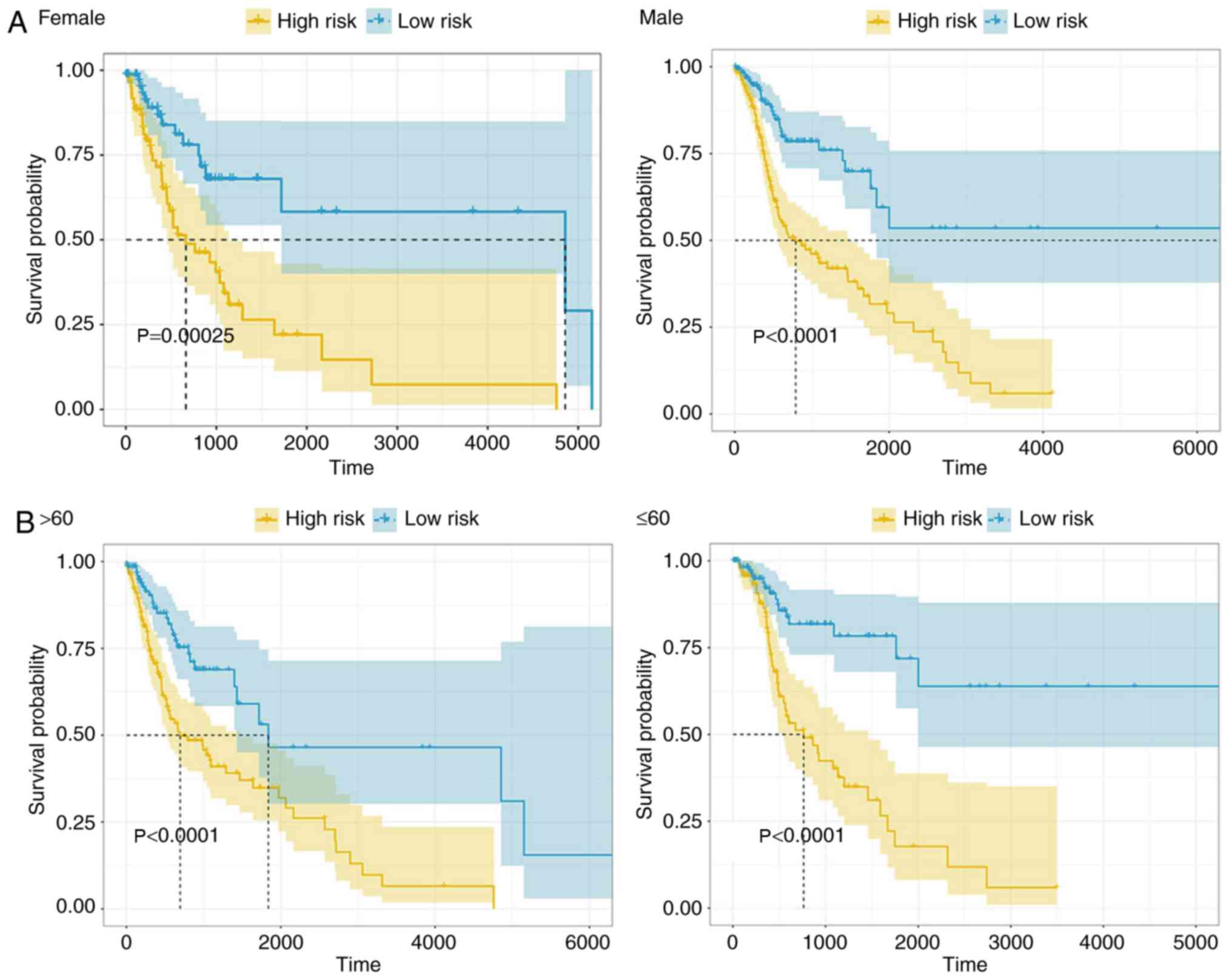
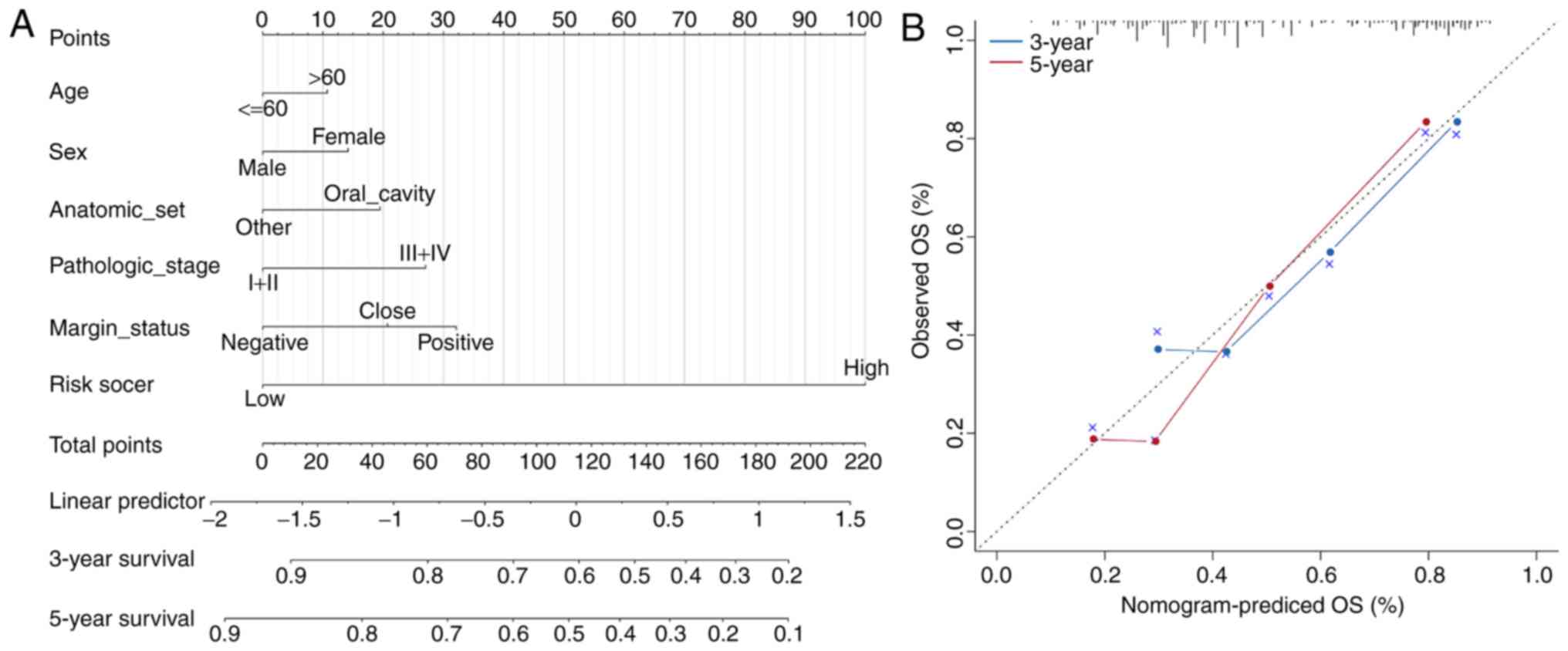
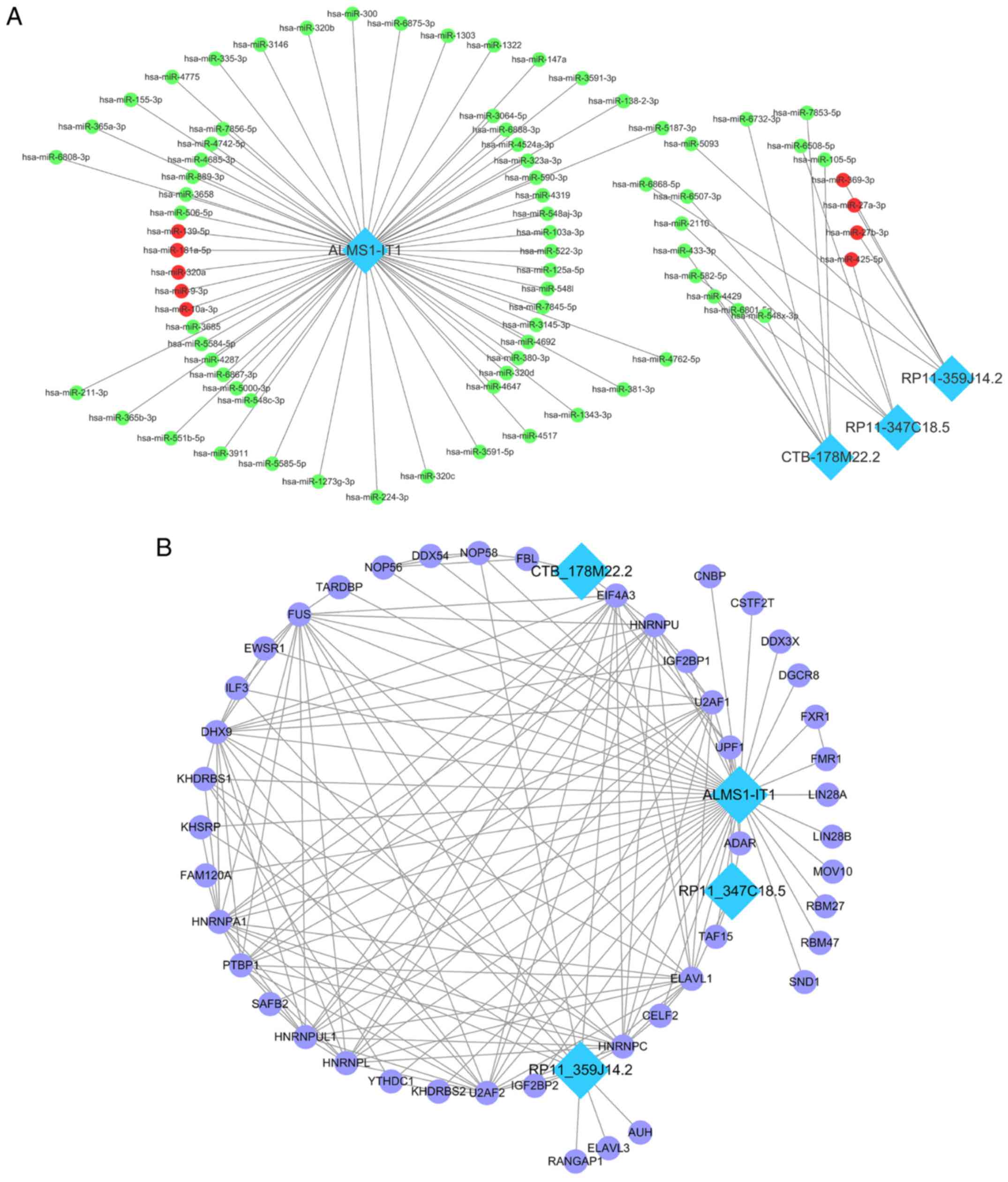
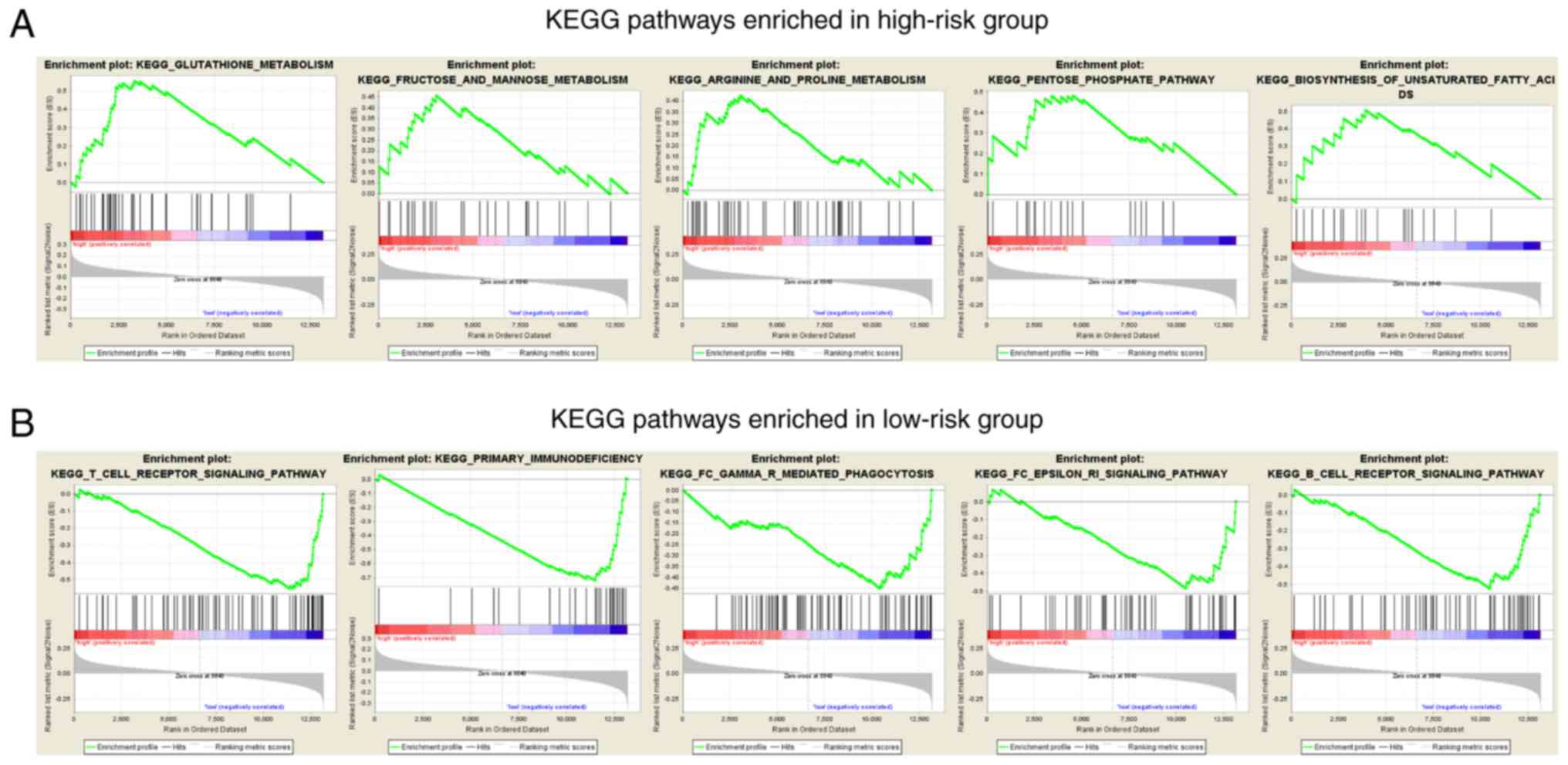
|
1
|
Zhang L, Zhang W, Wang YF, Liu B, Zhang
WF, Zhao YF, Kulkarni AB and Sun ZJ: Dual induction of apoptotic
and autophagic cell death by targeting survivin in head neck
squamous cell carcinoma. Cell Death Dis. 6:e17712015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liotta F, Querci V, Mannelli G,
Santarlasci V, Maggi L, Capone M, Rossi MC, Mazzoni A, Cosmi L,
Romagnani S, et al: Mesenchymal stem cells are enriched in head
neck squamous cell carcinoma, correlates with tumour size and
inhibit T-cell proliferation. Br J Cancer. 112:745–754. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Polanska H, Raudenska M, Gumulec J,
Sztalmachova M, Adam V, Kizek R and Masarik M: Clinical
significance of head and neck squamous cell cancer biomarkers. Oral
Oncol. 50:168–177. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rothenberg SM and Ellisen LW: The
molecular pathogenesis of head and neck squamous cell carcinoma. J
Clin Invest. 122:1951–1957. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Galot R, Le Tourneau C, Guigay J, Licitra
L, Tinhofer I, Kong A, Caballero C, Fortpied C, Bogaerts J,
Govaerts AS, et al: Personalized biomarker-based treatment strategy
for patients with squamous cell carcinoma of the head and neck:
EORTC position and approach. Ann Oncol. 29:2313–2327. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cech TR and Steitz JA: The noncoding RNA
revolution-trashing old rules to forge new ones. Cell. 157:77–94.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lai EC: Micro RNAs are complementary to
3′UTR sequence motifs that mediate negative post-transcriptional
regulation. Nat Genet. 30:363–364. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu F, Xing L and Zhang X and Zhang X: A
Four-Pseudogene classifier identified by machine learning serves as
a novel prognostic marker for survival of osteosarcoma. Genes
(Basel). 10:E4142019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu X, Tian X, Yu C, Shen C, Yan T, Hong
J, Wang Z, Fang JY and Chen H: A long non-coding RNA signature to
improve prognosis prediction of gastric cancer. Mol Cancer.
15:602016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qi P and Du X: The long non-coding RNAs, a
new cancer diagnostic and therapeutic gold mine. Mod Pathol.
26:155–165. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao G, Fu Y, Su Z and Wu R: How Long
Non-Coding RNAs and MicroRNAs mediate the endogenous RNA Network of
head and neck squamous cell carcinoma: A Comprehensive Analysis.
Cell Physiol Biochem. 50:332–341. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mao X, Qin X, Li L, Zhou J, Zhou M, Li X,
Xu Y, Yuan L, Liu QN and Xing H: A 15-long non-coding RNA signature
to improve prognosis prediction of cervical squamous cell
carcinoma. Gynecol Oncol. 149:181–187. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen H, Sun X, Ge W, Qian Y, Bai R and
Zheng S: A seven-gene signature predicts overall survival of
patients with colorectal cancer. Oncotarget. 8:95054–95065.
2017.PubMed/NCBI
|
|
17
|
Luo D, Deng B, Weng M, Luo Z and Nie X: A
prognostic 4-lncRNA expression signature for lung squamous cell
carcinoma. Artif Cells Nanomed Biotechnol. 46:1207–1214. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao X, Sun S, Zeng X and Cui L:
Expression profiles analysis identifies a novel three-mRNA
signature to predict overall survival in oral squamous cell
carcinoma. Am J Cancer Res. 8:450–461. 2018.PubMed/NCBI
|
|
19
|
Suer I, Guzel E, Karatas OF, Creighton CJ,
Ittmann M and Ozen M: MicroRNAs as prognostic markers in prostate
cancer. Prostate. 79:265–271. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang X, Feng H, Li Z, Li D, Liu S, Huang
H and Li M: Application of weighted gene co-expression network
analysis to identify key modules and hub genes in oral squamous
cell carcinoma tumorigenesis. Onco Targets Ther. 11:6001–6021.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xing L, Zhang X and Tong D: Systematic
profile analysis of prognostic alternative messenger RNA splicing
signatures and splicing factors in head and neck squamous cell
carcinoma. DNA Cell Biol. 38:627–638. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shen S, Wang G, Shi Q, Zhang R, Zhao Y,
Wei Y, Chen F and Christiani DC: Seven-CpG-based prognostic
signature coupled with gene expression predicts survival of oral
squamous cell carcinoma. Clin Epigenetics. 9:882017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xing L, Zhang X, Feng H, Liu S, Li D,
Hasegawa T, Guo J and Li M: Silencing FOXO1 attenuates
dexamethasone-induced apoptosis in osteoblastic MC3T3-E1 cells.
Biochem Biophys Res Commun. 513:1019–1026. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Amin MB, Greene FL, Edge SB, Compton CC,
Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR and
Winchester DP: The eighth edition AJCC Cancer Staging Manual:
Continuing to build a bridge from a population-based to a more
‘personalized’ approach to cancer staging. CA Cancer J Clin.
67:93–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Eytan DF, Blackford AL, Eisele DW and
Fakhry C: Prevalence of comorbidities and effect on survival in
survivors of human papillomavirus-related and human
papillomavirus-unrelated head and neck cancer in the United States.
Cancer. 125:249–260. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Coskun HH, Medina JE, Robbins KT, Silver
CE, Strojan P, Teymoortash A, Pellitteri PK, Rodrigo JP, Stoeckli
SJ, Shaha AR, et al: Current philosophy in the surgical management
of neck metastases for head and neck squamous cell carcinoma. Head
Neck. 37:915–926. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Marur S and Forastiere AA: Head and neck
squamous cell carcinoma: Update on epidemiology, diagnosis, and
treatment. Mayo Clin Proc. 91:386–396. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Guo W, Chen X, Zhu L and Wang Q: A
six-mRNA signature model for the prognosis of head and neck
squamous cell carcinoma. Oncotarget. 8:94528–94538. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jamali Z, Asl Aminabadi N, Attaran R,
Pournagiazar F, Ghertasi Oskouei S and Ahmadpour F: MicroRNAs as
prognostic molecular signatures in human head and neck squamous
cell carcinoma: A systematic review and meta-analysis. Oral Oncol.
51:321–331. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Quan J, Pan X, Zhao L, Li Z, Dai K, Yan F,
Liu S, Ma H and Lai Y: LncRNA as a diagnostic and prognostic
biomarker in bladder cancer: A systematic review and meta-analysis.
Onco Targets Ther. 11:6415–6424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Song P, Jiang B, Liu Z, Ding J, Liu S and
Guan W: A three-lncRNA expression signature associated with the
prognosis of gastric cancer patients. Cancer Med. 6:1154–1164.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
You X, Yang S, Sui J, Wu W, Liu T, Xu S,
Cheng Y, Kong X, Liang G and Yao Y: Molecular characterization of
papillary thyroid carcinoma: A potential three-lncRNA prognostic
signature. Cancer Manag Res. 10:4297–4310. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang JX, Song W, Chen ZH, Wei JH, Liao
YJ, Lei J, Hu M, Chen GZ, Liao B, Lu J, et al: Prognostic and
predictive value of a microRNA signature in stage II colon cancer:
A microRNA expression analysis. Lancet Oncol. 14:1295–1306. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang P, Jin M, Sun CH, Li YS, Wang X, Sun
YN, Tian LL and Liu M: A three-lncRNA expression signature predicts
survival in head and neck squamous cell carcinoma (HNSCC). Biosci
Rep. 38:BSR201815282018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu G, Zheng J, Zhuang L, Lv Y, Zhu G, Pi
L, Wang J, Chen C, Li Z, Liu J, et al: A prognostic 5-lncRNA
expression signature for head and neck squamous Cell Carcinoma. Sci
Rep. 8:152502018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang B, Wang H, Guo Z and Zhang X:
Prediction of head and neck squamous cell carcinoma survival based
on the expression of 15 lncRNAs. J Cell Physiol. 234:18781–18791.
2019.PubMed/NCBI
|
|
37
|
Wang Y, Ren F, Chen P, Liu S, Song Z and
Ma X: Identification of a six-gene signature with prognostic value
for patients with endometrial carcinoma. Cancer Med. 7:5632–5642.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang Z, Chen G, Wang Q, Lu W and Xu M:
Identification and validation of a prognostic 9-genes expression
signature for gastric cancer. Oncotarget. 8:73826–73836.
2017.PubMed/NCBI
|
|
39
|
Nohata N, Abba MC and Gutkind JS:
Unraveling the oral cancer lncRNAome: Identification of novel
lncRNAs associated with malignant progression and HPV infection.
Oral Oncol. 59:58–66. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pavlova NN and Thompson CB: The emerging
hallmarks of cancer metabolism. Cell Metab. 23:27–47. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
DeBerardinis RJ and Chandel NS:
Fundamentals of cancer metabolism. Sci Adv. 2:e16002002016.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Vazquez A, Kamphorst JJ, Markert EK, Schug
ZT, Tardito S and Gottlieb E: Cancer metabolism at a glance. J Cell
Sci. 129:3367–3373. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Woo SR, Corrales L and Gajewski TF: Innate
immune recognition of cancer. Annu Rev Immunol. 33:445–474. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Guglas K, Bogaczynska M, Kolenda T, Ryś M,
Teresiak A, Bliźniak R, Łasińska I, Mackiewicz J and Lamperska K:
lncRNA in HNSCC: Challenges and potential. Contemp Oncol (Pozn).
21:259–266. 2017.PubMed/NCBI
|