Introduction
According to Globocan 2018, 403,262 new cases of
renal cell carcinoma (RCC) were diagnosed in 185 countries and
these accounted for 2.2% of 36 types of cancer (1). The number of deaths associated with RCC
was 175,098, accounting for 1.8% of deaths associated with 36 types
of cancer (1). Papillary RCC (pRCC)
accounts for 10–20% of all RCC cases (2). At present, surgery is the preferred
therapeutic option for localized and locally advanced pRCC, and
several agents, including anti-vascular endothelial growth factor
drugs and mTOR inhibitors may be considered when treating advanced
and metastatic pRCC (2). At present,
pathological stage based primarily on clinicopathological data is
still the most important factor influencing the prognosis of renal
carcinoma, but it does not reflect the biological heterogeneity of
the cancer (3). In recent years,
with the development of molecular biological techniques and
bioinformatics, new biomarkers have the potential of becoming
effective and specific prognostic factors for different types of
cancer, including pRCC.
Long non-coding RNAs (lncRNAs) are RNA transcripts
that do not encode proteins and are >200 nucleotides (nt) in
length. lncRNAs are a diverse class of RNAs, distinct from mRNAs,
and are involved in a number of important biological phenomena,
including imprinting genomic loci, shaping chromosome conformation
and allosterically regulating enzymatic activity (4–6). lncRNAs
account for the largest portion of the mammalian non-coding
transcriptome (7). Specific lncRNA
expression patterns, such as overexpression, depletion, or
mutations of lncRNA genes are associated with numerous human
diseases (7,8). Altered expression of specific lncRNAs
may promote tumor formation, progression and metastasis in many
malignancies, including RCC (8–10). Based
on the features of cancer, lncRNAs have been suggested to serve
important roles in their tumorigenesis, progression, treatment, and
prognosis. However, the functions of most of the lncRNAs are not
well understood.
In the present study, lncRNA-based prognostic
biomarkers were identified and a prognostic model of pRCC was
constructed. The lncRNA expression profiles between pRCC tissues
and corresponding normal kidney tissues from The Cancer Genome
Atlas (TCGA) were compared. Key lncRNAs were screened by univariate
Cox regression and least absolute shrinkage and selection operator
(LASSO) regression, followed by multivariate Cox regression
analysis to establish the prognostic model using the obtained
lncRNA expression data and corresponding clinical data of patients
with pRCC. The Kaplan-Meier (KM) survival curves, ROC curves and
the C-index were determined to estimate the prognostic power of the
model. In addition, the hazard ratio (HR) and corresponding P-value
of each key lncRNA were calculated by multivariate Cox regression.
Based on results of the multivariate Cox regression analysis, KM
survival analysis for each significant key lncRNA was
performed.
The results of the present study may contribute to
the management of pRCC and improve our understanding of the
underlying molecular mechanisms in tumorigenesis and progression of
pRCC.
Materials and methods
lncRNA expression data and clinical
data of patients with pRCC
lncRNA expression data and corresponding clinical
data of patients with pRCC were obtained from the TCGA database
(https://portal.gdc.cancer.gov/)
(11). The version of the dataset
was: Data Release 14.0-December 18, 2018. A total of 32
cancer-adjacent normal tissues and 289 cancer tissues from patients
with pRCC were included. R software (version 3.4.4) (12), RStudio 1.2.1335-Windows 7+ (64-bit)
(13) and R packages were used to
prepare and analyze the corresponding data.
Differential expression analysis
The R package ‘edgeR’ (14) was utilized to screen differentially
expressed lncRNAs in pRCC tissues compared with normal kidney
samples. A false discovery rate (FDR) <0.05 and
|log2FC|>2 were used as the cutoff criteria for the
differential expression, and values beyond these cutoffs were
considered statistically significant. The differentially expressed
lncRNAs were plotted using a volcano plot and heatmap using the R
packages ‘ggplot2’ (cran.r-project.org/web/packages/ggplot2/index.html)
and ‘pheatmap’ (cran.r-project.org/web/packages/pheatmap/index.html).
Univariate Cox regression
To determine the association between the expression
of differentially expressed lncRNAs and overall survival (OS) rate,
univariate Cox regression was performed using the ‘survival’
package (cran.r-project.org/web/packages/survival/index.html).
P<0.05 was considered to indicate a statistically significant
difference. Survival data, including the survival time and survival
status were merged with the expression data of the differentially
expressed lncRNAs. Univariate Cox regression was then performed and
statistically significant lncRNAs were identified and used for
LASSO regression.
LASSO regression
To identify key lncRNAs with better performance
parameters and to avoid overfitting, only statistically significant
lncRNAs based on the results of the univariate Cox regression
analysis were used for further LASSO regression using the ‘glmnet’
(https://cran.r-project.org/web/packages/glmnet/index.html)
and ‘survival’ packages. Similarly, the survival and expression
data of the statistically significant lncRNAs from the results of
the univariate Cox regression analysis were merged. Using the R
packages ‘glmnet’ and ‘survival’, and the LASSO algorithm, the
lncRNAs involved in the prognosis of patients with pRCC were
selected by shrinkage of the regression coefficient, through
imposing a penalty proportional to their size (15). As a result, most potential indicators
were shrunk to zero and a relatively small number of indicators
with a weight ≠0 were left (17 lnRNAs).
Multivariate Cox regression
To further establish a prognostic model and to
determine the relationship between the key lncRNAs based on the
LASSO regression analysis results and the OS time, multivariate Cox
regression analysis was performed using the ‘survival’ package,
which calculated the risk score for each patient. All the patients
were divided into high- and low-risk groups based on the median
cutoff point of the risk scores (median, 0.76). In addition, the
HR, 95% CI and statistical significance of each key lncRNA was
calculated and illustrated using a forest plot via the R package
‘survminer’ (cran.r-project.org/web/packages/survminer/index.html).
P<0.05 was considered to indicate a statistically significant
difference. A heatmap was used to illustrate the expression levels
of the key lncRNAs in both the groups.
ROC curve and C-index
To evaluate the prognostic power of the model, the
3-year and 5-year ROC curves were generated and the area under the
curves (AUCs) were calculated using the ‘survival’ and ‘timeROC’
(cran.r-project.org/web/packages/timeROC/index.html)
packages. An AUC of <0.5 was considered insignificant, while an
AUC of 0.51–0.7, 0.71–0.9, and >0.9 were considered low,
moderate, and high accuracy for the prognostic model, respectively.
Furthermore, the C-index was calculated using the ‘survival’
package. The evaluation criterion of the C-index was similar to
that of the AUC.
KM survival analysis
The KM survival curve was performed for the high-
and low-risk groups using the ‘survival’ package. KM survival
curves were plotted individually for each key lncRNA obtained in
the multivariate Cox regression analysis.
Prediction of subcellular
location
As the function of lncRNA is associated with its
subcellular localization, the sequences of the prognostic
biomarkers were obtained from the lncRNAMap website (16). The obtained sequences were entered
into the lncLocator website to predict the subcellular localization
of the lncRNA (17). The results
were obtained as a score for each potential subcellular location
for each lncRNA, including the cytoplasm, nucleus, ribosome,
cytosol and exosome. Finally, a location was predicted for each
lncRNA, using a function of the website.
Results
Identification of differentially
expressed lncRNAs in pRCC
A total of 14,447 lncRNAs were extracted from the
database, and 8,044 lncRNAs were identified as expressed genes.
Moreover, 1,001 lncRNAs were differentially expressed in pRCC
tissues compared with normal tissues according to the screening
criterion (FDR <0.05 and |log2FC|>2). Of these,
546 were upregulated (log2FC>2) and 455 were
downregulated (log2FC<-2; Fig. 1A).
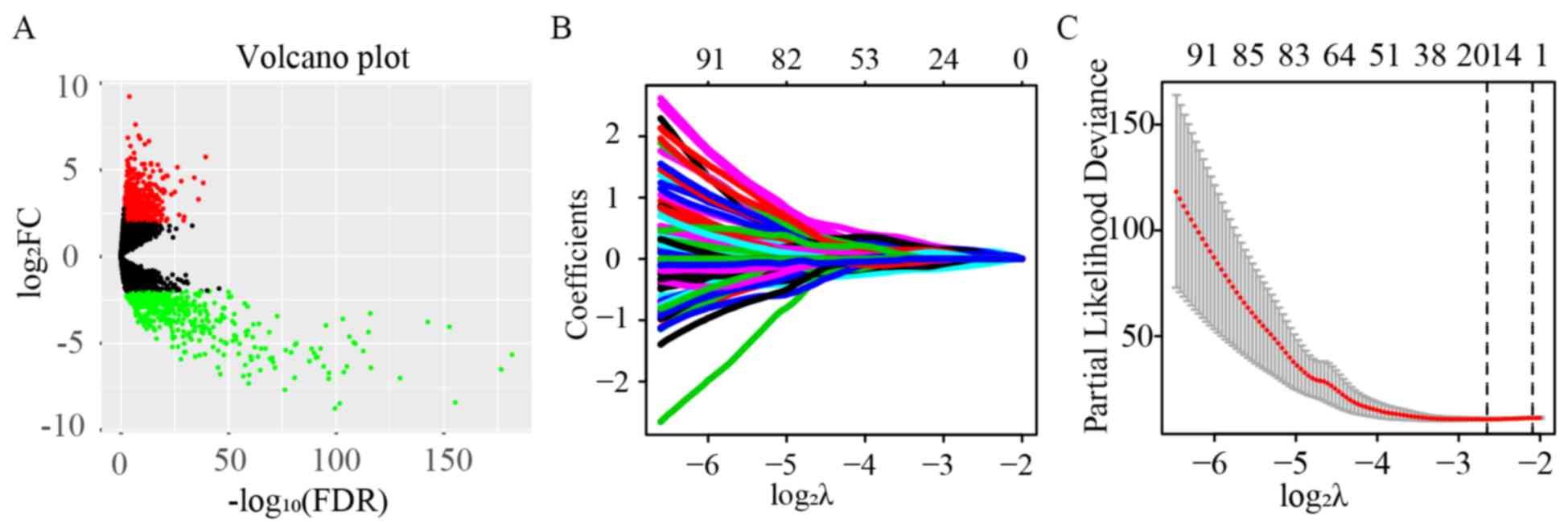 | Figure 1.Identification of 17 key lncRNA
signatures using univariate Cox and LASSO regression. (A) Volcano
plot of all lncRNAs. A total of 1,001 differentially expressed
lncRNAs out of 8,044 were identified in pRCC, including 546
upregulated and 455 downregulated lncRNAs (FDR<0.05 and
|log2FC|>2). (B) Tuning parameter and (C) variable
selection using the LASSO model. The numbers on the top of the
figures indicated the number of the candidate lncRNAs for the
corresponding lambda (λ) value in LASSO regression. A total of
348/1,001 significant lncRNAs from the result of the univariate Cox
regression were used for the LASSO regression, and 17 key lncRNAs
were identified by multivariate Cox regression. pRCC, papillary
renal cell carcinoma; lncRNAs, long non-coding RNAs; LASSO, least
absolute shrinkage and selection operator; FC, fold change; FDR,
false discovery rate. |
Establishment and evaluation of the
prognostic model
Univariate Cox regression analysis was first
performed for the 1,001 differentially expressed lncRNAs. Of these,
the differential expression of 348 was considered statistically
significant. LASSO regression was carried out on these 348 lncRNAs,
and 17 key lncRNAs were identified (Fig.
1B and C). Multivariate Cox regression was performed to
establish the prognostic model based on the 17 key lncRNAs. The
risk scores for all patients were calculated, and patients were
divided into high- and low-risk groups based on 0.76, the median
cutoff point of the scores. To evaluate the prognostic model, the
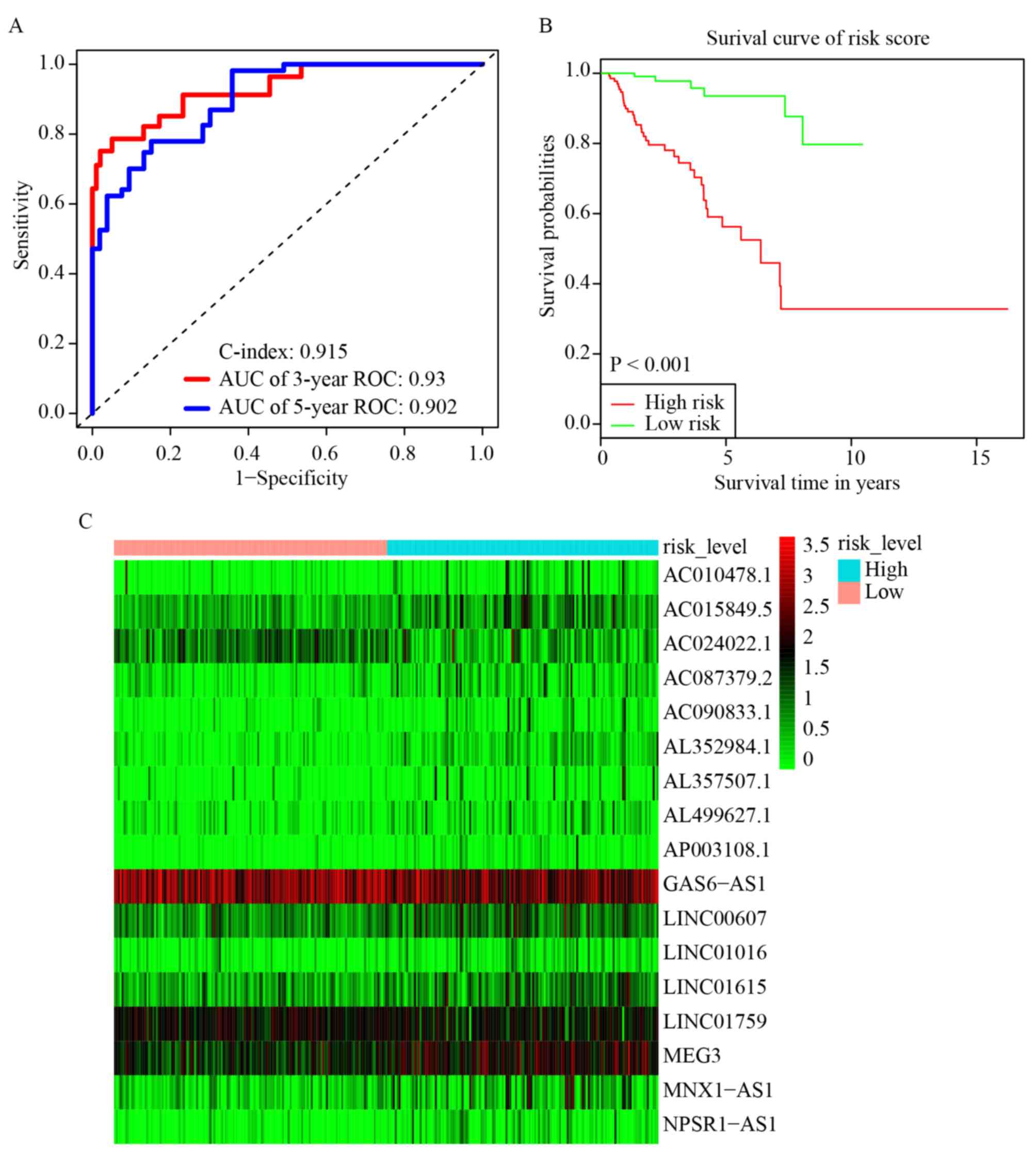
3-year and 5-year ROC curves were plotted and the C-index was
calculated. The AUC values were 0.93 (3-year ROC) and 0.902 (5-year
ROC), and the C-index was 0.915 (Fig.
2A). Furthermore, KM survival analysis revealed that the OS
rate was significantly worse in the high-risk group compared with
the low-risk group (P<0.001; Fig.
2B). The HR, 95% CI, and its statistical significance for each
of the 17 key lncRNAs were determined using multivariate Cox
regression, and the results are presented as a forest plot in
Fig. 3.
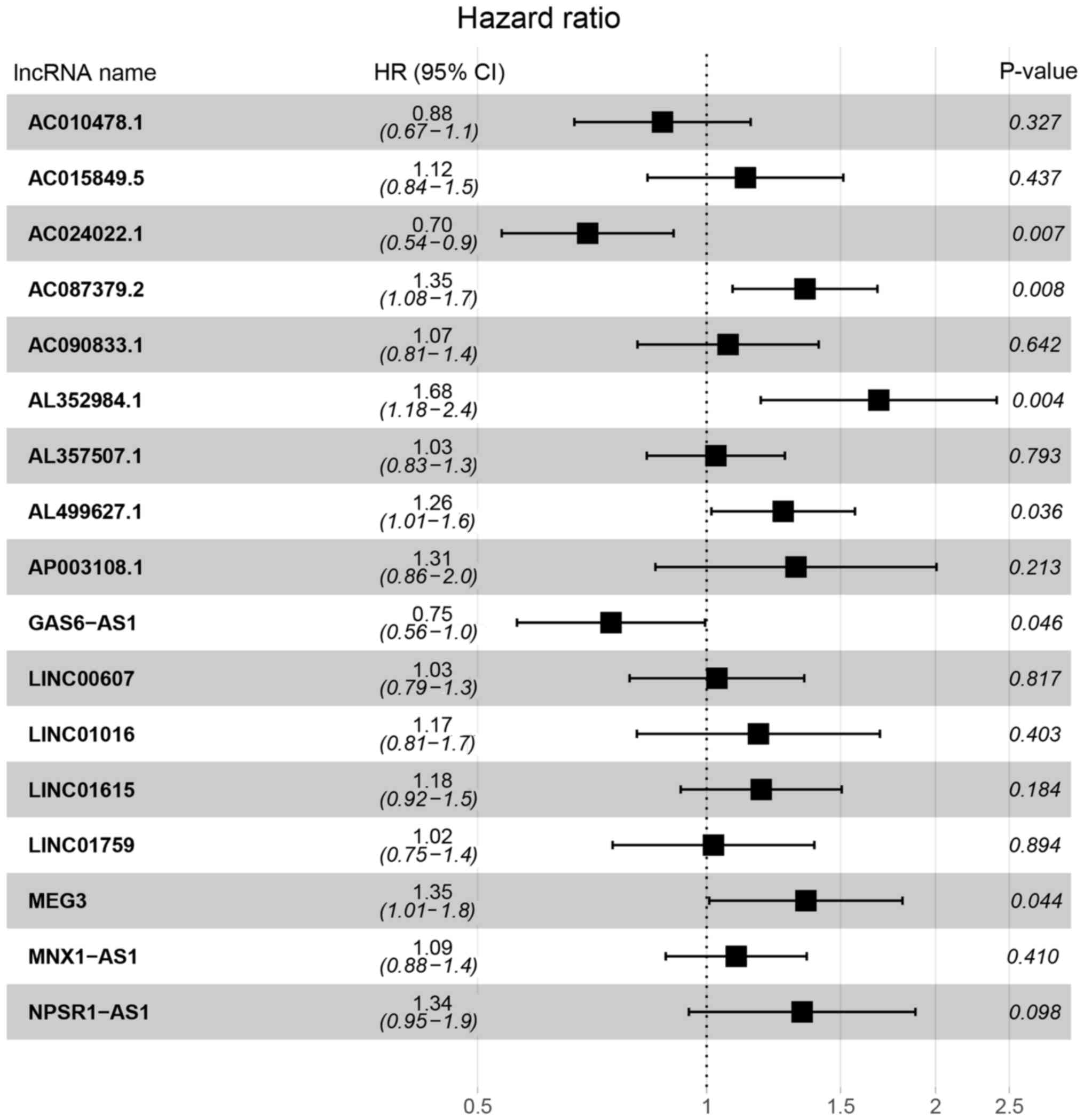 | Figure 3.HR and 95% CI of the 17 key lncRNAs by
the multivariate Cox regression. A total of 285 patients was
included in the analysis. Of the 17 key lncRNAs, six had a
significant influence on the OS rate of patients with pRCC,
including AC024022.1, AC087379.2, AL352984.1, AL499627.1, GAS6-AS1
and MEG3. HR, hazard ratio; CI, confidence interval; lncRNAs, long
non-coding RNAs; OS, overall survival; pRCC, papillary renal cell
carcinoma. |
KM survival analysis for key
lncRNAs
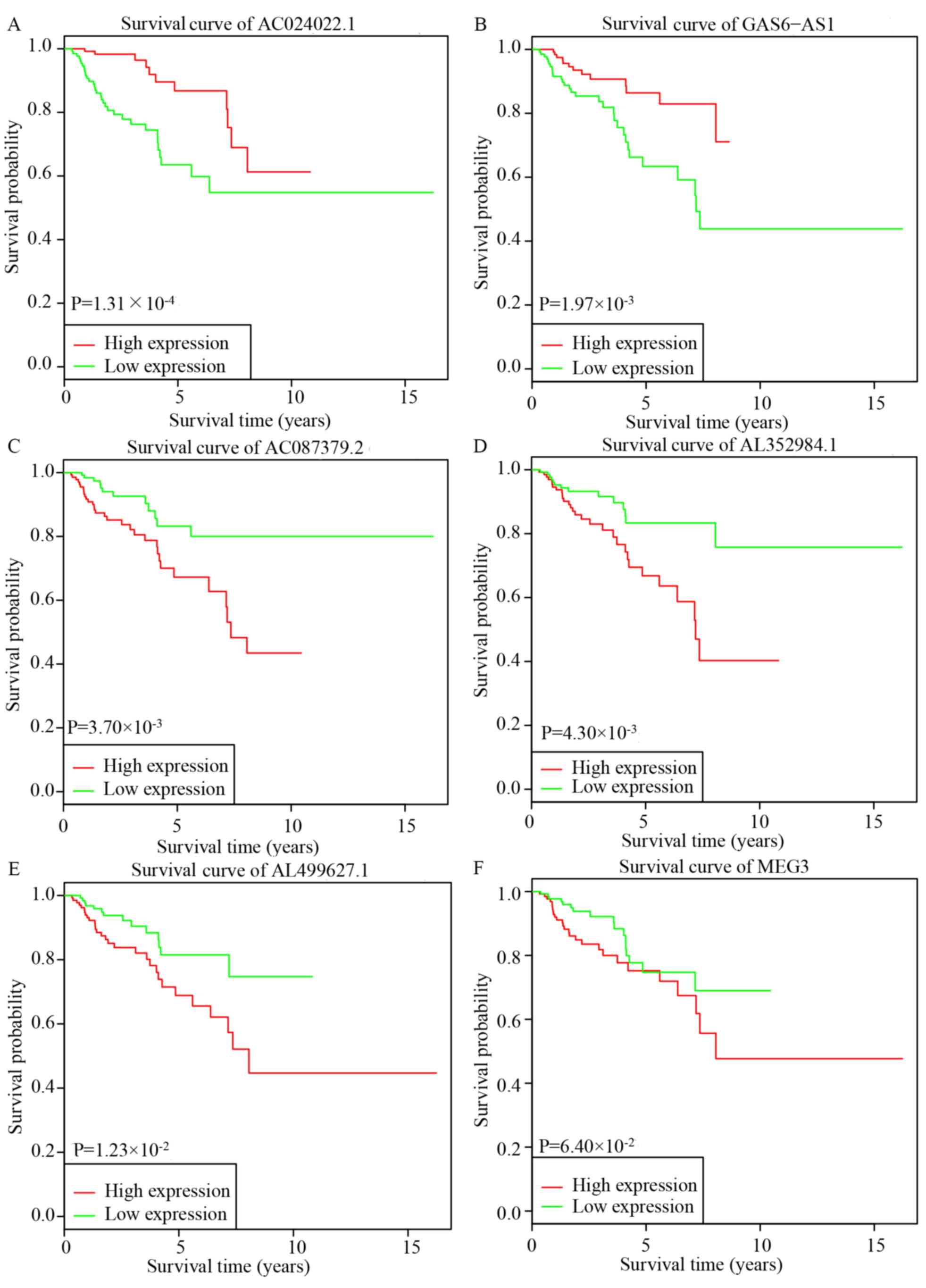
Based on the results of the multivariate Cox
regression, six lncRNAs, AC024022.1, AC087379.2, AL352984.1,
AL499627.1, GAS6-AS1 and MEG3, were used for further KM survival
analysis. AC024022.1 and GAS6-AS1 were identified as protective
prognostic biomarkers, while AC087379.2, AL352984.1 and AL499627.1
were identified as adverse prognostic biomarkers (Fig. 4A-E). MEG3 may additionally be a
potential adverse prognostic biomarker, although the difference was
not statistically significant (Fig.
4F).
Subcellular location prediction
lncLocator predicted that AC024022.1 and AC087379.2
were located in the cytoplasm and GAS6-AS1 was located in the
cytosol (Table I). The subcellular
localizations of AL352984.1 and AL499627.1 were not predicted as
lncRNAMap website did not possess a record of their sequences. The
sequence information of AC024022.1, AC087379.2 and GAS6-AS1
obtained from lncRNAMap are presented in Table S1.
 | Table I.Prediction of subcellular locations of
long non-coding RNAs. |
Table I.
Prediction of subcellular locations of
long non-coding RNAs.
| Location | AC024022.1 | AC087379.2 | GAS6-AS1 |
|---|
| Cytoplasm | 0.512660116 | 0.603843419 | 0.139349109 |
| Nucleus | 0.363217766 | 0.097721928 | 0.107388576 |
| Ribosome | 0.016871894 | 0.073857973 | 0.094522592 |
| Cytosol | 0.06045974 | 0.073769925 | 0.640534382 |
| Exosome | 0.046790483 | 0.150806755 | 0.01820534 |
| Predicted
location | Cytoplasm | Cytoplasm | Cytosol |
Discussion
Transcription is classified into protein-coding and
non-coding transcripts. Coding genes are estimated to account for
<2% of the human transcripts, while non-coding RNA transcripts
account for ~98% of transcriptome (18,19).
Among the non-coding transcripts, lncRNAs are a loosely classified
group of long RNA transcripts (>200 nucleotides in length) with
no apparent protein-coding potential (4). However, lncRNAs not only have similar
structures as protein-coding genes such as introns and exons, but
also possess a wide range of biological functions involved in a
variety of cellular activities (19). For instance, lncRNAs are involved in
cell proliferation, survival, apoptosis and movement by regulating
gene expression at the epigenetic, transcriptional, and
post-transcriptional levels (20,21). The
functions of lncRNA are primarily executed by serving as a: i)
Molecular signal that can respond to intracellular or extracellular
signals and act as a regulator for specific signal pathways; ii)
molecular decoy that may combine certain RNAs or proteins that may
be removed from a particular subcellular location, thus abrogating
or attenuating their normal function; iii) molecular guides for
proteins to a specific mRNA or chromosome locus, influencing
genetic transcription, mRNA stability and translation; and iv)
molecular scaffold, which can bind multiple molecules for specific
functions (20). The subcellular
localization of lncRNA can be used to predict its functions
(20). For example, if a lncRNA is
located in the nucleus, it may be involved in chromatin regulation,
gene transcription and alternative splicing of transcripts, whereas
if it is in the cytoplasm, it may serve as a competing endogenous
(ce)RNA or regulate mRNA stability or translation.
At present, there is increasing evidence that
aberrations within lncRNAs such as overexpression, deficiency or
mutations drive important cancer phenotypes (18), including tumor formation, progression
and metastasis in many types of cancer (7,8,10,22,23). In
addition, certain aberrant lncRNAs are additionally associated with
increasing the malignant biological behaviors of cancer cells,
clinicopathological features and poor prognosis of cancers
(23–25). In clear cell RCC, some aberrant
lncRNAs, such as PVT1, TUG1, NEAT1, and PANDAR, can serve as
prognostic indicators (26–29). However, to the best of our knowledge,
there are no previous studies investigating the association between
lncRNA and the prognosis of pRCC.
In the present study, to identify potentially
valuable lncRNAs for improving prognosis of patients with pRCC,
existing lncRNA expression data along with corresponding clinical
data in TCGA was analyzed to identify the key lncRNAs using
univariate Cox regression and LASSO regression analyses. A
prognostic model was then constructed by multivariate Cox
regression based on the identified key lncRNAs. All the patients
were divided into high- and low-risk groups and the KM survival
curves, ROC curves and C-index calculation were performed, and the
prognostic power of the model was estimated. In addition, the HR
and 95% CI of each key lncRNA were also calculated by multivariate
Cox regression analysis. Based on these results, KM survival
analysis was performed for each significant key lncRNA. A total of
1,001 differentially expressed lncRNAs were identified, of which
546 were upregulated and 455 were downregulated. Furthermore,
univariate Cox regression and LASSO regression analyses identified
17 key lncRNAs, which were used to establish the prognostic model.
Furthermore, the results of the KM survival analysis revealed that
patients in the high-risk group had significantly lower OS rate
compared with the low risk group. Based on the multivariate Cox
regression analysis, six lncRNAs were identified as potential
prognostic biomarkers. The prognostic values of five lncRNAs,
AC024022.1, AC087379.2, AL352984.1, AL499627.1, and GAS6-AS1 (all
P<0.05), were validated by KM survival analysis.
Subsequently, the nucleotide database was retrieved
from the lncRNA map. The lncRNA AC024022.1 (ENSG00000250781) is the
sequence of ‘Homo sapiens BAC clone RP11-63A11’. The lncRNA
AC087379.2 (ENSG00000254695) is defined as ‘Homo sapiens
chromosome 11, clone RP11-396020’. The lncRNA AL352984.1
(ENSG00000258386) is the sequence of Homo sapiens BAC
R-187E13 of library RPCI-11 from chromosome 14. The lncRNA
AL499627.1 (ENSG00000260542) is the sequence from clone RP13-379O24
on human chromosome 20. GAS6- AS1 (ENSG00000233695) is defined as
Homo sapiens GAS6 antisense RNA-1. Decreased expression of
lncRNA GAS6-AS1 predicted a poor prognosis in patients with
non-small cell lung cancer (NSCLC) (30), consistent with the present study.
Decreased expression of GAS6-AS1 predicted a worse OS rate,
suggesting that GAS6-AS1 is a protective biomarker for the
prognosis of pRCC. Correct subcellular localization of lncRNA
transcripts is important for their functions. Therefore, the
subcellular locations of the lncRNA transcripts were predicted
using lncLocator (17) based on the
sequences acquired from lncRNAMap (16). It was predicted that AC024022.1
(ENSG00000250781) and AC087379.2 (ENSG00000254695) were likely
located in the cytoplasm, and GAS6-AS1 (ENSG00000233695) were
likely located in the cytosol. The subcellular localization of
AL352984.1 (ENSG00000258386) and AL499627.1 (ENSG00000260542) were
not predicted because their sequence information was not present on
lncRNAMap. As previously mentioned, if a lncRNA was located in the
cytoplasm or cytosol, it may serve as a ceRNA, and regulate mRNA
stability or translation (20).
Therefore, we hypothesized that lncRNAs AC024022.1
(ENSG00000250781), AC087379.2 (ENSG00000254695), and GAS6-AS1
(ENSG00000233695) acted as ceRNA, and served a role in regulating
the stability or translation of mRNA. Unfortunately, to the best of
our knowledge, there were no previous studies for these lncRNAs,
and further studies are required to investigate the features,
functions, and mechanisms of them.
In summary, a 17-lncRNA signature was constructed by
mining the TCGA database, and identified two protective lncRNAs,
AC024022.1 (ENSG00000250781) and GAS6-AS1 (ENSG00000233695), and
three potentially tumorigenic lncRNAs, AC087379.2
(ENSG00000254695), AL352984.1 (ENSG00000258386) and AL499627.1
(ENSG00000260542). AC024022.1 (ENSG00000250781) and AC087379.2
(ENSG00000254695) might be located in the cytoplasm while GAS6-AS1
(ENSG00000233695) might be located in the cytosol. The present
study identified novel lncRNAs, which may serve as biomarkers for
the prognosis of pRCC and these findings may contribute to the
management or further investigation into the underlying mechanisms
of pRCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was funded by The National Natural
Science Foundation of China (grant nos. 81711530048 and 81572515),
Beijing Municipal Science & Technology Commission (grant no.
Z151100003915105).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the TCGA repository (https://portal.gdc.cancer.gov/). The version of
the dataset was: Data Release 14.0-December 18, 2018.
Authors' contributions
LM and CL contributed to the design of the study,
acquisition of funding, and general supervision. FY contributed to
data analysis and wrote the manuscript. LG and GZ contributed to
acquisition and interpretation of data, and revised the manuscript
critically for important intellectual content. YS offered guidance,
interpreted the data for the work, and reviewed the article for
publication. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Courthod G, Tucci M, Di Maio M and
Scagliotti GV: Papillary renal cell carcinoma: A review of the
current therapeutic landscape. Crit Rev Oncol Hematol. 96:100–112.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cheng P: A prognostic 3-long noncoding RNA
signature for patients with gastric cancer. J Cell Biochem.
119:9261–9269. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Martens-Uzunova ES, Bottcher R, Croce CM,
Jenster G, Visakorpi T and Calin GA: Long noncoding RNA in
prostate, bladder, and kidney cancer. Eur Urol. 65:1140–1151. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jin Y, Feng SJ, Qiu S, Shao N and Zheng
JH: LncRNA MALAT1 promotes proliferation and metastasis in
epithelial ovarian cancer via the PI3K-AKT pathway. Eur Rev Med
Pharmacol Sci. 21:3176–3184. 2017.PubMed/NCBI
|
|
10
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Deng M, Bragelmann J, Schultze JL and
Perner S: Web-TCGA: an online platform for integrated analysis of
molecular cancer data sets. BMC Bioinformatics. 17:722016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
R Core Team. R, . A language and
environment for statistical computing. R Foundation for Statistical
Computing; Vienna: 2012, http://www.r-project.org/
|
|
13
|
RStudio Team (2015), . RStudio: Integrated
Development for R. RStudio, Inc., Boston, MA, 2015. http://www.rstudio.com/
|
|
14
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tibshirani R: Regression shrinkage and
selection via the lasso: A retrospective. J Royal Statist Soc
Series B. 73:273–282. 2011. View Article : Google Scholar
|
|
16
|
Chan WL, Huang HD and Chang JG: lncRNAMap:
A map of putative regulatory functions in the long non-coding
transcriptome. Comput Biol Chem. 50:41–49. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cao Z, Pan X, Yang Y, Huang Y and Shen HB:
The lncLocator: A subcellular localization predictor for long
non-coding RNAs based on a stacked ensemble classifier.
Bioinformatics. 34:2185–2194. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shen Y, Wang Z, Loo LW, Ni Y, Jia W, Fei
P, Risch HA, Katsaros D and Yu H: LINC00472 expression is regulated
by promoter methylation and associated with disease-free survival
in patients with grade 2 breast cancer. Breast Cancer Res Treat.
154:473–482. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee JT: Epigenetic regulation by long
noncoding RNAs. Science. 338:1435–1439. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu Y, Yang J, Li Q, Xu B, Lian Y and Miao
L: LINC00152: A pivotal oncogenic long non-coding RNA in human
cancers. Cell Prolif. 50:2017. View Article : Google Scholar
|
|
23
|
Yue B, Qiu S, Zhao S, Liu C, Zhang D, Yu
F, Peng Z and Yan D: LncRNA-ATB mediated E-cadherin repression
promotes the progression of colon cancer and predicts poor
prognosis. J Gastroenterol Hepatol. 31:595–603. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ellinger J, Gevensleben H, Muller SC and
Dietrich D: The emerging role of non-coding circulating RNA as a
biomarker in renal cell carcinoma. Expert Rev Mol Diagn.
16:1059–1065. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang HW, Xie H, Ma X, Zhao F and Gao Y:
Upregulation of LncRNA PANDAR predicts poor prognosis and promotes
cell proliferation in cervical cancer. Eur Rev Med Pharmacol Sci.
21:4529–4535. 2017.PubMed/NCBI
|
|
26
|
Bao X, Duan J, Yan Y, Ma X, Zhang Y, Wang
H, Ni D, Wu S, Peng C, Fan Y, et al: Upregulation of long noncoding
RNA PVT1 predicts unfavorable prognosis in patients with clear cell
renal cell carcinoma. Cancer Biomark. 21:55–63. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang PQ, Wu YX, Zhong XD, Liu B and Qiao
G: Prognostic significance of overexpressed long non-coding RNA
TUG1 in patients with clear cell renal cell carcinoma. Eur Rev Med
Pharmacol Sci. 21:82–86. 2017.PubMed/NCBI
|
|
28
|
Xu Y, Tong Y, Zhu J, Lei Z, Wan L, Zhu X,
Ye F and Xie L: An increase in long non-coding RNA PANDAR is
associated with poor prognosis in clear cell renal cell carcinoma.
BMC Cancer. 17:3732017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ning L, Li Z, Wei D, Chen H and Yang C:
LncRNA, NEAT1 is a prognosis biomarker and regulates cancer
progression via epithelial-mesenchymal transition in clear cell
renal cell carcinoma. Cancer Biomark. 19:75–83. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Han L, Kong R, Yin DD, Zhang EB, Xu TP, De
W and Shu YQ: Low expression of long noncoding RNA GAS6-AS1
predicts a poor prognosis in patients with NSCLC. Med Oncol.
30:6942013. View Article : Google Scholar : PubMed/NCBI
|