Introduction
Liver cancer is one of the most frequently diagnosed
human malignancies and is also one of the leading causes of
cancer-associated mortality worldwide (1). As risk factor screening programs have
become more popular, liver cancer incidence rates have declined
over the past decade in a number of Asian countries (2). However, the incidence of liver in the
majority of developed countries is increasing (2). In spite of efforts made on the
treatment of liver cancer (3),
survival of patients with liver cancer remains poor (4,5), largely
due to the high prevalence of cancer metastasis by the time of
diagnosis before radical treatment (6).
Cancer cells are characterized by accelerated energy
metabolism compared with normal cells (7). In effect, inhibition of energy
metabolism is considered to be a promising target for cancer
treatment (8). Glucose transporter 1
(GLUT1) is a solute carrier that facilitates the transport of
glucose across mammalian cell plasma membrane (9). A growing body of literatures has
indicated that GLUT1 also contributes to the development of
different types of cancer, partially through the interactions with
long non-coding RNAs (lncRNAs) (10,11),
which are a subgroup of non-coding RNAs with critical roles in
cancer biology (12). It is has been
previously reported that long intergenic non-protein coding RNA
1638 (LINC01638) promotes breast cancer (13), while its roles in other types of
cancers remain unclear. The present study indicated that LINC01638
lncRNA promoted cancer cell proliferation in hepatocellular
carcinoma (HCC), a major type of liver cancer, potentially by
increasing cancer cell glucose uptake and promoting GLUT1
expression.
Materials and methods
Patients, human tissues and HCC cell
lines
A total of 74 patients with HCC admitted to and
treated at the Sixth People's Hospital of Qingdao (Qindao, China)
between May 2016 and 2018 were selected as research subjects.
Inclusion criteria were as follows: HCC diagnosed by pathological
examinations; patients with normal function of other major organs;
patients with a complete medical record and patients, who had
understood the experimental protocol. Exclusion criteria were as
follows: Patients combined with other diseases, including chronic
diseases other than liver diseases; patients who received treatment
within 3 months prior to admission. Liver biopsy was performed on
all patients, and tumor tissues as well as adjacent healthy
tissues, were collected. Tissues were stored in liquid nitrogen
prior to use. The 74 patients included 40 males and 34 females, and
age ranged between 36 and 66 years (mean age, 49.1±5.6 years).
There were 8 cases of stage I, 18 cases of stage II, 30 cases of
stage III and 18 cases of stage IV according to the American Joint
Committee on Cancer staging system (14). The 74 patients included 29
HBV-positive cases, 17 HCV-positive cases, 12 both-positive cases
and 16 both-negative cases. This study was approved by the ethics
committee of Sixth People's Hospital of Qingdao (Qingdao, China).
Written informed consent was obtained from all patients.
The present study included two HCC cell lines,
SNU-398 and SNU-182. The cell lines and RPMI-1640 medium were
purchased from American Type Culture Collection (ATCC). Cells were
cultured in RPMI-1640 medium supplemented with 10% heat-inactivated
FBS (ATCC) in an incubator at 37°C with 5% CO2.
Reverse transcription-quantitative PCR
(RT-qPCR)
To determine the expression of LINC01638 lncRNA and
GLUT1, total RNA was extracted using TRIzol reagent (Thermo Fisher
Scientific, Inc.), RT was performed using Applied Biosystems™
High-Capacity cDNA Reverse Transcription kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.) and the reverse transcription
protocol was 20 min at 55°C and 10 min at 80°C, PCR reaction
systems were prepared using a SYBR® Green Quantitative
RT-qPCR kit (Sigma-Aldrich; Merck KGaA). Primers of LINC01638
lncRNA, GLUT1 and β-actin endogenous control were designed and
synthesized by Sangon Biotech Co., Ltd. Primer sequences were as
follows: 5′-CCTCTGGAATACATCAGCAC-3′ (forward) and
5′-GGTGGAGACTGTAGTGAGCC-3′ (reverse) for LINC01638;
5′-AGGTGATCGAGGAGTTCTAC-3′ (forward) and 5′-TCAAAGGACTTGCCCAGTTT-3′
(reverse) for GLUT1; 5′-GAGACCTTCAACACCCCAGCC-3′ (forward) and
5′-AATGTCACGCACGATTTCCC-3′ (reverse) for β-actin. All PCR reactions
were performed on an S1000™ Thermal Cycler (Bio-Rad Laboratories
Inc.). The thermocycling conditions were: 95°C for 50 sec, followed
by 40 cycles of 95°C for 10 sec and 55°C for 40 sec. Expression of
LINC01638 lncRNA and GLUT1 was normalized to β-actin endogenous
control using 2−ΔΔCq method (15). Similar expression results were
obtained using 18S rRNA as the endogenous control (data not
shown).
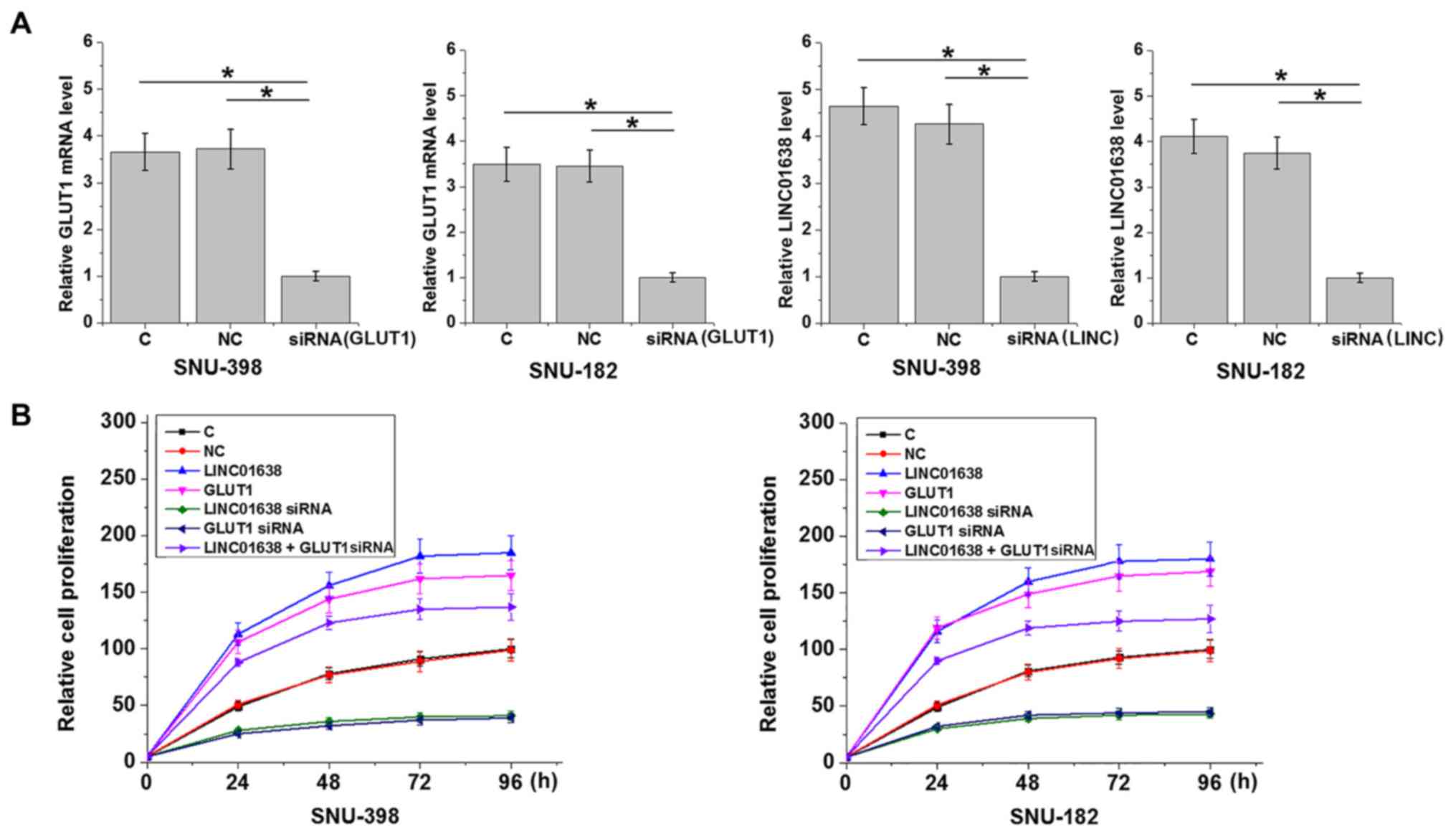
Cell transfection
LINC01638 lncRNA and GLUT1 expressing vectors
(pcDNA3.1) and empty vectors were designed and constructed by
Sangon Biotech Co., Ltd. GLUT1 small interfering RNA (siRNA;
5′-CCAAGAGTGTGCTAAAGAATT-3′), LINC01638 siRNA
(5′-CATACATACAACTCCAAAAAGT-3′), as well as negative control siRNA
(5′-ACAATGAGTCGTAGCATGG-3′) were designed and synthesized by
Shanghai GenePharma Co., Ltd. SNU-398 and SNU-182 cell lines were
cultured overnight to reach 70–80% confluency and all cell
transfections were performed using Lipofectamine® 2000
(cat. no. 11668-019; Invitrogen; Thermo Fisher Scientific Inc.).
All operations were performed in strict accordance with the
manufacturer's protocols. Doses of vectors and siRNAs were 10 and
35 nM, respectively. Cells only treated with
Lipofectamine® 2000 were the control cells. Empty vector
or negative control siRNA transfection was the negative control.
Transfection efficacy was determined by RT-qPCR. In cases of
co-transfection, expression vector (10 nM) and siRNA (35 nM) were
mixed with cells at the same time.
Glucose uptake assay
LINC01638 lncRNA and GLUT1 expression was detected
by RT-qPCR at 24 h after transfection. Cells were harvested for
glucose uptake assay only when GLUT1/LINC01638 overexpression was
>200% and knockdown had reached 50%. Cells of both SNU-398 and
SNU-182 cell lines were harvested and washed with PBS buffer. A
total of 1×106 cells were subsequently dissolved in
Krebs-Ringer-HEPES (KRH) buffer (25 mM HEPES pH 7.4, 1.3 mM
KH2PO4, 120 mM NaCl, 1.2 mM MgSO4,
1.3 mM CaCl2 and 5 mM KCl). A total of 1 µCi of
(3H)-2-deoxyglucose was added and cells were incubated at 37°C for
30 min to initiate glucose uptake. Cells were washed with ice-cold
KRH buffer to stop glucose uptake. Finally, cells were incubated
with lysis buffer (10 mM Tris-HCl pH 8.0, 0.2% SDS) to induce cell
lysis, and radioactivity was measured using liquid scintillation
spectrometry (LS6500 Liquid Scintillation Counter; Beckman Coulter,
Inc.). Disintegrations per minute was used to represent glucose
uptake.
Cell proliferation assay
LINC01638 lncRNA and GLUT1 expression was detected
by RT-qPCR at 24 h after transfection. Cells were harvested for
cell proliferation analysis only in cases of LINC01638 lncRNA, and
when GLUT1 overexpression rate was >200% and knockdown rate had
reached 50%. In brief, cells were harvested, and single cell
suspensions were prepared. Cell density was adjusted to
4×104 cells/ml. Cell suspensions were then transferred
to a 96-well plate, 0.1 ml per well. Cells were cultivated in an
incubator at 37°C with 5% CO2, followed by the addition
of Cell Counting Kit-8 solution (10 µl; Sigma-Aldrich; Merck KGaA)
24, 48, 72 and 96 h later. Cells were subsequently cultivated for
an additional 4 h, and optical density values were measured at a
wavelength of 450 nm.
Western blot analysis
LINC01638 lncRNA and GLUT1 expression was detected
by RT-qPCR at 24 h following transfection. Cells were harvested for
western blot analysis only when GLUT1/LINC01638 overexpression was
>200% and knockdown rate had reached 50%. To detect the
expression of GLUT1, total protein was extracted using RIPA buffer
(Thermo Fisher Scientific, Inc.). Protein concentrations were
measured using a bicinchoninic acid assay (Thermo Fisher
Scientific, Inc.). Following denaturation, protein samples (30 µg
per well) were separated by SDS-PAGE on 12% gels, followed by gel
transfer to polyvinylidene difluoride membranes. Membranes were
blocked in 5% non-fat milk for 2 h at room temperature, followed by
incubation with primary antibodies at 4°C for 18 h: Rabbit
anti-human GLUT1 (dilution, 1:1,400; cat. no. ab32551; Abcam) and
GAPDH (dilution, 1:1,400; cat. no. ab8245; Abcam). Cells were
further incubated with goat anti-rabbit horseradish peroxidase (IgG
H&L; cat. no. ab6721; Abcam) for 2 h at 24°C. Enhanced
chemiluminescence detection reagent (Sigma-Aldrich; Merck KGaA) was
used to develop signals. Signals were normalized using ImageJ 1.8.0
software (National Institutes of Health).
Statistical analysis
All experiments in this study were performed in
triplicate (biological replicates) and mean ± standard deviation
was calculated. All statistical analysis was performed using SPSS
19.0 software (IBM Corp., Armonk, NY, USA). Correlations between
levels of LINC01638 lncRNA and GLUT1 in tumor tissues and adjacent
healthy tissues were performed by Pearson's correlation
coefficient. Comparisons of levels of LINC01638 lncRNA and GLUT1
between tumor tissue and adjacent healthy tissues were performed by
paired t-test. Comparisons among three groups were performed by
one-way ANOVA followed by Tukey's post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
LINC01638 lncRNA and GLUT1 mRNA are
upregulated in tumor tissues compared with adjacent healthy tissues
of patients with HCC
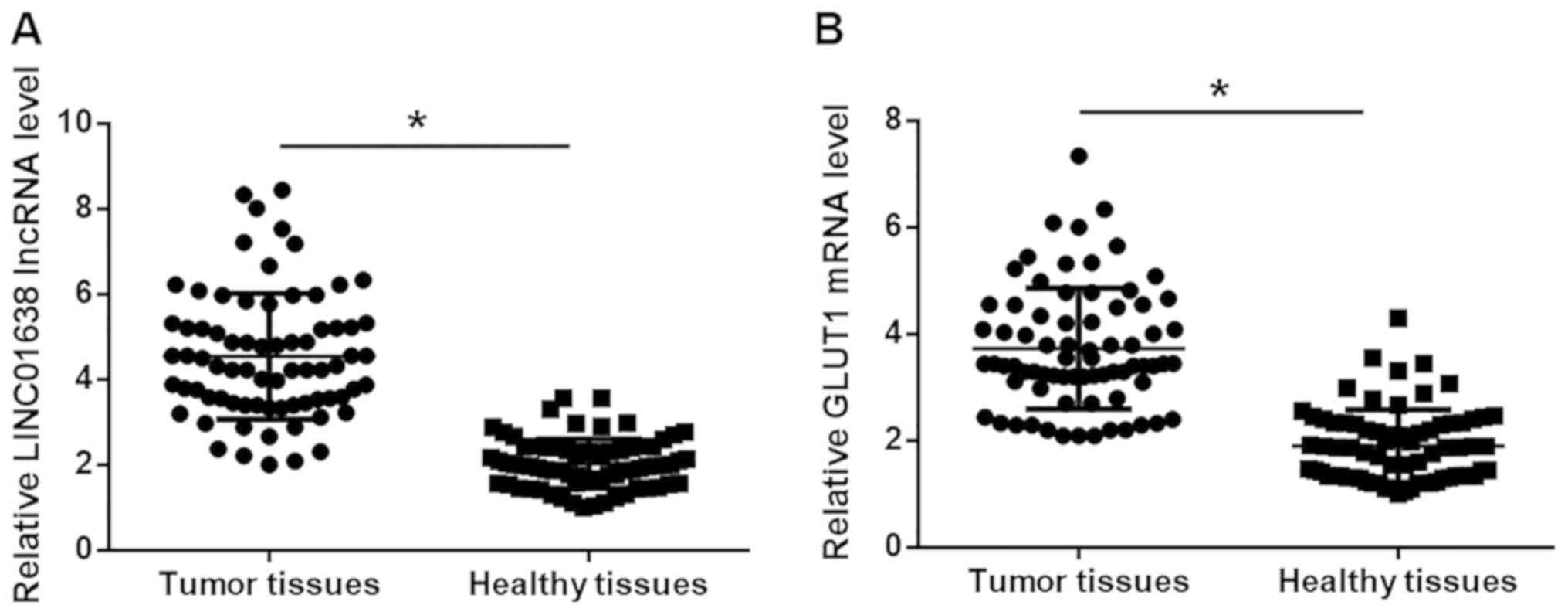
Expression levels of LINC01638 lncRNA and GLUT1 mRNA
in tumor tissues and adjacent healthy tissues of 74 HCC patients
were detected by RT-qPCR. Compared with adjacent healthy tissues,
expression levels of LINC01638 lncRNA (Fig. 1A) and GLUT1 mRNA (Fig. 1B) were significantly higher in tumor
tissues (P<0.05).
Expression levels of LINC01638 lncRNA
and GLUT1 are positively correlated in tumor tissues
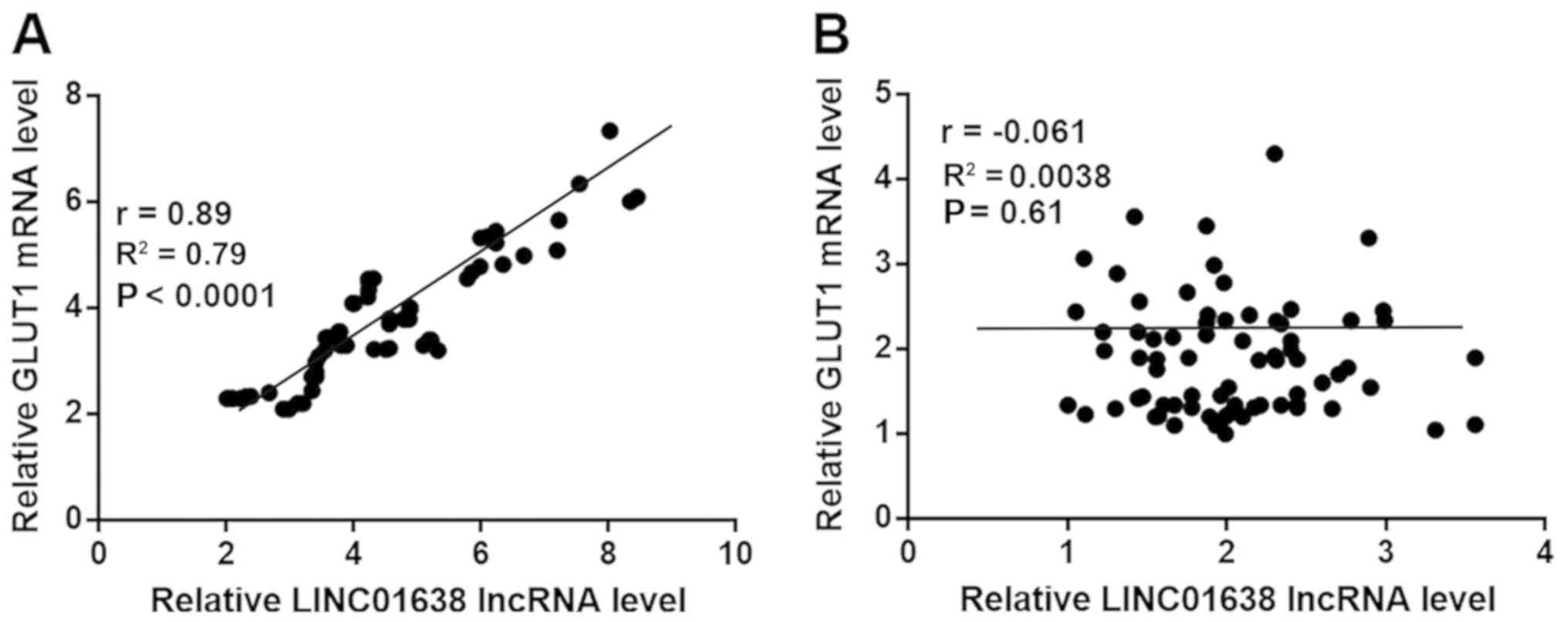
Correlation between expression levels of LINC01638
lncRNA and GLUT1 in tumor tissues and adjacent healthy tissues were
performed by Pearson's correlation coefficient. The expression
levels of LINC01638 lncRNA and GLUT1 were significantly and
positively correlated in tumor tissues (Fig. 2A); however, there was no significant
correlation between expression levels of LINC01638 lncRNA and GLUT1
in adjacent healthy tissues (Fig.
2B).
LINC01638 lncRNA and GLUT1 promotes
glucose uptake in HCC cells
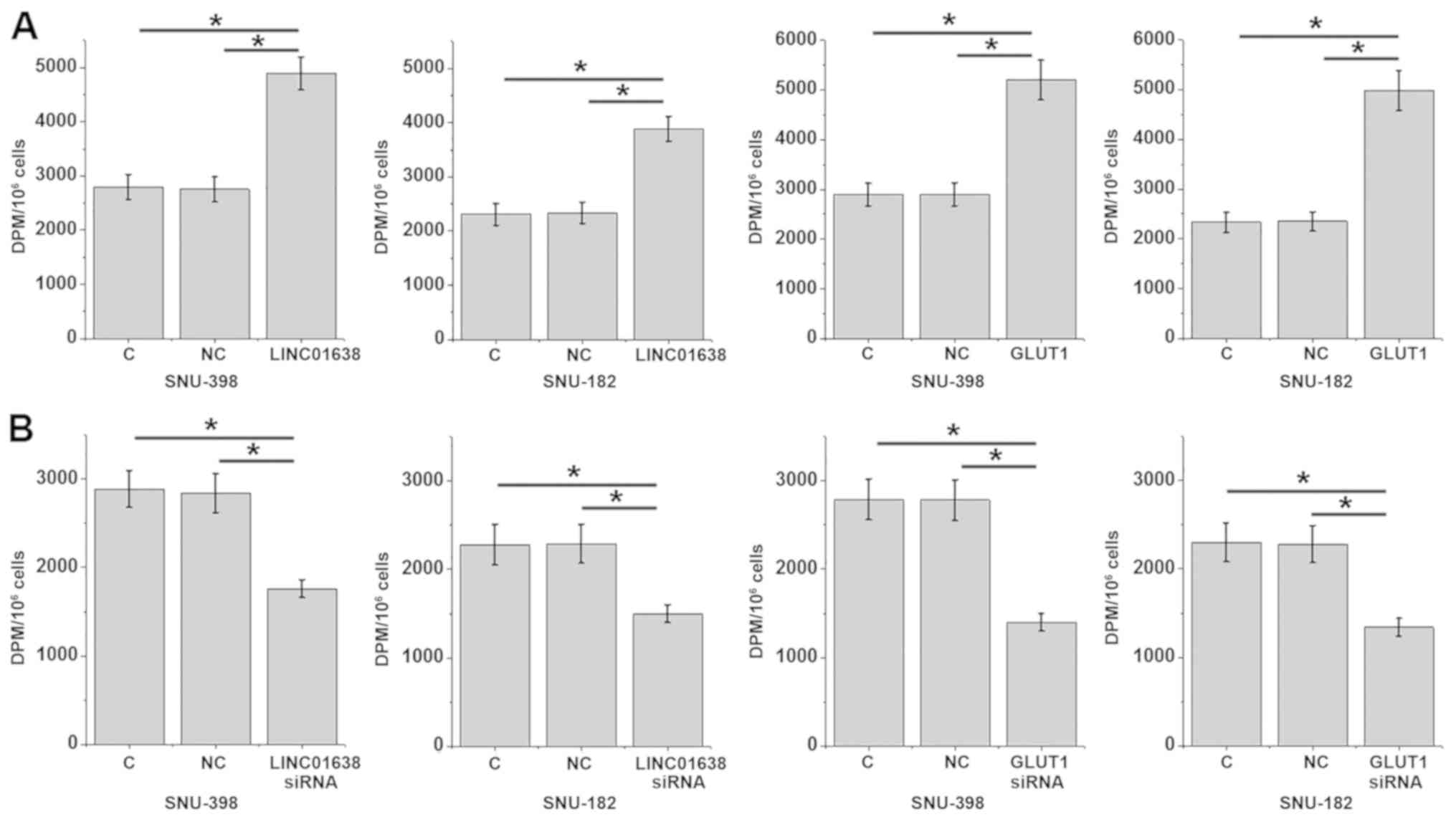
Glucose uptake assay indicated that, compared with
the control and the negative control groups, overexpression of
LINC01638 lncRNA and GLUT1 led to a significant increase in glucose
uptake (Fig. 3A), while LINC01638
lncRNA and GLUT1-knockdown led to a significant inhibition in
glucose uptake (Fig. 3B) in SNU-398
and SNU-182 cells (P<0.05).
LINC01638 lncRNA is a potential
activator of GLUT1 in HCC cells
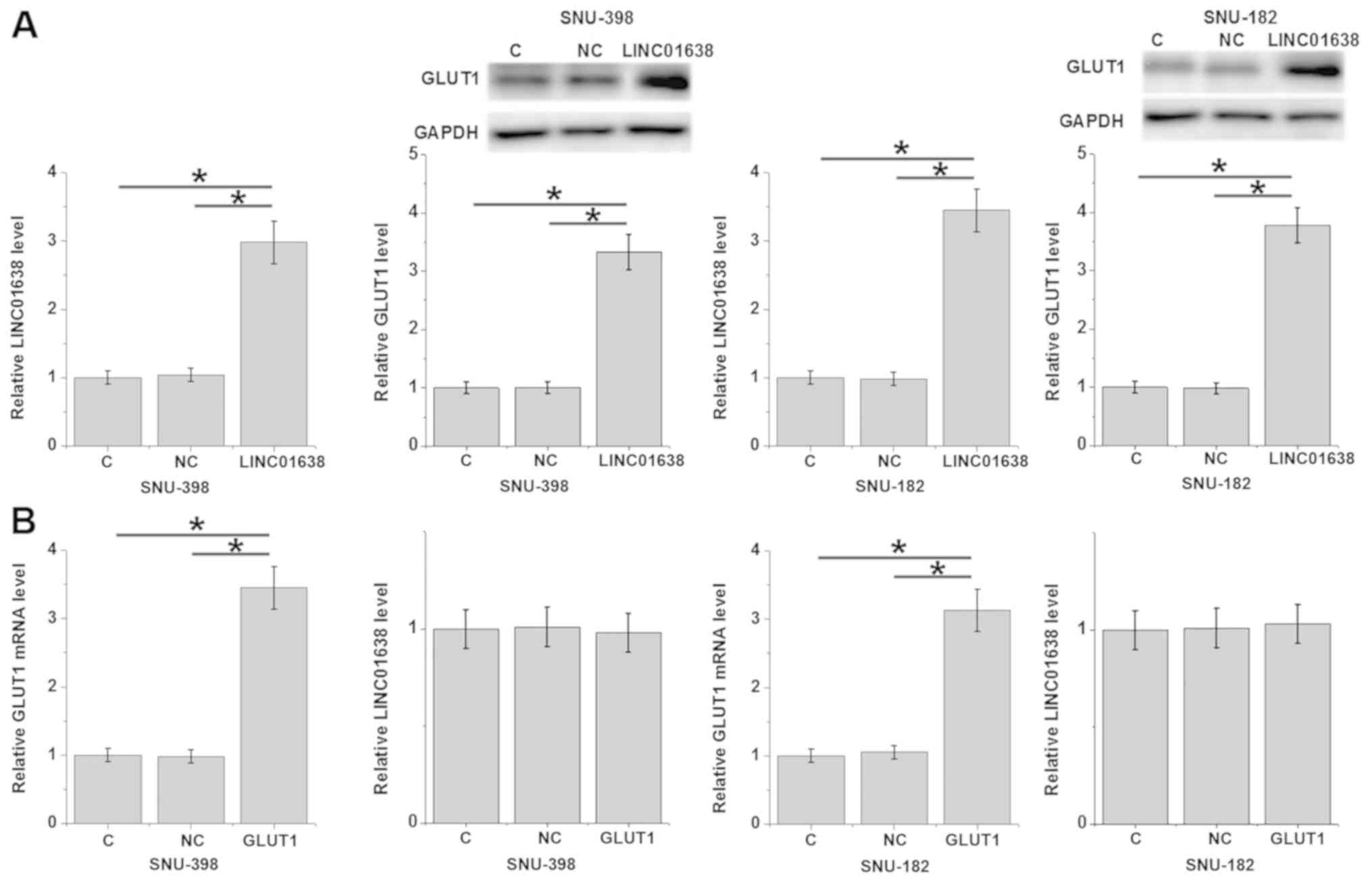
Compared with the control and the negative control
groups, overexpression of LINC01638 lncRNA led to significantly
upregulated GLUT1 expression in SNU-398 and SNU-182 cells (Fig. 4A; P<0.05). By contrast, GLUT1
overexpression did not significantly affect LINC01638 lncRNA
expression in SNU-398 and SNU-182 cells (Fig. 4B).
LINC01638 lncRNA promotes the
proliferation of HCC cells through GLUT1
Compared with the control and the negative control
groups, overexpression of LINC01638 lncRNA and GLUT1 led to
increased proliferation, while LINC01638 lncRNA and GLUT1 siRNA
silencing led to inhibited proliferation of HCC cells (Fig. 5). In addition, co-transfection
experiments revealed that GLUT1 siRNA silencing attenuated the
enhancing effects of LINC01638 lncRNA overexpression on cancer cell
proliferation (Fig. 5).
Discussion
LINC01638 is a recently identified lncRNA with a
function only characterized in breast cancer (13). To the best of our knowledge, the
involvement of LINC01638 lncRNA in other types of cancer remains
unknown. In the present study, LINC01638 lncRNA was upregulated in
HCC, and the upregulation of LINC01638 lncRNA promoted glucose
uptake in cancer cells and accelerated cancer cell proliferation.
It was also indicated that the actions of LINC01638 lncRNA in HCC
are likely mediated by the upregulation of GLUT1.
GLUT1, as a key player in cancer biology, is usually
upregulated during the development of different types of cancer,
including HCC (16). Consistent with
previous studies, the present study also indicated that GLUT1 was
upregulated in HCC tissues compared with adjacent healthy tissues.
The upregulation of GLUT1 promotes cancer cell glucose uptake and
affects cancer cell behaviors, including proliferation (17). In accordance with the aforementioned,
the present study also demonstrated that GLUT1 was a positive
regulator of glucose uptake in HCC cells and HCC cell
proliferation. These data further confirmed the oncogenic role of
GLUT1 in HCC.
A large set of lncRNAs are differentially expressed
during the development and progression of HCC (18). In the present study the functionality
of LINC01638 lncRNA was characterized, and LINC01638 also
demonstrated to be upregulated in HCC. lncRNAs have key roles in
glucose metabolism (19). In the
present study, LINC01638 lncRNA was shown to promote glucose uptake
in HCC cells, demonstrating the regulatory role of LINC01638 lncRNA
in cancer cell energy metabolism. In addition, this study also
suggested that LINC01638 lncRNA is a positive regulator of HCC cell
proliferation. Therefore, these data suggest that LINC01638 lncRNA
may be an oncogene in HCC.
It has been frequently observed that GLUT1 has a
role in cancer biology through interaction with lncRNAs (20). For example, lncRNAs regulate
GLUT1-mediated glycolysis to mediate tumor metastasis (20). The present study indicated that
LINC01638 lncRNA may be an upstream activator of GLUT1 in HCC
cells. In addition, the upregulation of GLUT1 by LINC01638 lncRNA
may be involved in the regulation of HCC cell proliferation.
However, the data also suggested that the interaction between
LINC01638 lncRNA and GLUT1 is likely indirect, due to the
significant correlation between LINC01638 lncRNA and GLUT1 only in
tumor tissues, without correlation in adjacent healthy tissues.
The present study used two cell lines derived from
patients with HCC with hepatitis B virus (HBV) infection.
Preliminary data indicated that LINC01638 expression was not
affected by HBV and hepatitis C virus (HCV) infections (data not
shown). This suggests that LINC01638 may participate in HCC through
a HBV or HCV-independent pathway. Therefore, these two cell lines
are appropriate for this study. In conclusion, LINC01638 lncRNA may
be an oncogenic lncRNA in HCC. LINC01638 lncRNA may promote HCC by
upregulating glucose uptake and promoting GLUT1 expression.
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the author on reasonable request.
Authors' contributions
HW designed the experiments. XC and LW performed the
experiments and collected the data. HW drafted the manuscript. All
authors approved the manuscript.
Ethics approval and consent to
participate
The present study was approved by The Ethics
Committee of Sixth People's Hospital of Qingdao (Qingdao,
China).
Patient consent for publication
Patients provided informed consent for publication
of the present study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang Y, Ren JS, Shi JF, Li N, Wang YT, Qu
C, Zhang Y and Dai M: International trends in primary liver cancer
incidence from 1973 to 2007. BMC Cancer. 15:942015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bruix J, Han KH, Gores G, Llovet JM and
Mazzaferro V: Liver cancer: Approaching a personalized care. J
Hepatol 62 (1 Suppl). S144–S156. 2015. View Article : Google Scholar
|
|
4
|
Momin BR, Pinheiro PS, Carreira H, Li C
and Weir HK: Liver cancer survival in the United States by race and
stage (2001–2009): Findings from the CONCORD-2 study. Cancer. 123
(Suppl 24):S5059–S5078. 2017. View Article : Google Scholar
|
|
5
|
Zeng H, Zheng R, Guo Y, Zhang S, Zou X,
Wang N, Zhang L, Tang J, Chen J, Wei K, et al: Cancer survival in
China, 2003–2005: A population-based study. Int J Cancer.
136:1921–1930. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee MH, Kim EJ, Lee H, Kim HM, Chang MJ,
Park SY, Hong KS, Kim JS and Sessler JL: Liposomal texaphyrin
theranostics for metastatic liver cancer. J Am Chem Soc.
138:16380–16387. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Moreno-Sánchez R, Marín-Hernández A,
Saavedra E, Pardo JP, Ralph SJ and Rodríguez-Enríquez S: Who
controls the ATP supply in cancer cells? Biochemistry lessons to
understand cancer energy metabolism. Int J Biochem Cell Biol.
50:10–23. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pattni BS, Jhaveri A, Dutta I, Baleja JD,
Degterev A and Torchilin V: Targeting energy metabolism of cancer
cells: Combined administration of NCL-240 and 2-DG. Int J Pharm.
532:149–156. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Olson AL and Pessin JE: Structure,
function, and regulation of the mammalian facilitative glucose
transporter gene family. Annu Rev Nutr. 16:235–256. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gwak HR, Haegeman G, Tsang BK and Song YS:
Cancer-specific interruption of glucose metabolism by resveratrol
is mediated through inhibition of Akt/GLUT1 axis in ovarian cancer
cells. Mol Carcinog. 54:1529–1540. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu X and Gan B: lncRNA NBR2 modulates
cancer cell sensitivity to phenformin through GLUT1. Cell Cycle.
15:3471–3481. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Luo L, Tang H, Ling L, Li N, Jia X, Zhang
Z, Wang X, Shi L, Yin J, Qiu N, et al: LINC01638 lncRNA activates
MTDH-Twist1 signaling by preventing SPOP-mediated c-Myc degradation
in triple-negative breast cancer. Oncogene. 37:6166–6179. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chun YS, Pawlik TM and Vauthey JN: 8th
edition of the AJCC cancer staging manual: Pancreas and
hepatobiliary cancers. Ann Surg Oncol. 25:845–847. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Amann T, Maegdefrau U, Hartmann A, Agaimy
A, Marienhagen J, Weiss TS, Stoeltzing O, Warnecke C, Schölmerich
J, Oefner PJ, et al: GLUT1 expression is increased in
hepatocellular carcinoma and promotes tumorigenesis. Am J Pathol.
174:1544–1552. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sawayama H, Iwatsuki M, Yoshida N, Baba Y,
Sakamoto Y, Kinoshita K, Nakamura K, Kuroda D, Tsugio E, Toihata T,
et al: Glucose transporter 1 is associated with proliferation and
prognosis on esophageal squamous cell carcinoma. Cancer Res.
77:44072017.
|
|
18
|
Zhu J, Liu S, Ye F, Shen Y, Tie Y, Zhu J,
Jin Y, Zheng X, Wu Y and Fu H: The long noncoding RNA expression
profile of hepatocellular carcinoma identified by microarray
analysis. PLoS One. 9:e1017072014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ruan X: Long non-coding RNA central of
glucose homeostasis. J Cell Biochem. 117:1061–1065. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Y, Zhang X, Wang Z, Hu Q, Wu J, Li Y,
Ren X, Wu T, Tao X, Chen X, et al: lncRNA-p23154 promotes the
invasion-metastasis potential of oral squamous cell carcinoma by
regulating Glut1-mediated glycolysis. Cancer Lett. 434:172–183.
2018. View Article : Google Scholar : PubMed/NCBI
|