Introduction
Acute myeloid leukemia (AML) is the most prevalent
malignant myeloid tumor, primarily occurring in elderly patients
and characterized by the accumulation of blast cells and the
blockage of myeloid differentiation in bone marrow (1). Molecular, genetic and cytogenetic
abnormalities cause clonal expansion of early hematopoietic
progenitor cells and obstruct hematopoiesis of normal bone marrow
(2). Although several patients with
AML respond to the induction chemotherapy, the perseverance of
residuary blast cells causes an unusual relapse rate in the bone
marrow (3,4). Consequently, it is required to
elucidate the pathogenesis and discover novel therapeutic targets
for AML.
MicroRNAs (miRNAs or miR) are small endogenous
non-coding RNA molecules consisting of 21–25 nucleotides (5). miRNAs bind to the 3′-untranslated
region (UTR) of their target genes, resulting in decreased mRNA
expression or transcriptional inhibition of protein synthesis
(6). miRNAs have been used as
diagnostic and prognosis biomarkers due to their role as oncogenes
(7) or tumor suppressors (8) in cancer. Evidence has demonstrated that
various miRNAs are closely associated with AML, including miR-143,
miR-34a and miR-126 (9–11). miR-21 has been demonstrated to serve
an essential role in various types of cancer. For instance, miR-21
acts as a predictor and prognostic factor for trastuzumab therapy
in human epidermal growth factor receptor 2-positive metastatic
breast cancer (12). In addition, a
previous study demonstrated that miR-21 is overexpressed in
nucleophosmin 1-mutant acute myeloid leukemias (13). However, the regulatory mechanism of
miR-21 in AML progression remains unknown.
Krüppel-like factor 5 (KLF5), a zinc-finger
transcription factor, functions as a tumor suppressor or an
oncogene (14,15). It serves an important role in cell
proliferation and metastasis by regulating expression of several
downstream target genes, such as tumor necrosis factor α-induced
protein 2, hypoxia inducible factor-1α, FYN proto-oncogene
(16–18). A previous study suggested that KLF5
functions as a tumor suppressor in AML (19). However, the role of KLF5 in AML
progression is not fully understood.
In the present study, it was suggested that KLF5 may
be a potential direct target of miR-21 in AML cells, with a binding
site at the 3′-UTR. The study aimed to investigate the role of
miR-21 on AML cells and its underlying molecular mechanisms.
Materials and methods
Cell culture
The human AML cell lines HL-60 and SKM-1 and the
bone marrow stromal cell line HS-5 were obtained from the Cell Bank
of Type Culture Collection of the Chinese Academy of Sciences. Each
cell line was cultured in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) with 10% fetal bovine serum (HyClone; GE
Healthcare Life Sciences) and 1% penicillin/streptomycin (Gibco;
Thermo Fisher Scientific, Inc.). Cells were maintained in a
humidified incubator at 37°C with 5% CO2.
Reverse transcription-quantitative PCR
(RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to isolate the total RNA from the
harvested HL-60, SKM-1 and HS-5 cells. cDNA was generated using a
PrimeScript™ RT-qPCR (Takara Biotechnology Co, Ltd.). The reverse
transcription reaction conditions are as follows: 37°C for 15 min,
85°C for 5 sec and finally at 4°C. An Applied Biosystems 7500
real-time PCR system (Thermo Fisher Scientific, Inc.) was used to
perform the qPCR using a SYBR premix ex Taq™ kit (Takara Bio, Inc.)
according to the manufacturer's protocol. The PCR thermocycling
conditions were as follows: 95°C for 30 sec, followed by 40 cycles
of 95°C for 5 sec and 60°C for 34 sec. The relative expression
levels of miR-21 and KLF5 mRNA were analyzed with GAPDH and U6 as
internal references, respectively, using a TaqMan microRNA assay
(Ambion; Thermo Fisher Scientific, Inc). Analysis of relative gene
expression data was performed using the real-time quantitative PCR
and the 2−ΔΔCq method. A total of three repeats were
performed. The primer sequences were: U6 forward,
5′-CTCGCTTCGGCAGCACATATACT-3′; U6 reverse,
5′-ACGCTTCACGAATTTGCGTGTC-3′; GAPDH forward,
5′-TCAACGACCACTTTGTCAAGCTCA-3′; GAPDH reverse,
5′-GCTGGTGGTCCAGGGGTCTTACT-3′; miR-21 forward,
5′-TGCGCTAGCTTATCAGACTGAT-3′; miR-21 reverse,
5′-CCAGTGCAGGGTCCGAGGTATT-3′; KLF5 forward,
5′-AGCTACAATACGCTTGGCCT-3′; and KLF5 reverse,
5′-ATGTGTGTTACGCACGGTCT-3′.
Western blot analysis
Protein was extracted from the cells and tissues
using RIPA lysis buffer [1% NP40, 0.1% sodium dodecyl sulfate
(SDS), 100 µg/ml phenylmethylsulfonyl fluoride and 0.5% sodium
deoxycholate in PBS] on ice. The supernatants were collected
following centrifugation at 12,000 x g at 4°C for 20 min.
The bicinchoninic acid method (Thermo Fisher
scientific, Inc.) was used to determine the protein concentrations
in extracts from cultured cells or resected tumors. Subsequently,
10 µg of protein samples were separated by 10% SDS-PAGE and
transferred to PVDF membranes (EMD Millipore). Following blocking
with 5% skimmed milk at room temperature for 2 h, the membranes
were incubated with primary antibodies against KLF5 (1:1,000; cat.
no. ab137676; Abcam), the proliferation marker protein Ki67
(1:1,000; cat. no. ab15580; Abcam) and GAPDH (1:3,000; cat. no.
ab9485; Abcam) overnight at 4°C. The membranes were then incubated
with secondary antibody, goat anti-rabbit IgG H&L (horseradish
peroxidase-conjugated; 1:5,000; cat. no. ab205718; Abcam).
Antibodies bound to the target proteins were visualized using the
Enhanced Chemiluminescence Western Blotting Detection reagent
(Santa Cruz Biotechnology, Inc.). The protein bands were quantified
using the LAS3000 imaging system (Fujifilm Corporation).
Cell transfection
miR-21 mimics (mimic), miR-21 inhibitor (inhibitor)
and scramble negative control (s-MiR) were synthesized by GE
Healthcare Dharmacon, Inc. To overexpress or knockdown miR-21,
SKM-1 and HL-60 cells were transfected with miR-21 mimics,
inhibitor or negative control (all 50 nM) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Their sequences were as follows: miR-21 mimic
sense, 5′-UAGCUUAUCAGACUGAUGUUGA-3′; miR-21 mimic antisense,
5′-AACAUCAGUCUGAUAAGCUAUU-3′; s-MiR sense,
5′-UUCUCCGAACGUGUCACGUTT-3′; s-MiR antisense,
5′-ACGUGACACGUUCGGAGAATT-3′; and miR-21 inhibitor,
5′-UCAACAUCAGUCUGAUAAGCUA-3′. Subsequent experiments were conducted
following 24 h post-transfection.
Cell proliferation assays
HL-60 and SKM-1 cells transfected with miR-21 mimic,
inhibitor and negative control were added to 96-well plates
(5×103 cells/ml) at 0–5 days post-transfection and
incubated with the CCK-8 reagent for 4 h at 37°C. Cell
proliferation was determined using the Cell Counting Kit-8 assay
(Beyotime Institute of Biotechnology) according to the
manufacturer's instructions. The absorbance was recorded at 450 nm
using an ELISA plate reader (BioTek Instruments, Inc.).
Luciferase reporter assay
The potential targets of miR-21 were predicted using
TargetScan 7.2 (http://www.targetscan.org/vert_72/) and miRBase
(http://www.Microrna.org/microrna/home.do). Using the
QuikChange lightning site-directed mutagenesis kit (Stratagene;
Agilent Technologies, Inc.), the present study constructed a
version of the 3′-UTR sequence of the KLF5 gene that had the
potential miR-21 binding sites mutated (MT). The wild-type (WT) or
MT KLF5 3′-UTR fragments were then inserted downstream of the
firefly luciferase gene in the pGL3 vector (Promega Corporation).
The reporter plasmids were co-transfected with miR-21 mimic into
AML cells for 36 h in 96-well plates using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. Dual
luciferase assays (Promega Corporation) were performed to analyze
luciferase activity, normalized to Renilla luciferase activity,
using a GloMax fluorescence reader (Promega Corporation).
Tumour formation in nude mice
Male athymic nude mice (n=20; weights, 12–15 g; age,
4 weeks) were purchased from the Animal Center of the Chinese
Academy of Science and housed in sterile laminar flow cabinets,
maintained with temperatures between 18 and 28°C, and ventilated
air flow. The mice were exposed to a light/dark cycle of 12/12 h
and were given continuous supplies of food, as well as disinfected
and filtered drinking water. Mice (at the age of 6 weeks old) were
randomly divided into two groups and inoculated subcutaneously with
1×106 HL-60 cells (in 100 µl PBS) transfected with s-MiR
or miR-21 mimic. A slide calliper was used to measure the tumour
width and length every 5 days, in order to calculate the tumour
volume. After 35 days, the animals were sacrificed, the tumours
were removed and the tumour volume was calculated using the
following formula: Volume=1/2 x length × width. Western blot and
RT-qPCR analyses were performed to evaluate the KLF5 expression
levels in the resected tumours. All animal care and procedures were
approved by the Guizhou Provincial People's Hospital for Animal
Experiments committee.
Statistical analysis
Statistical analysis was performed using SPSS v18.0
(SPSS Inc.). Student's t-test or one-way analysis of variance,
followed by Tukey's post hoc test, was used to compare the
difference between two or more groups. Results were presented as
mean ± SEM. Experiments were performed in triplicate. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of miR-21 and KLF5 in AML
cells
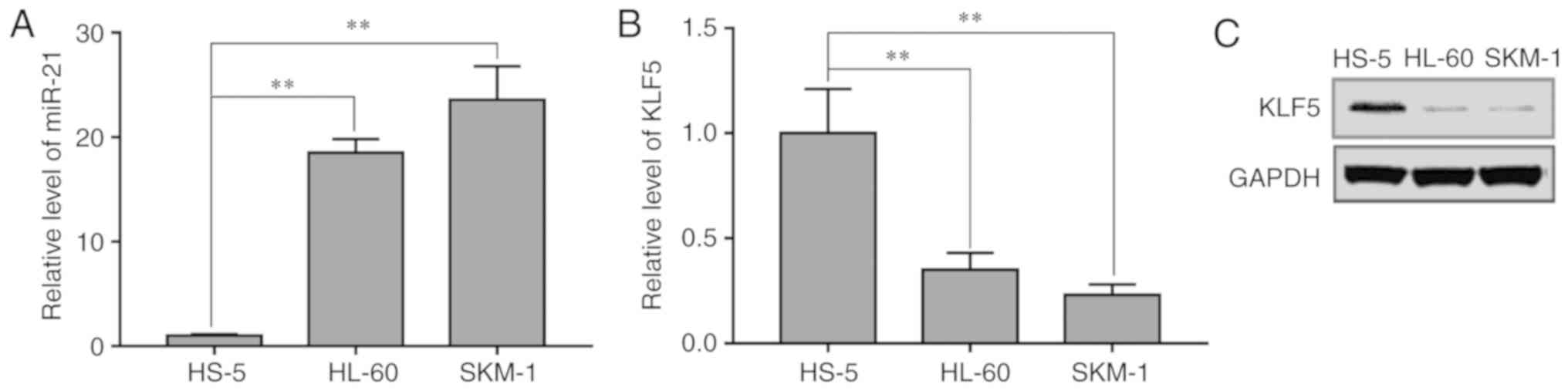
To explore the relative expression levels of miR-21
and KLF5 in SKM-1, HL-60 and HS-5 cells, RT-qPCR analysis was
performed. The results demonstrated that miR-21 expression levels
were increased in the AML SKM-1 and HL-60 cells compared with that
in the normal bone marrow stromal HS-5 cells (Fig. 1A). KLF5 mRNA (Fig. 1B) and protein (Fig. 1C) expression levels were decreased in
HL-60 and SKM-1 cells compared with that in HS-5 cells. These data
suggested that KLF5 and miR-21 may be associated with the
development of AML.
KLF5 is a direct target of miR-21 in
AML cells
To further elucidate the function of miR-21 in AML,
TargetScan 7.2 and miRBase database analysis was performed. The
results demonstrated that the 3′-UTR of KLF5 contained a putative
binding sequence for miR-21 (Fig.
2A). Using a dual luciferase activity assay, it was
demonstrated that miR-21 overexpression by mimic transfection
significantly suppressed luciferase activity in SKM-1 and HL-60
cells transfected with the KLF5-3′-UTR-WT construct (Fig. 2B); by contrast, the KLF5-3′-UTR-MT
construct had no effect (Fig. 2B).
The efficiency of the miR-21 mimic and a miR-21 inhibitor was
confirmed by RT-qPCR (Fig. 2C).
Notably, the KLF5 mRNA expression levels in SKM-1 and HL-60 cells
were significantly decreased following miR-21 overexpression, while
they were significantly enhanced following miR-21 inhibition
(Fig. 2D). These results suggested
that miR-21 could downregulate KLF5 expression by directly
targeting its 3′-UTR.
miR-21 promotes proliferation in AML
cells
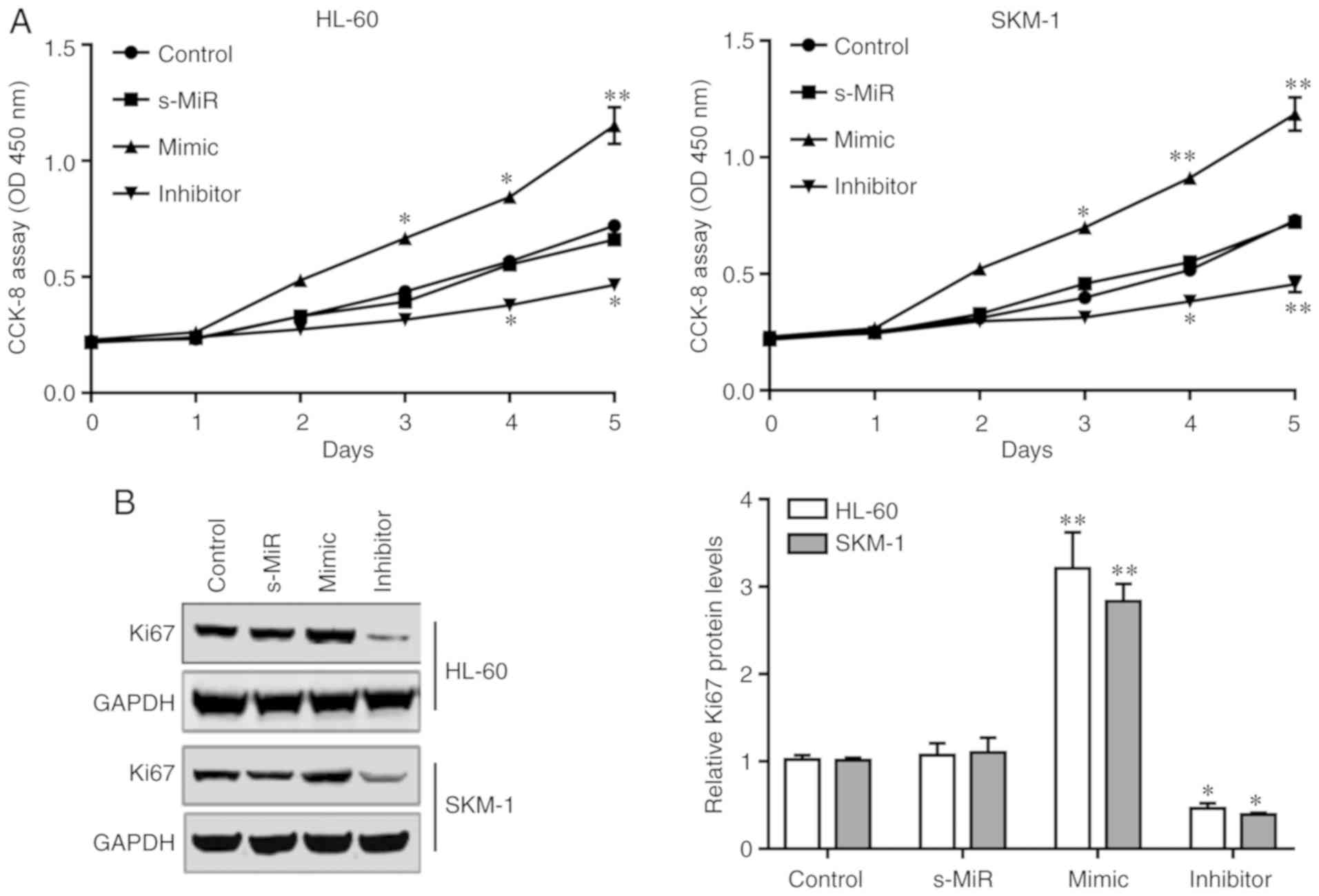
CCK-8 assays were performed to examine the effect of
miR-21 on the proliferation of SKM-1 and HL-60 cells. As presented
in Fig. 3A, transfection with miR-21
mimic significantly increased the total cell numbers of HL-60 and
SKM-1 cells compared with that in the s-MiR group. By contrast, the
total cell numbers were significantly reduced following
transfection with the miR-21 inhibitor. In addition, western blot
analysis demonstrated that the protein expression levels of the
proliferation marker Ki67 were increased following transfection of
the HL-60 and SKM-1 cells with miR-21 mimic (Fig. 3B). By contract, Ki67 protein
expression levels were significantly reduced following transfection
with the miR-21 inhibitor (Fig. 3B).
These results indicated that high levels of endogenous miR-21 may
promote AML cell proliferation by inhibiting KLF5.
miR-21 promotes AML tumor growth in
vivo
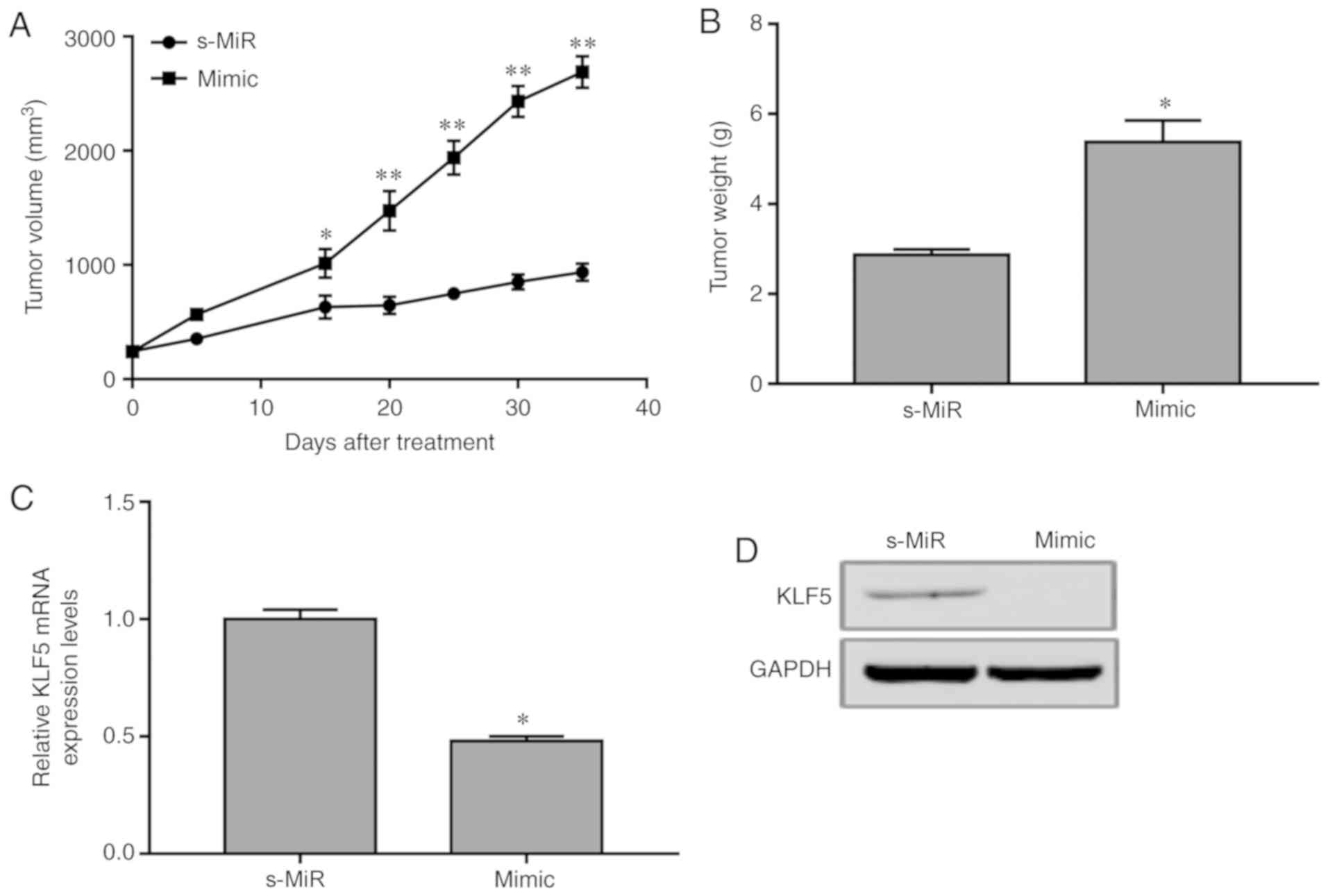
To validate the biological role of miR-21, an AML
mouse model was established by subcutaneously injecting HL-60 cells
transfected with miR-21 mimic or scramble s-MiR control into nude
mice. The tumor volume was determined every 5 days. At 35 days,
mice were sacrificed and the final tumor weights were evaluated. A
significant increase was detected in the xenograft tumors
overexpressing miR-21 compared with those in the s-MiR group
(Fig. 4A). In addition, the weights
of the tumors from animals injected with miR-21-overexpressing
cells were significantly increased compared with those in the s-MiR
group (Fig. 4B). Furthermore, miR-21
overexpression resulted in a decrease in KLF5 protein and mRNA
expression levels in the xenograft tumors (Fig. 4C and D). Consequently, overexpression
of miR-21 downregulated KLF4 expression and promoted AML
tumorigenesis in vivo.
Discussion
Differential expression of miRNAs has been reported
in various studies on patients with AML, including of miR-143,
miR-34a, miR-126, miR-21 and miR-425 (9–11,13,20).
The potential mechanisms underlying the effects of miRNAs in AML
progression require further elucidation. Previous reports
demonstrated that miR-21 is significantly upregulated in AML
(13), which may identify this miRNA
as a potential novel biomarker in the early diagnosis of AML.
It is well established that miRNAs are involved in
tumorigenesis by modulating the expression of their target genes
(7). Preceding studies revealed that
miR-21 functions as a tumor suppressor or an oncogene in gastric
carcinoma (21), colorectal
carcinoma (22), hepatocellular
carcinoma (23) and acute myeloid
leukemias (13). In the present
study, it was demonstrated that miR-21 was significantly
upregulated and KLF5 was significantly downregulated in AML cells,
compared with normal bone marrow cells. TargetScan 7.2 and miRBase
databases predicted that KLF5 may be a target of miR-21. Subsequent
dual luciferase reporter analysis confirmed that KLF5 was a direct
target of miR-21. Furthermore, miR-21 overexpression resulted in
KLF5 downregulation, while miR-21 inhibition resulted in KLF5
upregulation in AML cells in vitro. Finally, miR-21
overexpression promoted AML cell proliferation in vitro and
enhanced AML tumor growth in vivo.
In conclusion, the present results provided
experimental evidence that miR-21 promoted AML tumorigenesis by
promoting cell proliferation and by directly regulating KLF5
expression in vitro and in vivo. Thus, miR-21 may
serve as a potential diagnostic marker and therapeutic target in
AML.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in the published article.
Authors' contributions
ChaL and LW were involved in the study concept and
design, and analysis and interpretation of data. HY, TC and GL were
involved in acquisition of data, analysis and interpretation of
data, statistical analysis, and drafting of the manuscript. ChoL,
JY and ZD performed bioinformatics analysis. SY, LJW and YH
performed the luciferase reporter assay. AL and BY performed the
cell proliferation assay and western blotting. JM and LJW revised
the manuscript, and performed analysis and interpretation of
data.
Ethics approval and consent to
participate
All animal care and procedures were approved by
Guizhou Provincial People's Hospital for Animal Experiments
committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Graubert TA and Mardis ER: Genomics of
acute myeloid leukemia. Cancer J. 17:487–491. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bosshard R, O'Reilly K, Ralston S, Chadda
S and Cork D: Systematic reviews of economic burden and
health-related quality of life in patients with acute myeloid
leukemia. Cancer Treat. Rev. 69:224–232. 2018.
|
|
3
|
Yang X and Wang J: Precision therapy for
acute myeloid leukemia. J Hematol Oncol. 11:32018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mao J, Li S, Zhao H, Zhu Y, Hong M, Zhu H,
Qian S and Li J: Effects of chidamide and its combination with
decitabine on proliferation and apoptosis of leukemia cell lines.
Am J Transl Res. 10:2567–2578. 2018.PubMed/NCBI
|
|
5
|
McGuire A, Brown JA and Kerin MJ:
Metastatic breast cancer: The potential of miRNA for diagnosis and
treatment monitoring. Cancer Metastasis Rev. 34:145–155. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang C, Bai G, Zhu W, Bai D and Bi G:
Identification of miRNA-mRNA network associated with acute myeloid
leukemia survival. Med Sci Monit. 23:4705–4714. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu L, Cai X, Liu E, Tian X and Tian C:
MicroRNA-18a promotes proliferation and metastasis in
hepatocellular carcinoma via targeting KLF4. Oncotarget.
8:68263–68269. 2017.PubMed/NCBI
|
|
8
|
Tian C, Wu H, Li C, Tian X, Sun Y, Liu E,
Liao X and Song W: Downreguation of FoxM1 by miR-214 inhibits
proliferation and migration in hepatocellular carcinoma. Gene Ther.
25:312–319. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hartmann JU, Bräuer-Hartmann D, Kardosova
M, Wilke F, Schödel C, Gerloff D, Katzerke C, Krakowsky R, Namasu
CY, Bill M, et al: MicroRNA-143 targets ERK5 in granulopoiesis and
predicts outcome of patients with acute myeloid leukemia. Cell
Death Dis. 9:8142018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang Y, Zou Y, Lin L, Ma X and Chen H:
Identification of serum miR-34a as a potential biomarker in acute
myeloid leukemia. Cancer Biomark. 22:799–805. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ding Q, Wang Q, Ren Y, Zhu HQ and Huang Z:
MicroRNA-126 attenuates cell apoptosis by targeting TRAF7 in acute
myeloid leukemia cells. Biochem Cell Biol. 25:(Epub ahead of
print). 2018.
|
|
12
|
Badr M, Said H, Louka ML, Elghazaly HA,
Gaballah A and Atef Abd El Mageed M: MicroRNA-21 as a predictor and
prognostic factor for trastuzumab therapy in human epidermal growth
factor receptor 2-positive metastatic breast cancer. J Cell
Biochem. 120:3459–3466. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Riccioni R, Lulli V, Castelli G, Biffoni
M, Tiberio R, Pelosi E, Lo-Coco F and Testa U: miR-21 is
overexpressed in NPM1-mutant acute myeloid leukemias. Leuk Res.
39:221–228. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li Y, Sui X, Hu X and Hu Z: Overexpression
of KLF5 inhibits puromycin-induced apoptosis of podocytes. Mol Med
Rep. 18:3843–3849. 2018.PubMed/NCBI
|
|
15
|
Ma Y, Wang Q, Liu F, Ma X, Wu L, Guo F,
Zhao S, Huang F and Qin G: KLF5 promotes the tumorigenesis and
metastatic potential of thyroid cancer cells through the NF-κB
signaling pathway. Oncol Rep. 40:2608–2618. 2018.PubMed/NCBI
|
|
16
|
Jia L, Zhou Z, Liang H, Wu J, Shi P, Li F,
Wang Z, Wang C, Chen W, Zhang H, et al: KLF5 promotes breast cancer
proliferation, migration and invasion in part by upregulating the
transcription of TNFAIP2. Oncogene. 35:2040–2051. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gong T, Cui L, Wang H, Wang H and Han N:
Knockdown of KLF5 suppresses hypoxia-induced resistance to
cisplatin in NSCLC cells by regulating HIF-1α-dependent glycolysis
through inactivation of the PI3K/Akt/mTOR pathway. J Transl Med.
16:1642018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Du C, Gao Y, Xu S, Jia J, Huang Z, Fan J,
Wang X, He D and Guo P: KLF5 promotes cell migration by
up-regulating FYN in bladder cancer cells. FEBS Lett. 590:408–418.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Diakiw SM, Kok CH, To LB, Lewis ID, Brown
AL and D'Andrea RJ: The granulocyte-associated transcription factor
Krüppel-like factor 5 is silenced by hypermethylation in acute
myeloid leukemia. Leuk Res. 36:110–116. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang C, Shao T, Zhang H, Zhang N, Shi X,
Liu X, Yao Y, Xu L, Zhu S, Cao J, et al: MiR-425 expression
profiling in acute myeloid leukemia might guide the treatment
choice between allogeneic transplantation and chemotherapy. J
Transl Med. 16:2672018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu X, Abraham JM, Cheng Y, Wang Z, Wang
Z, Zhang G, Ashktorab H, Smoot DT, Cole RN, Boronina TN, et al:
Synthetic circular RNA functions as a miR-21 sponge to suppress
gastric carcinoma cell proliferation. Mol Ther Nucleic Acids.
13:312–321. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ding T, Cui P, Zhou Y, Chen C, Zhao J,
Wang H, Guo M, He Z and Xu L: Antisense oligonucleotides against
miR-21 inhibit the growth and metastasis of colorectal carcinoma
via the DUSP8 pathway. Mol Ther Nucleic Acids. 13:244–255. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Z, Yang H and Ren L: MiR-21 promoted
proliferation and migration in hepatocellular carcinoma through
negative regulation of Navigator-3. Biochem Biophys Res Commun.
464:1228–1234. 2015. View Article : Google Scholar : PubMed/NCBI
|