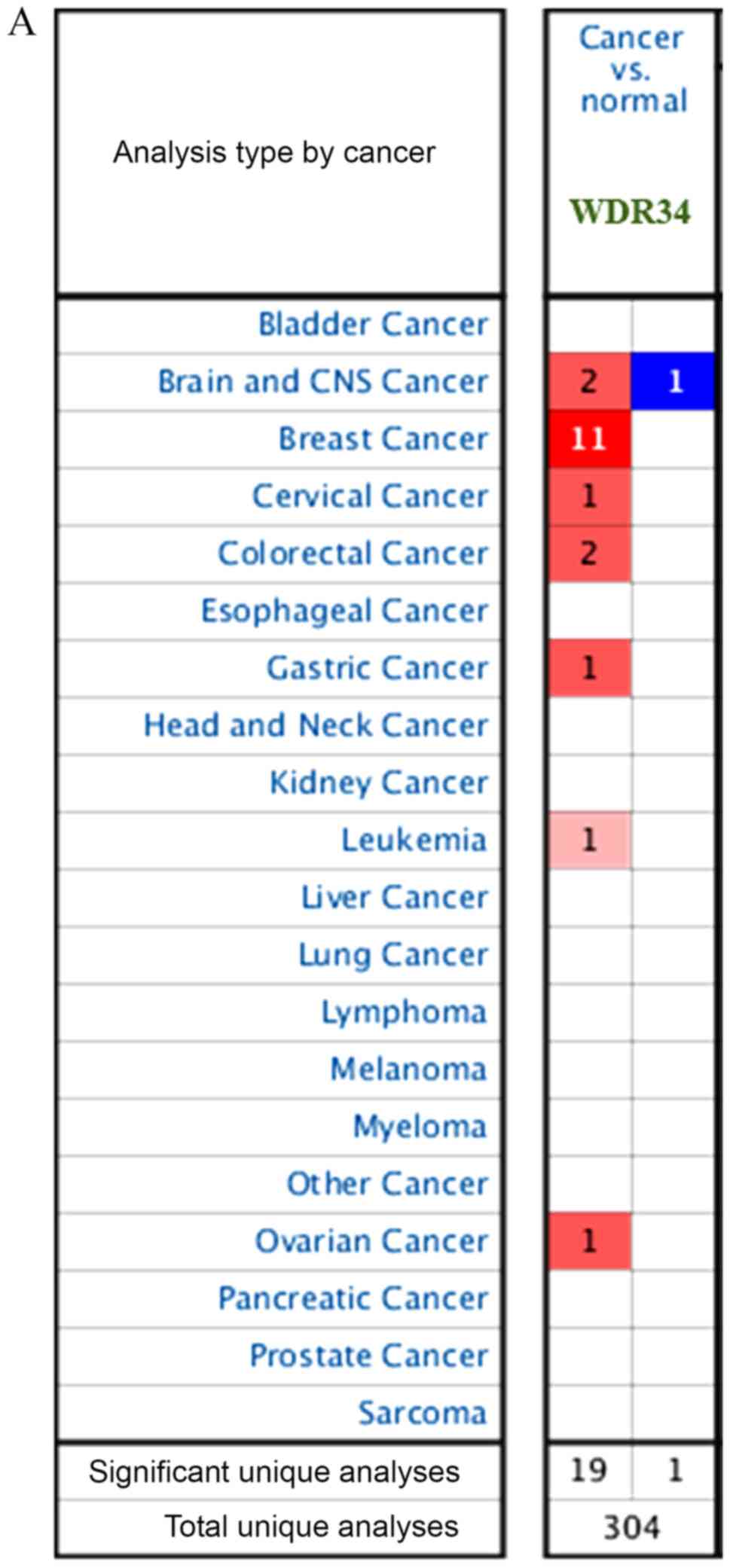
A total of 304 unique analyses were gathered from
different types of cancer in the Oncomine database. The results of
20 analyses demonstrated statistical significance; 19 analyses
consisted of high expression and one analysis consisted of reduced
expression. Notably, the mRNA expression of WDR34 in breast cancer
was the highest among different cancer types (Fig. 1A).
Using data from a previous study by Curtis et
al (19), it was demonstrated
that WDR34 mRNA expression was increased 2.034- and 2.103-fold in
breast cancer tissues compared with normal tissues (Fig. 1C and D). A consistent result was
identified in another dataset derived from TCGA, consisting of 593
breast cancer samples; WDR34 mRNA expression was increased
2.199-fold in breast cancer tissues when compared with normal
tissues (Fig. 1E). To demonstrate
the reliability of the study, a meta-analysis of 27 studies from 10
datasets obtained from the Oncomine database was performed. It was
revealed that the expression of WDR34 mRNA in breast cancer tissues
was significantly higher compared with that in normal controls
(P=0.002; Fig. 1B).
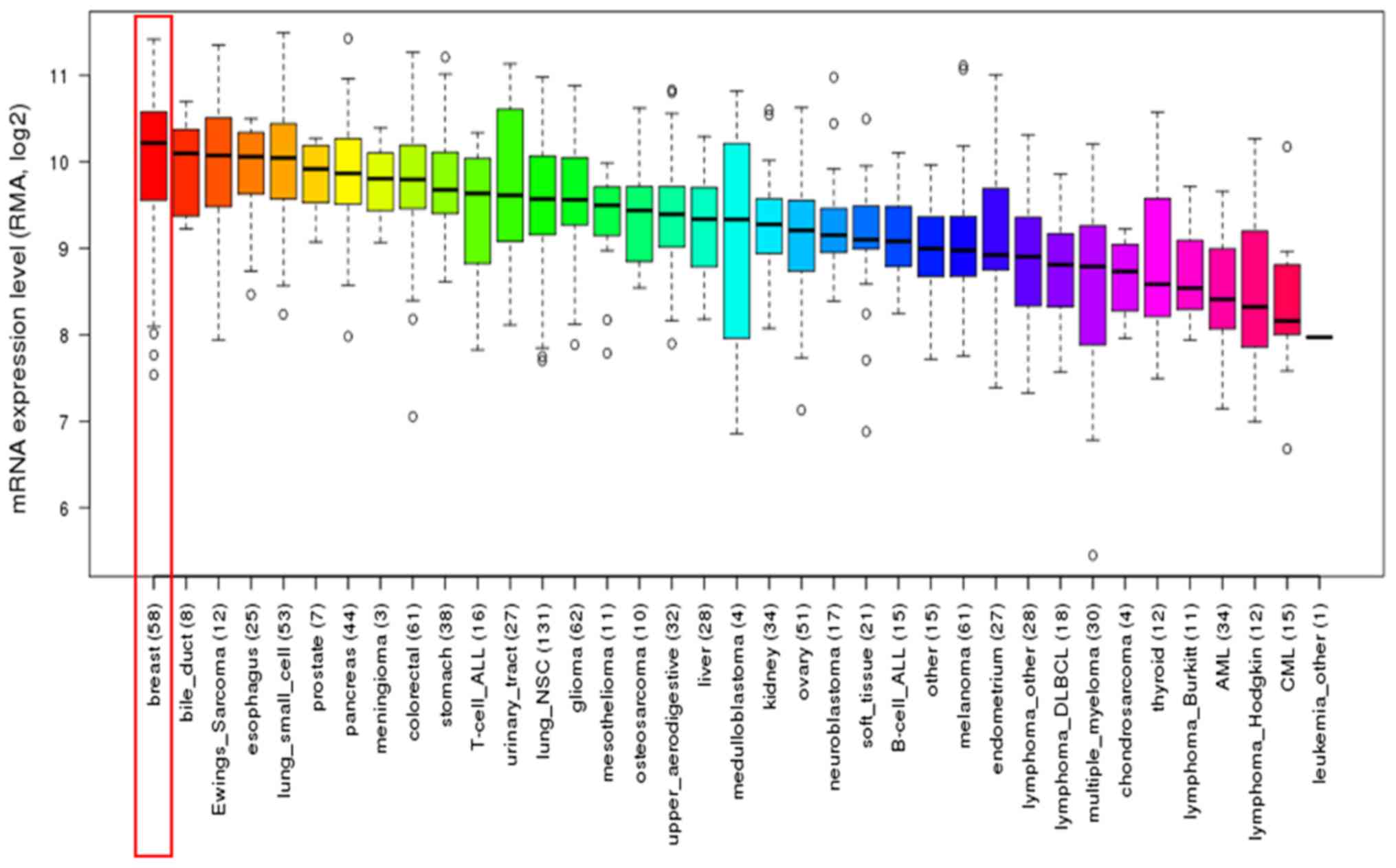
Furthermore, upregulation of WDR34 mRNA was
identified in breast cancer cell lines using CCLE analysis. This
result was consistent with that obtained from the breast cancer
tissues (Fig. 2).
WDR34 mRNA expression is associated
with molecular subtypes of breast cancer
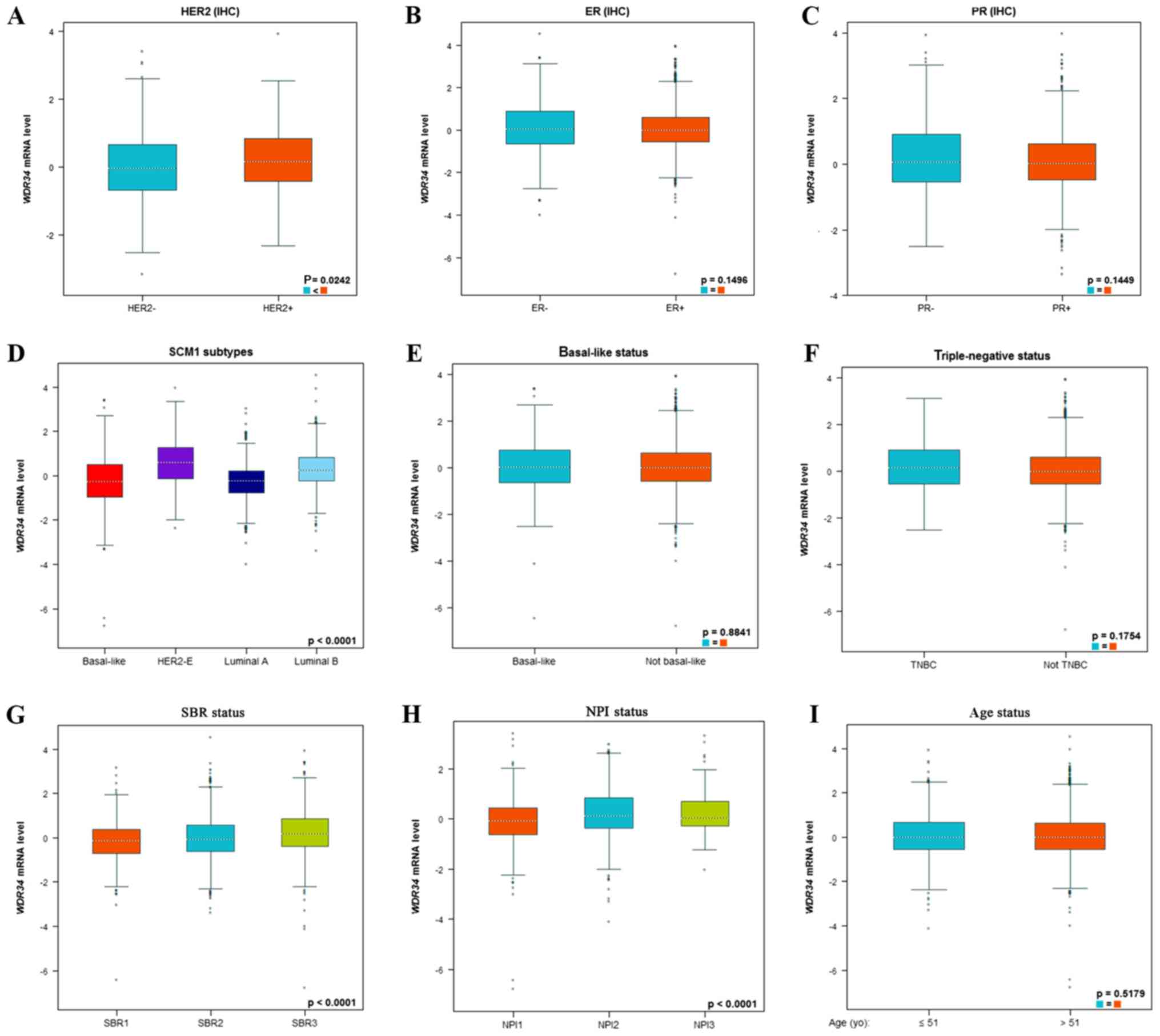
In the BC-GenExMiner database, the expression level
of WDR34mRNA was analyzed in different types of breast cancer
tissues. The expression level of WDR34 mRNA in patients with human
epidermal growth factor receptor 2 (HER2)-positive breast cancer
was increased compared with patients with HER2-negative breast
cancer (P=0.0242; Fig. 3A). However,
there were no statistically significant differences observed
regarding the WDR34 mRNA expression level in patients with estrogen
receptor (ER)-positive or ER-negative breast cancer (Fig. 3B). Similar results were obtained for
patients with progesterone receptor (PR)-positive and PR-negative
breast cancer (Fig. 3C).
Breast cancer subtypes were determined according to
SCM1 classification using the bc-GenExMiner. Notably, WDR34 mRNA
expression in the HER2 subtype was significantly increased compared
with the luminal A and luminal B subtypes (P<0.0001;
Dunnett-Tukey-Kramer's test; Fig.
3D). Furthermore, WDR34 mRNA expression was not significantly
different between patients with basal-like and non-basal-like
breast cancer (P=0.8841; Fig. 3E).
Similarly, WDR34 mRNA expression was not significantly different
between patients with triple negative breast cancer (TNBC) and
non-TNBC patients (P=0.1754; Fig.
3F).
Regarding the Scarff Bloom and Richardson (SBR)
grade status, higher WDR34 mRNA expression was significantly
associated with a more advanced SBR grade (P<0.0001;
Dunnett-Tukey-Kramer's test; Fig.
3G). Regarding the Nottingham Prognostic Index (NPI) status, a
higher NPI level was significantly associated with increased WDR34
mRNA expression (P<0.0001; Dunnett-Tukey-Kramer's test; Fig. 3H). When age was taken into account,
it was identified that WDR34 mRNA expression was not significantly
elevated with increasing age (P=0.5179; Fig. 3I).
High expression of WDR34 mRNA is
correlated with high expression of forkhead box M1 (FOXM1) and
PTTG1 regulator of sister chromatid separation securin (PTTG1)
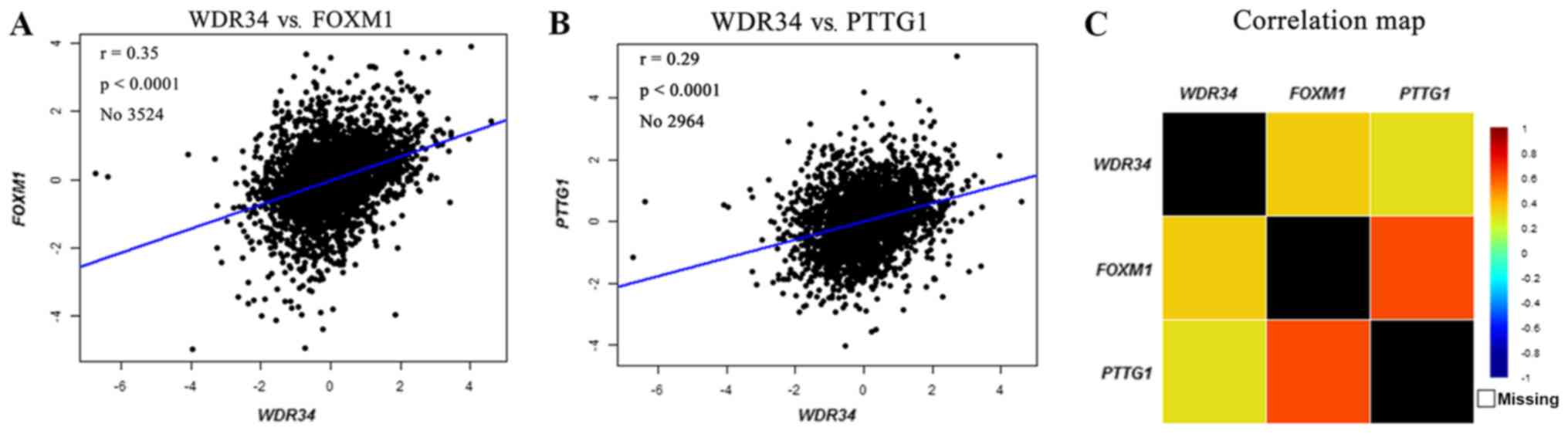
It was demonstrated that WDR34 mRNA expression was
upregulated in breast cancer. Therefore, an in-depth exploration on
whether WDR34 was associated with other potential gene biomarkers
was performed. In the bc-GenExMiner, mRNA correlation analysis
indicated that high WDR34 mRNA expression was positively correlated
with the expression of FOXM1 (r=0.35; P<0.0001; Fig. 4A) and PTTG1 (r=0.29; P<0.0001;
Fig. 4B). A correlation map for all
patients was produced for WDR34, FOXM1 and PTTG1 (Fig. 4C).
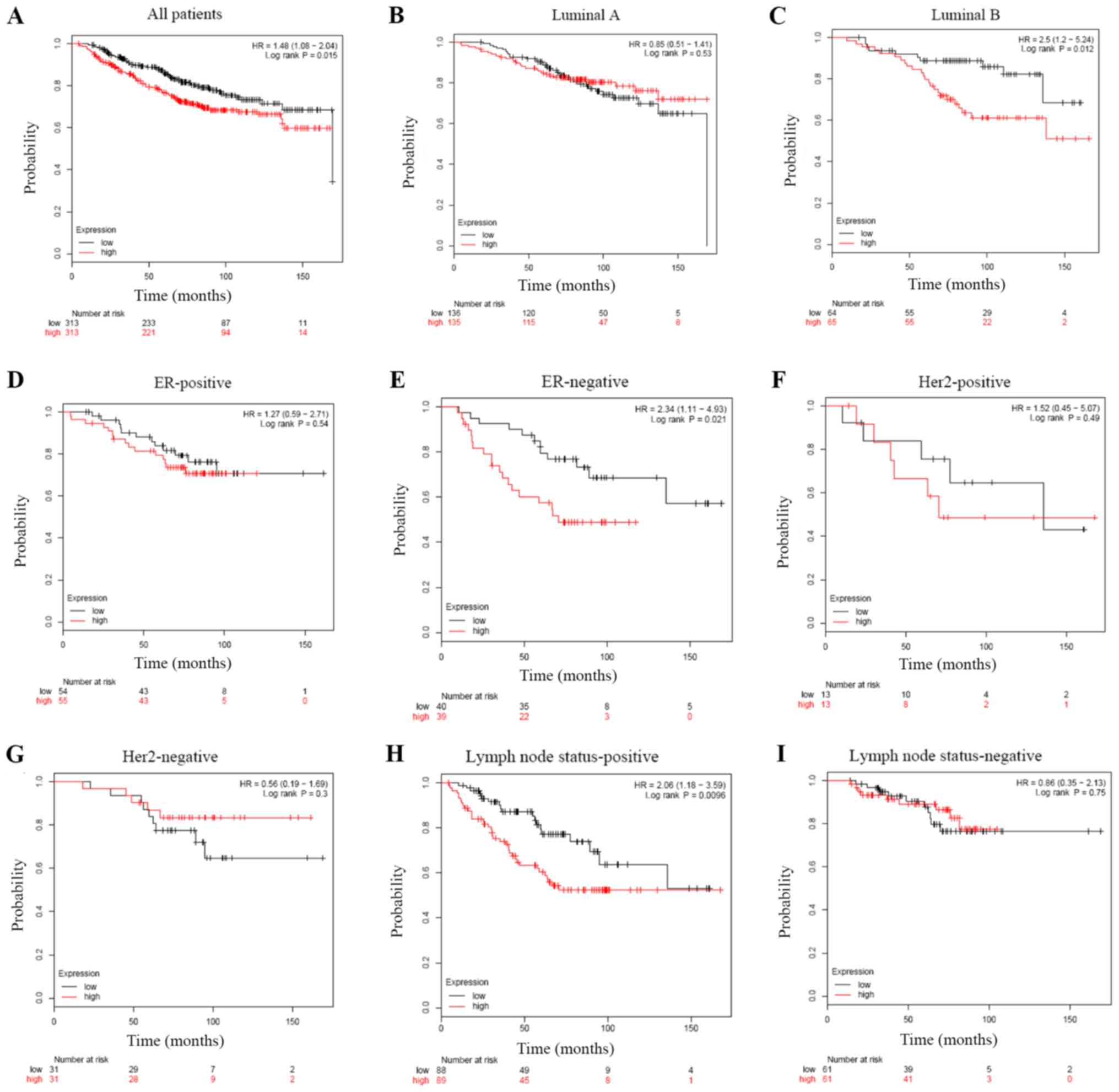
Increased WDR34 mRNA expression
indicates poor OS in patients with breast cancer, particularly in
luminal B, lymph node status-positive and ER-negative
subgroups
The potential prognostic value of WDR34 mRNA
expression in patients with breast cancer was further assessed. The
current study indicated that high WDR34 mRNA expression was
associated with shorter OS in patients with breast cancer (HR=1.48;
P=0.015; Fig. 5A). Sub-analysis
indicated that high WDR34 mRNA expression was associated with
shorter OS in luminal B (HR=2.50; P=0.012; Fig. 5C), but not luminal A (HR=0.85;
P=0.53; Fig. 5B). Furthermore, high
WDR34 mRNA expression was correlated with shorter OS in patients
with ER-negative breast cancer (HR=2.34; P=0.021; Fig. 5E), but not in those with ER-positive
breast cancer (HR=1.27; P=0.54; Fig.
5D). In patients with breast cancer with a positive lymph node
status, a significant correlation was identified between high WDR34
mRNA expression and OS (HR=2.06; P=0.0096; Fig. 5H); however, a correlation was not
indicated in patients with breast cancer with a negative lymph node
status (HR=0.86; P=0.75; Fig. 5I).
Significant differences were not observed in breast cancer subtypes
including HER2-positive (HR=1.52; P=0.49; Fig. 5F), HER2-negative (HR=0.56; P=0.30;
Fig. 5G) and basal subgroups
(HR=1.10; P=0.76; data not shown).
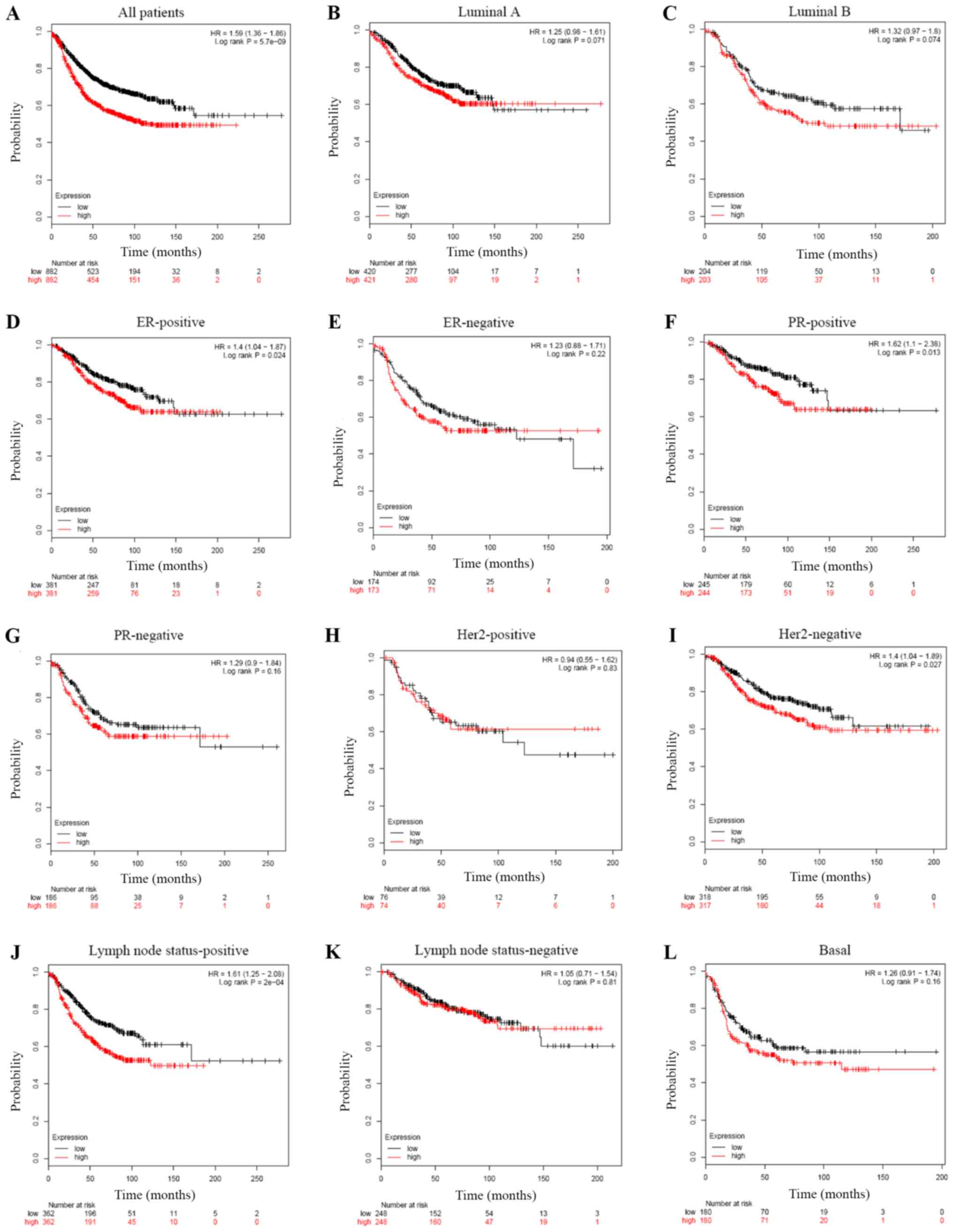
Elevated WDR34 mRNA expression is
correlated with shorter RFS in patients with breast cancer,
particularly in ER-positive, HER2-negative and PR-positive
subgroups
High WDR34 mRNA expression was significantly
associated with shorter RFS in breast cancer patients (HR=1.59;
P=5.7×10−09; Fig. 6A).
Sub-analysis on different subtypes of breast cancer was conducted,
which indicated that high WDR34 mRNA expression was correlated with
shorter RFS in patients with ER-positive (HR=1.40; P=0.024;
Fig. 6D), but not in ER-negative
breast cancer (HR=1.23; P=0.22; Fig.
6E). Moreover, high WDR34 mRNA expression was correlated with
shorter RFS in patients with PR-positive (HR=1.62; P=0.013;
Fig. 6F), HER2-negative (HR=1.40;
P=0.027; Fig. 6I) and lymph node
status-positive (HR=1.61; P=2×10−04; Fig. 6J) breast cancer subtypes, but not in
PR-negative (HR=1.29; P=0.16; Fig.
6G), HER2-positive (HR=0.94; P=0.83; Fig. 6H), lymph node status-negative
(HR=1.05; P=0.81; Fig. 6K), luminal
A (HR=1.25; P=0.071; Fig. 6B),
luminal B (HR=1.32; P=0.074; Fig.
6C) or basal (HR=1.26; P=0.16; Fig.
6L) breast cancer subtypes.
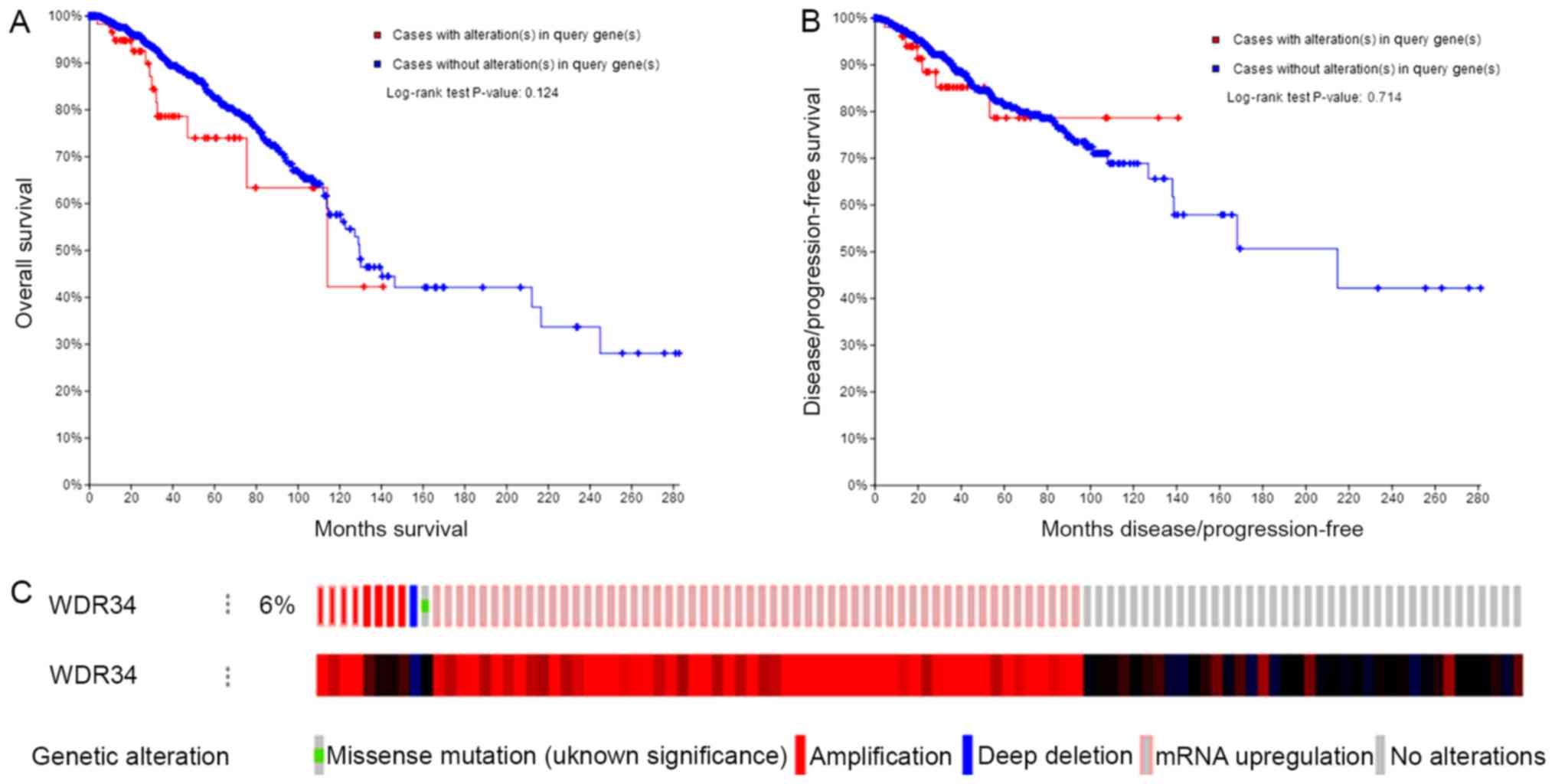
WDR34 mRNA mutations do not affect OS
or DFS in patients with breast cancer
A total of 66/1,093 (6%) sequenced patients with
invasive breast carcinoma exhibited alterations of WDR34. The
Kaplan-Meier plot and log-rank test identified that the alterations
of WDR34 had no significant influence on the OS or DFS of patients
with breast cancer (Fig. 7).
Discussion
Breast cancer is the most commonly diagnosed
malignancy among females, and is a heterogeneous disease with
distinct molecular subtypes (20).
Several molecular subtypes of breast cancer have been identified,
including luminal A, luminal B, HER2-positive and basal-like
subtypes (21–23). The development of breast cancer
resistance to chemoradiotherapy or targeted therapy, as well as
distant metastasis, remains a challenge. Therefore, there is a
requirement for the identification of potential target genes or
proteins that may be beneficial for the treatment of breast cancer
(24,25).
The WDR protein family is involved in a variety of
cellular processes, including cell cycle progression, signal
transduction, apoptosis and gene regulation (7). Receptor for activated C kinase 1, a
member of the WDR protein family, was significantly upregulated in
breast cancer, non-small-cell lung cancer, pulmonary
adenocarcinoma, glioma and esophageal squamous-cell carcinoma
(26). Based on Oncomine database
analysis, the current study demonstrated, to the best of our
knowledge for the first time, that high WDR34 mRNA expression
occurred in breast cancer tissues and breast cancer cell lines.
Furthermore, co-expression analysis, as evidenced by bc-GenExMiner,
indicated that high WDR34 expression was positively associated with
the expression levels of FOXM1 and PTTG1 (27). Previous studies indicated that FOXM1
is overexpressed in breast cancer and is strongly associated with
resistance to targeted therapies and chemotherapy (28). Elevated expression of FOXM1 has been
reported in a variety of human tumors, including those of the
breast (29). The mean expression
level of FOXM1 was previously reported to be the highest in the
TNBC subtype, which was associated with poor prognosis and reduced
survival time in patients with breast cancer (29,30).
Notably, PTTG1 is an oncogene that is important for the progression
of mitosis in the metaphase-anaphase transition (31,32).
Additionally, PTTG1 overexpression is associated with malignancy,
particularly thyroid, breast and colorectal carcinoma (32). A previous study indicated that PTTG1
is highly expressed in patients with breast cancer and that the
expression levels were correlated with the degree of malignancy in
breast cancer cell lines. Furthermore, the study demonstrated that
PTTG1 enhanced the migratory and invasive properties of breast
cancer cells by inducing epithelial-to-mesenchymal transition
(33).
Survival analysis of WDR34 expression levels in the
current study demonstrated that high WDR34 expression was
associated with poor OS in patients with breast cancer,
particularly in the luminal B, lymph node status-positive and
ER-negative subgroups, when compared with controls. These results
suggested that WDR34 serves an important role in the tumor
progression of hormone-sensitive breast cancer. In addition, it was
revealed that high WDR34 expression frequently predicted shorter
RFS in patients with breast cancer, particularly in ER-positive,
HER2-negative and PR-positive subgroups. The aforementioned results
suggested that patients with breast cancer with high WDR34
expression had an increased risk of mortality. Previous studies
indicated that the WDR protein coronin-3, which contains five WD
motifs, is associated with various invasive tumors, including
melanoma, human diffuse glioma, liver cancer, breast cancer and
gastric cancer (34–36). Furthermore, high expression of
coronin-3 was associated with increased tumor malignancy and a more
advanced clinical stage. F-box and WD repeat domain containing 7
(FBXW7) recognizes and binds to substrates via eight WD motifs at
the C-terminus (37,38). However, knockout of FBXW7 with short
hairpin RNA increases the KLF5 gene and Fbw7 targets the KLF5
protein for ubiquitin-mediated proteasomal degradation and
suppresses breast cancer cell proliferation (39). Taken together, these results
suggested that overexpression of WDR34 may be used as a biomarker
of poor prognosis in patients with breast cancer in the future.
Previous studies have reported that HER2-positive
breast cancer is frequently more aggressive and is associated with
an increased risk of disease recurrence and mortality compared with
other breast cancer subtypes (23,40,41). At
present, a substantial number of HER2-targeting agents have been
introduced for the clinical treatment of breast cancer (42). One of these agents, trastuzumab
(Herceptin), has several shortcomings, including its high cost and
side effects such as cardiotoxicity (43). The present study indicated that the
WDR34 mRNA expression level was higher in patients with
HER2-positive breast cancer compared with patients with
HER2-negative breast cancer. Furthermore, the WDR34 mRNA expression
level was higher in patients with HER2-positive breast cancer
compared with patients with the luminal A and luminal B subtypes.
Thus, it may be inferred that increased levels of WDR34 mRNA may
result in more aggressive breast cancer. However, it remains
unknown whether the upregulation of WDR34 may be used as a target
for novel agents for the treatment of HER2-positive breast
cancer.
The NPI is widely used to predict survival in
patients with breast cancer, and a higher NPI is associated with a
worse prognosis (44,45). The present study indicated that a
higher NPI level was significantly associated with the expression
level of WDR34 mRNA. In other words, WDR34 mRNA upregulation
predicted a poorer prognosis in patients with breast cancer. In
addition, high WDR34 mRNA expression was associated with a higher
SBR grade, which typically suggests growing and spreading tumors
(46).
Previous studies have reported that mutations of
WDR34 mRNA are primarily associated with short-rib polydactyly
syndrome type III and severe asphyxiating thoracic dysplasia
(8,9). Loss-of-function mutations of FBXW7,
another WDR protein, have been identified in colorectal cancer
(18%), uterine endometrial carcinoma (15%) and uterine
carcinosarcoma (40%), suggesting that the interactions between
FBXW7 and its substrates are interrupted by disrupting the
structural integrity of the WDR domain (47,48).
Notably, compounds targeting the WDR domain of FBXW7 are expected
to antagonize binding of cyclin E and phenocopy the oncogenic
effect of mutations that are recurrent in cancer (47). Although gene alterations of WDR34
occurred in ~6% of sequenced patients with invasive breast
carcinoma, no influence on OS or DFS was observed in the present
study. Thus, it can be inferred that a WDR34 mRNA mutation did not
significantly influence the prognosis of breast cancer. However,
future studies on the correlation between WDR34 mRNA mutation and
breast cancer are required.
The present study had a number of limitations.
Firstly, the study only analyzed the high expression of WDR34 in
patients with breast cancer compared with normal samples using
comprehensive bioinformatics analysis. Functional verification was
not carried out through cohort research and molecular mechanism
research. Secondly, the Oncomine database is a cancer microarray
data-mining platform, which can be used to identify new prognostic
biomarkers. However, gene chip technology has a certain degree of
false-positive results that are likely to influence the accuracy of
the current study. Thirdly, the number of samples in the OS
analysis using the Kaplan-Meier plotter was small, which possibly
influenced the outcome. Finally, since the survival analysis was
based on an online database (the Kaplan-Meier Plotter), integrated
data could not be acquired to perform multivariate analysis.
In conclusion, to the best of our knowledge, this
study demonstrated for the first time that the WDR34 mRNA
expression was significantly increased in breast cancer tissues
compared with normal tissues. Furthermore, high WDR34 mRNA
expression was associated with poor OS and shorter RFS in patients
with breast cancer, and may be an attractive prognostic prediction
biomarker and promising therapeutic target for breast cancer.
However, the role of WDR34 mRNA expression in the genesis and
progression of breast cancer requires further basic and clinical
study.
Acknowledgements
The authors would like to thank Professor Li Zhang
for providing careful guidance in writing this paper. The authors
would also like to thank Wen-Jie Shi for helping with dataset
usage.
Funding
This study was supported by the Chongming Committee
of Science and Technology (Shanghai, China; grant no.
CKY2018-11).
Availability of data and materials
The datasets used during the present study may be
obtained from the corresponding author upon reasonable request.
Authors' contributions
LZ and DJH designed the experiment, provided
financial support, revised the manuscript and gave final approval
of the version to be published. DJH and WJS performed the
statistical analysis and wrote the paper. MY collected data and
revised the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kadalayil L, Khan S, Nevanlinna H,
Fasching PA, Couch FJ, Hopper JL, Liu J, Maishman T, Durcan L,
Gerty S, et al: Germline variation in ADAMTSL1 is associated with
prognosis following breast cancer treatment in young women. Nat
Commun. 8:16322017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Si W, Shen J, Du C, Chen D, Gu X, Li C,
Yao M, Pan J, Cheng J, Jiang D, et al: A miR-20a/MAPK1/c-Myc
regulatory feedback loop regulates breast carcinogenesis and
chemoresistance. Cell Death Differ. 25:406–420. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nicolini A, Ferrari P and Duffy MJ:
Prognostic and predictive biomarkers in breast cancer: Past,
present and future. Semin Cancer Biol. 52:56–73. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Smith TF: Diversity of WD-repeat proteins.
Subcell Biochem. 48:20–30. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
van Nocker S and Ludwig P: The WD-repeat
protein superfamily in Arabidopsis: Conservation and divergence in
structure and function. BMC Genomics. 4:502003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gao D, Wang R, Li B, Yang Y, Zhai Z and
Chen DY: WDR34 is a novel TAK1-associated suppressor of the
IL-1R/TLR3/TLR4-induced NF-kappaB activation pathway. Cell Mol Life
Sci. 66:2573–2584. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huber C, Wu S, Kim AS, Sigaudy S,
Sarukhanov A, Serre V, Baujat G, Le Quan Sang KH, Rimoin DL, Cohn
DH, et al: WDR34 mutations that cause short-rib polydactyly
syndrome type III/severe asphyxiating thoracic dysplasia reveal a
role for the NF-κB pathway in cilia. Am J Hum Genet. 93:926–931.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
You SH, Lee YS, Lee CP, Lin CP, Lin CY,
Tsai CL, Chang YL, Cheng PJ, Wang TH and Chang SD: Identification
of a c.544C>T mutation in WDR34 as a deleterious recessive
allele of short rib-polydactyly syndrome. Taiwan J Obstet Gynecol.
56:857–862. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu C, Li J, Peterson A, Tao K and Wang B:
Loss of dynein-2 intermediate chain Wdr34 results in defects in
retrograde ciliary protein trafficking and Hedgehog signaling in
the mouse. Hum Mol Genet. 26:2386–2397. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yamamoto JI, Kasamatsu A, Okubo Y,
Nakashima D, Fushimi K, Minakawa Y, Kasama H, Shiiba M, Tanzawa H
and Uzawa K: Evaluation of tryptophan-aspartic acid
repeat-containing protein 34 as a novel tumor-suppressor molecule
in human oral cancer. Biochem Biophys Res Commun. 495:2469–2474.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mares J, Szakacsova M, Soukup V, Duskova
J, Horinek A and Babjuk M: Prediction of recurrence in low and
intermediate risk non-muscle invasive bladder cancer by real-time
quantitative PCR analysis: cDNA microarray results. Neoplasma.
60:295–301. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang Y, Zhang L, Kong F, Zhang M, Lv H,
Liu G, Liao M, Feng R, Li J and Zhang R: MCPerm: A Monte Carlo
permutation method for accurately correcting the multiple testing
in a meta-analysis of genetic association studies. PLoS One.
9:e892122014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jézéquel P, Campone M, Gouraud W,
Guérin-Charbonnel C, Leux C, Ricolleau G and Campion L:
bc-GenExMiner: An easy-to-use online platform for gene prognostic
analyses in breast cancer. Breast Cancer Res Treat. 131:765–775.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jézéquel P, Frénel JS, Campion L,
Guérin-Charbonnel C, Gouraud W, Ricolleau G and Campone M:
bc-GenExMiner 3.0: New mining module computes breast cancer gene
expression correlation analyses. Database (Oxford).
2013:bas0602013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lánczky A, Nagy Á, Bottai G, Munkácsy G,
Szabó A, Santarpia L and Győrffy B: miRpower: A web-tool to
validate survival-associated miRNAs utilizing expression data from
2178 breast cancer patients. Breast Cancer Res Treat. 160:439–446.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Curtis C, Shah SP, Chin SF, Turashvili G,
Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et
al: The genomic and transcriptomic architecture of 2,000 breast
tumours reveals novel subgroups. Nature. 486:346–352. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Goda AA, Siddique AB, Mohyeldin M, Ayoub
NM and El Sayed KA: The maxi-K (BK) channel antagonist penitrem a
as a novel breast cancer-targeted therapeutic. Mar Drugs. 16(pii):
E1572018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Goldhirsch A, Glick JH, Gelber RD, Coates
AS, Thurlimann B and Senn HJ; Panel members, : Meeting highlights:
International expert consensus on the primary therapy of early
breast cancer 2005. Ann Oncol. 16:1569–1583. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sorlie T, Tibshirani R, Parker J, Hastie
T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et
al: Repeated observation of breast tumor subtypes in independent
gene expression data sets. Proc Natl Acad Sci USA. 100:8418–8423.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Perou CM, Sorlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang C, Sun X, Wang K, Wang Y, Yang F and
Wang H: Breast cancer targeted chemotherapy based on
doxorubicin-loaded bombesin peptide modified nanocarriers. Drug
Deliv. 23:2697–2702. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mahaddalkar T and Lopus M: From natural
products to designer drugs: Development and molecular mechanisms
action of novel anti-microtubule breast cancer therapeutics. Curr
Top Med Chem. 17:2559–2568. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li JJ and Xie D: RACK1, a versatile hub in
cancer. Oncogene. 34:1890–1898. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Giuliano AE, Connolly JL, Edge SB,
Mittendorf EA, Rugo HS, Solin LJ, Weaver DL, Winchester DJ and
Hortobagyi GN: Breast Cancer-Major changes in the American Joint
Committee on Cancer eighth edition cancer staging manual. CA Cancer
J Clin. 67:290–303. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
O'Regan RM and Nahta R: Targeting forkhead
box M1 transcription factor in breast cancer. Biochem Pharmacol.
154:407–413. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Song X, Fiati Kenston SS, Zhao J, Yang D
and Gu Y: Roles of FoxM1 in cell regulation and breast cancer
targeting therapy. Med Oncol. 34:412017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee JJ, Lee HJ, Son BH, Kim SB, Ahn JH,
Ahn SD, Cho EY and Gong G: Expression of FOXM1 and related proteins
in breast cancer molecular subtypes. Int J Exp Pathol. 97:170–177.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nakachi I, Helfrich BA, Spillman MA,
Mickler EA, Olson CJ, Rice JL, Coldren CD, Heasley LE, Geraci MW
and Stearman RS: PTTG1 levels are predictive of saracatinib
sensitivity in ovarian cancer cell lines. Clin Transl Sci.
9:293–301. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Repo H, Gurvits N, Löyttyniemi E, Nykänen
M, Lintunen M, Karra H, Kurki S, Kuopio T, Talvinen K, Söderström M
and Kronqvist P: PTTG1-interacting protein (PTTG1IP/PBF) predicts
breast cancer survival. BMC Cancer. 17:7052017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yoon CH, Kim MJ, Lee H, Kim RK, Lim EJ,
Yoo KC, Lee GH, Cui YH, Oh YS, Gye MC, et al: PTTG1 oncogene
promotes tumor malignancy via epithelial to mesenchymal transition
and expansion of cancer stem cell population. J Biol Chem.
287:19516–19527. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ren G, Tian Q, An Y, Feng B, Lu Y, Liang
J, Li K, Shang Y, Nie Y, Wang X and Fan D: Coronin 3 promotes
gastric cancer metastasis via the up-regulation of MMP-9 and
cathepsin K. Mol Cancer. 11:672012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Iizaka M, Han HJ, Akashi H, Furukawa Y,
Nakajima Y, Sugano S, Ogawa M and Nakamura Y: Isolation and
chromosomal assignment of a novel human gene, CORO1C, homologous to
coronin-like actin-binding proteins. Cytogenet Cell Genet.
88:221–224. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Thal D, Xavier CP, Rosentreter A, Linder
S, Friedrichs B, Waha A, Pietsch T, Stumpf M, Noegel A and Clemen
C: Expression of coronin-3 (coronin-1C) in diffuse gliomas is
related to malignancy. J Pathol. 214:415–424. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li M, Ouyang L, Zheng Z, Xiang D, Ti A, Li
L, Dan Y, Yu C and Li W: E3 ubiquitin ligase FBW7α inhibits
cholangiocarcinoma cell proliferation by downregulating c-Myc and
cyclin E. Oncol Rep. 37:1627–1636. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang J, Wang H, Peters M, Ding N, Ribback
S, Utpatel K, Cigliano A, Dombrowski F, Xu M, Chen X, et al: Loss
of Fbxw7 synergizes with activated AKT signaling to promote c-Myc
dependent cholangiocarcinogenesis. J Hepatol. pii:S0168–8278,
30342-30343. 2019.
|
|
39
|
Zhao D, Zheng HQ, Zhou Z and Chen C: The
Fbw7 tumor suppressor targets KLF5 for ubiquitin-mediated
degradation and suppresses breast cell proliferation. Cancer Res.
70:4728–4738. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Krishnamurti U and Silverman JF: HER2 in
breast cancer: A review and update. Adv Anat Pathol. 21:100–107.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang K, Hong R, Kaping L, Xu F, Xia W,
Qin G, Zheng Q, Lu Q, Zhai Q, Shi Y, et al: CDK4/6 inhibitor
palbociclib enhances the effect of pyrotinib in HER2-positive
breast cancer. Cancer Lett. 447:130–140. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ahmed S, Sami A and Xiang J: HER2-directed
therapy: Current treatment options for HER2-positive breast cancer.
Breast Cancer. 22:101–116. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mercogliano MF, De Martino M, Venturutti
L, Rivas MA, Proietti CJ, Inurrigarro G, Frahm I, Allemand DH, Deza
EG, Ares S, et al: TNFα-induced mucin 4 expression elicits
trastuzumab resistance in HER2-positive breast cancer. Clin Cancer
Res. 23:636–648. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hearne BJ, Teare MD, Butt M and Donaldson
L: Comparison of Nottingham Prognostic Index and Adjuvant Online
prognostic tools in young women with breast cancer: Review of a
single-institution experience. BMJ Open. 5:e0055762015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Albergaria A, Ricardo S, Milanezi F,
Carneiro V, Amendoeira I, Vieira D, Cameselle-Teijeiro J and
Schmitt F: Nottingham Prognostic Index in triple-negative breast
cancer: A reliable prognostic tool? BMC Cancer. 11:2992011.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kabbage M, Trimeche M, Ben Nasr H, Hammann
P, Kuhn L, Hamrita B and Chahed K: Tropomyosin-4 correlates with
higher SBR grades and tubular differentiation in infiltrating
ductal breast carcinomas: An immunohistochemical and
proteomics-based study. Tumour Biol. 34:3593–3602. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Song R, Wang ZD and Schapira M: Disease
association and druggability of WD40 repeat proteins. J Proteome
Res. 16:3766–3773. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yuan Y, Qi G, Shen H, Guo A, Cao F, Zhu Y,
Xiao C, Chang W and Zheng S: Clinical significance and biological
function of WD repeat domain 54 as an oncogene in colorectal
cancer. Int J Cancer. 144:1584–1595. 2019.PubMed/NCBI
|