Introduction
Lung cancer (LC) is one of the most malignant
neoplasms, and increases in its morbidity and mortality rates have
made it a main cause of human mortality. In the last 50 years,
research has revealed a marked increase in the incidence and
mortality rates of LC (1). The
incidence and mortality rates of LC in men ranked first among all
malignant tumor groups, and the incidence in women ranked second
most common worldwide in 2018 (2).
The onset of LC is insidious and it has a poor prognosis.
Mediastinal lymph node metastasis (MLNM) during treatment is one of
the most important factors affecting the postoperative survival of
patients (3).
The MLNM process of LC includes the growth of a
primary tumor, angiogenesis and exfoliation of tumor cells, which
invade the tissue matrix, and make cancer cells survive in the
blood circulation and amass into tiny tumor thrombi (4). Previous studies in China have reported
that the 5-year survival rate of patients with LC without MLNM
could be >60%, but was only 15–42% with MLNM (5,6).
Numerous previous studies (7–11) have
demonstrated that the pathophysiological process of the development
of LC with MLNM (MM LC) is associated with the mutation and
abnormal expression of genes, including C-X-C motif chemokine
receptor 4 (CXCR4), vascular endothelial growth factor-C (VEGF-C),
vascular endothelial growth factor receptor-3 (VEGFR-3), ADAM
metallopeptidase domain (ADAM) and vascular endothelial growth
factor-D (VEGF-D). In a previous study by Na et al (9) of 46 patients with LC, abnormal
expression of CXCR4 in the nucleus was markedly associated with
MLNM. In addition, multiple previous studies have suggested that
co-expression of VEGFR-3 and VEGF-C (7), high expression levels of ADAM family
members (8), downregulation of
VEGF-D expression (10), and high
expression levels of VEGF-C (11)
may be involved in the MLNM of LC. These genes may be used as
prognostic factors or targets for gene therapy. However, in
individualized applications, the diagnostic or therapeutic value of
a single gene remains uncertain. Due to the lack of timely
detection, dynamic monitoring and effective control of the
occurrence of MLNM, the poor survival rate of MM LC still cannot be
effectively controlled. Therefore, novel signaling pathways and
molecular targets should be investigated and screened to develop
novel diagnostic and therapeutic methods.
Microarray technology allows simultaneous analysis
of alterations in the expression levels of multiple genes to obtain
gene sets that can predict MLNM in LC with high accuracy. Clinical
application of these gene sets is expected to maximize the survival
period and narrow the surgical range (whether lymph node resection
is required) for the benefit of patients. Previously, a number of
studies (12–17) have performed bioinformatic analyses
to investigate differentially expressed genes (DEGs) in various
types of cancer, as well as their roles in different pathways,
molecular functions and biological processes. Kikuchi et al
(12) used microarray technology to
obtain a set of genes which could predict MLNM and drug
sensitivity. Li et al (13)
applied the same method to identify the significant genes in the
carcinogenesis and progression of hepatocellular carcinoma.
Furthermore, multiple previous studies have also used microarray
technology to obtain gene expression profiles to predict lymph node
metastasis of other malignant tumors, including esophageal cancer
(14), oral squamous cell carcinoma
(15,16) and cervical cancer (17).
Therefore, the purpose of the present study was to
download and analyze two expression profiling datasets of human
samples from the Gene Expression Omnibus (GEO) database, and to
identify DEGs between LC samples without MLNM (non-MM LC) and MM LC
samples. Subsequently, Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathway and Gene Ontology (GO) analysis were carried out.
Protein-protein interaction (PPI) network analysis and
co-expression network analyses were used to demonstrate the
molecular pathogenesis underlying carcinogenesis and development of
MM LC. Overall, 11 hub genes and 308 DEGs, which may be potential
molecular targets or biomarkers for MM LC, were identified.
Materials and methods
Access to public data
The GEO (http://www.ncbi.nlm.nih.gov/geo) is an open platform
for storing genetic data (18). In
total, two expression profiling datasets [GSE23822 (19) (GPL6947 platform) and GSE13213
(20) (GPL6480 platform)] were
obtained from GEO. The GSE23822 dataset contained four non-MM LC
samples and four MM LC samples. Similarly, GSE13213 consisted of 22
non-MM LC samples and 22 MM LC samples.
Identification of DEGs using
GEO2R
GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) is an
interactive online tool for the identification of DEGs from GEO
series (21). GEO2R was used to
identify DEGs between MM LC and non-MM LC tissue samples. If one
probe set did not have the homologous gene, or if one gene had
numerous probe sets, the data were removed. The rules of
statistical significance were that P≤0.01 and fold change ≥1.5.
Functional annotation of DEGs by KEGG
and GO analysis
Database for Annotation, Visualization and
Integrated Discovery (DAVID; version 6.8, http://david.ncifcrf.gov/home.jsp), is an online
analysis tool suite with the functions of integrated discovery and
annotation (22). GO (http://geneontology.org) is widely used in
bioinformatics, and covers three aspects of biology; biological
process (BP), cellular component (CC) and molecular function (MF)
(23). KEGG (https://www.kegg.jp) is one of the most commonly used
biological information databases in the world (24). OmicShare (http://www.omicshare.com/tools), an open data analysis
platform, was used to perform GO analysis (25). To analyze the biological pathway
information of DEGs, the DAVID online tool was implemented.
P<0.05 was considered to indicate a statistically significant
difference.
Construction of the PPI network and
identification of the significant module
Search Tool for the Retrieval of Interacting Genes
(http://string.embl.de/), an online open tool, was
applied to construct the PPI network of DEGs, and Cytoscape was
used to present the network (26).
Cytoscape (version 3.6.1) is a free visualization software
(27). A confidence score >0.4
was considered as the criterion of judgment. Subsequently, the
Molecular Complex Detection (28)
(MCODE; version 1.5.1; a plug-in of Cytoscape), was used to
identify the most important module of the network map. The criteria
for MCODE analysis were as follows: i) Degree cut-off=2; ii) MCODE
scores >5; iii) max depth=100; iv) node score cut-off=0.2; and
v) k-score=2 (29). Subsequently,
following the KEGG and GO analysis using the DAVID database and
OmicShare website, functional annotation for genes of these modules
was performed.
Analysis and identification of hub
genes
The degrees were set (degrees ≥10), and the hub
genes were excavated. A co-expression network of these hub genes
was obtained from cBioPortal (http://www.cbioportal.org) (30). Furthermore, the Biological Networks
Gene Oncology tool (BiNGO; version 3.0.3) was used to analyze and
visualize the CCs, BPs and MFs of the hub genes (31). The clustering analysis of hub genes
was performed using OmicShare (https://www.omicshare.com/tools/Home/Soft/getsoft/type/index)
(25). The mean value of amount of
gene expression was defined as the cut-off value for the high or
low expression level. Additionally, Kaplan-Meier Plotter
(http://kmplot.com/analysis/index.php?p=background), an
online analysis tool, was utilized to perform survival analysis for
the hub genes. The Kaplan Meier plotter (32) is capable of assessing the effect of
54,000 genes on survival in 21 different types of cancer. The
largest datasets include breast (n=6,234), ovarian (n=2,190), lung
(n=3,452) and gastric (n=1,440) cancer. The miRNA subsystems
include 11k samples from 20 different cancer types. Primary purpose
of the tool is a meta-analysis based discovery and validation of
survival biomarkers. The P-value was achieved by using a log-rank
test. University of California Santa Cruz (UCSC) Xena (https://xena.ucsc.edu/welcome-to-ucsc-xena/) was used
to securely analyze and visualize the hub genes in the scope of
public genomic datasets. The expression profiles of actin β (ACTB)
and integrin subunit β1 (ITGB1) were analyzed and visualized using
the online database Gene Expression Profiling Interactive Analysis
(GEPIA; http://gepia.cancer-pku.cn/).
Results
Screening of DEGs in MM LC
samples
Following analysis of the datasets (GSE23822 and
GSE13213) with GEO2R, the difference between MM LC and non-MM LC
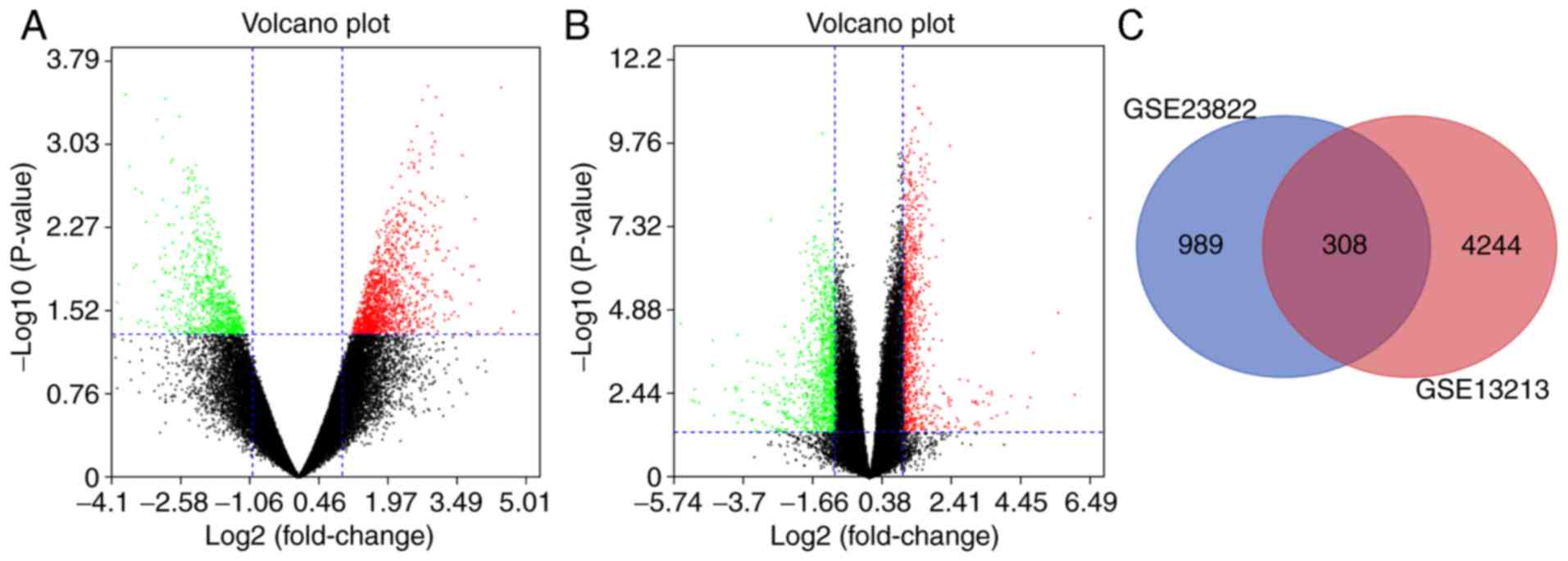
tissues was presented in volcano plots (Fig. 1A and B). The analyses of GSE23822 and
GSE13213 identified 1,297 and 4,552 DEGs, respectively (Fig. 1C). The Venn diagram demonstrated that
the commonality between the two datasets included 308 DEGs,
including the most upregulated genes [Erb-b2 receptor tyrosine
kinase 2 (ERBB2) and ITGB1] and the most downregulated genes [ACTB,
RAB5B, member RAS oncogene family (RAB5B) and intersectin 2
(ITSN2)] (data not shown).
Functional annotation of DEGs by KEGG
and GO analyses
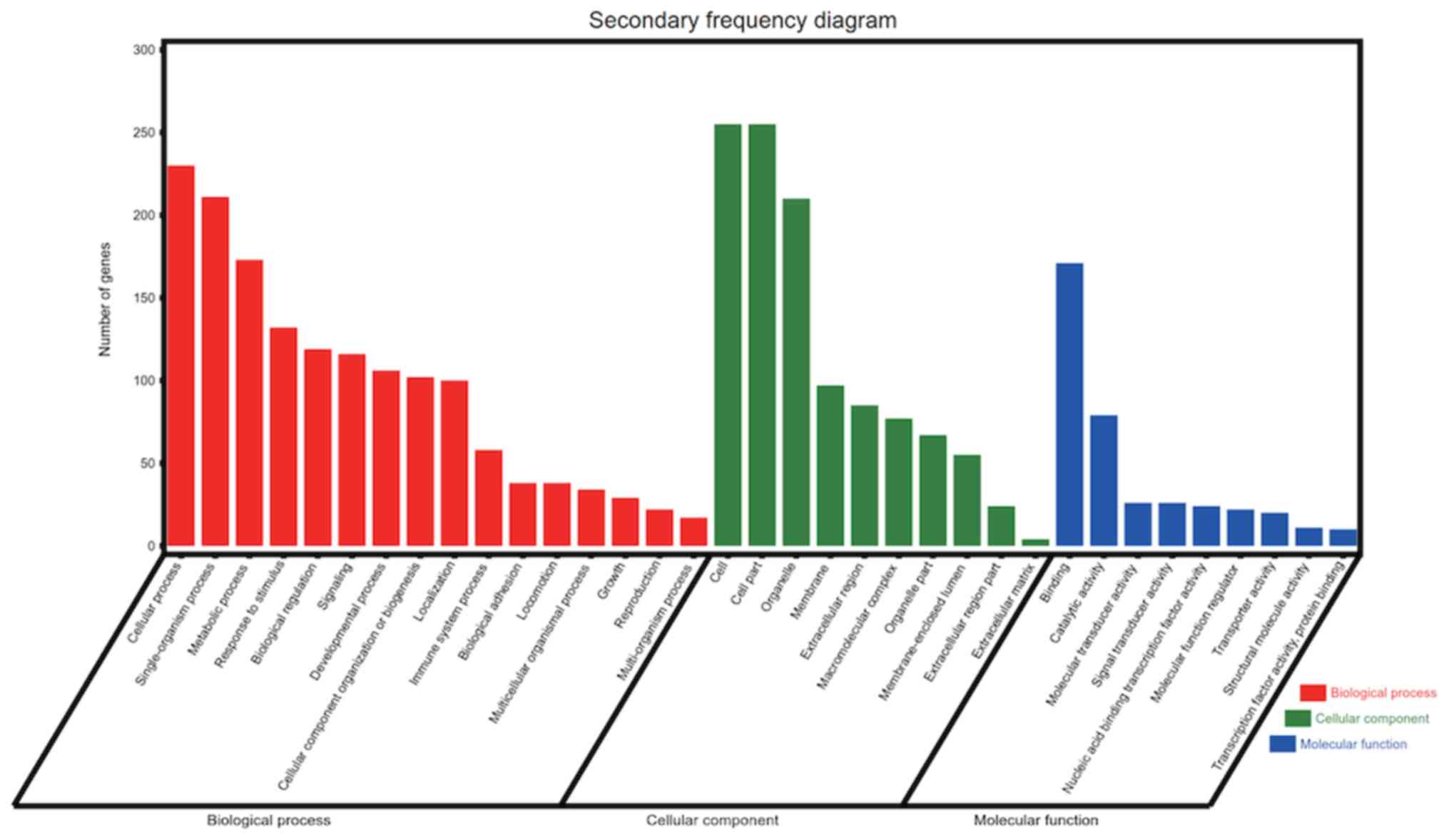
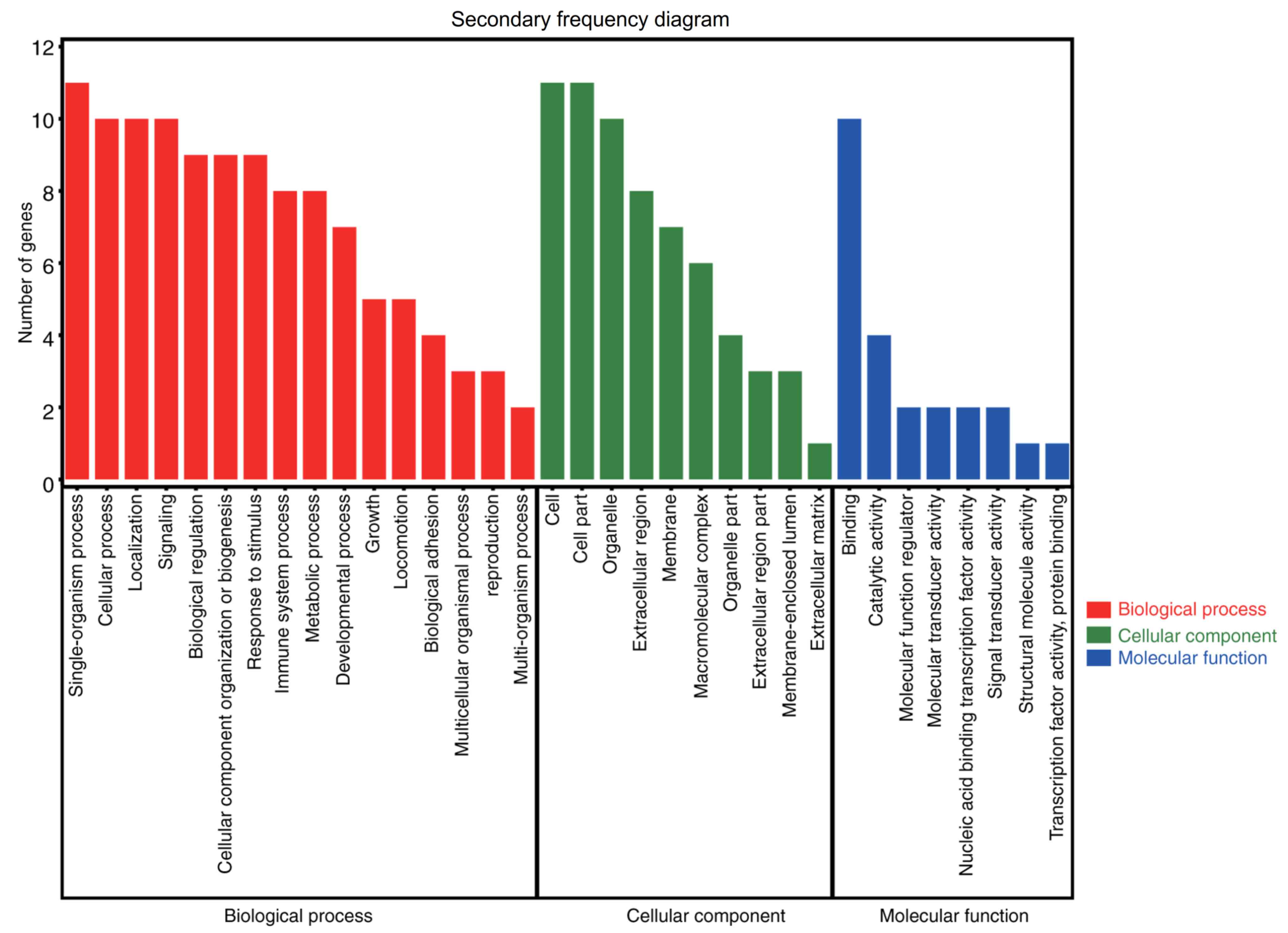
The results of the GO analysis demonstrated that
variations in the BP were primarily enriched in the GO terms
‘cellular process’, ‘metabolic process’, ‘response to stimulus’,
‘biological regulation’, ‘signaling’ and ‘localization’,
‘single-organism process’, ‘developmental process’, ‘cellular
component organization or biogenesis’, and so on. Alterations in CC
were mainly enriched in the GO terms ‘cell’, ‘membrane’,
‘extracellular region’ and ‘organelle part’. The variations in MF
were enriched in the GO terms ‘binding’, ‘catalytic activity’,
‘molecular transducer activity’, ‘signal transducer activity’ and
‘molecular function regulator’ (Fig.
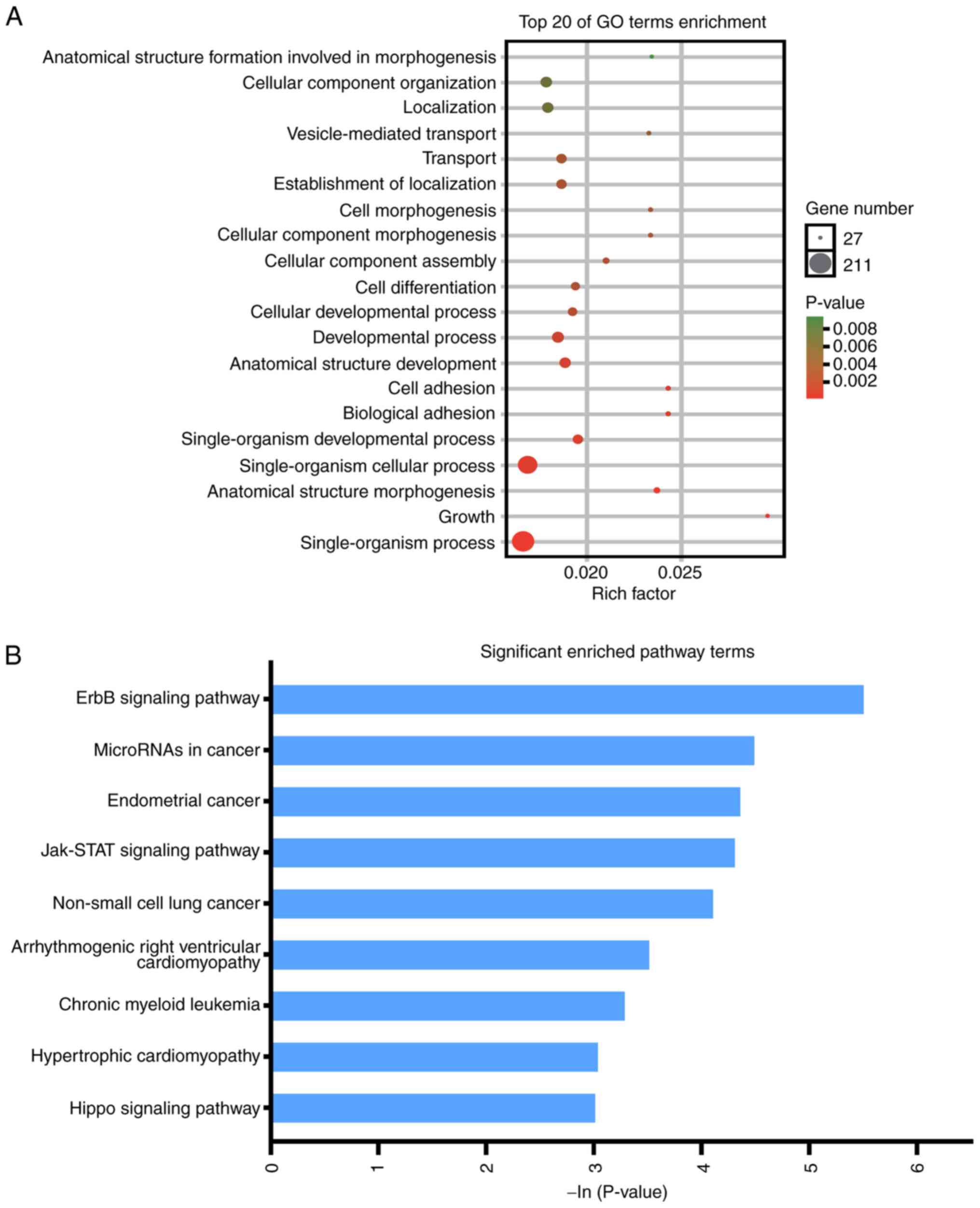
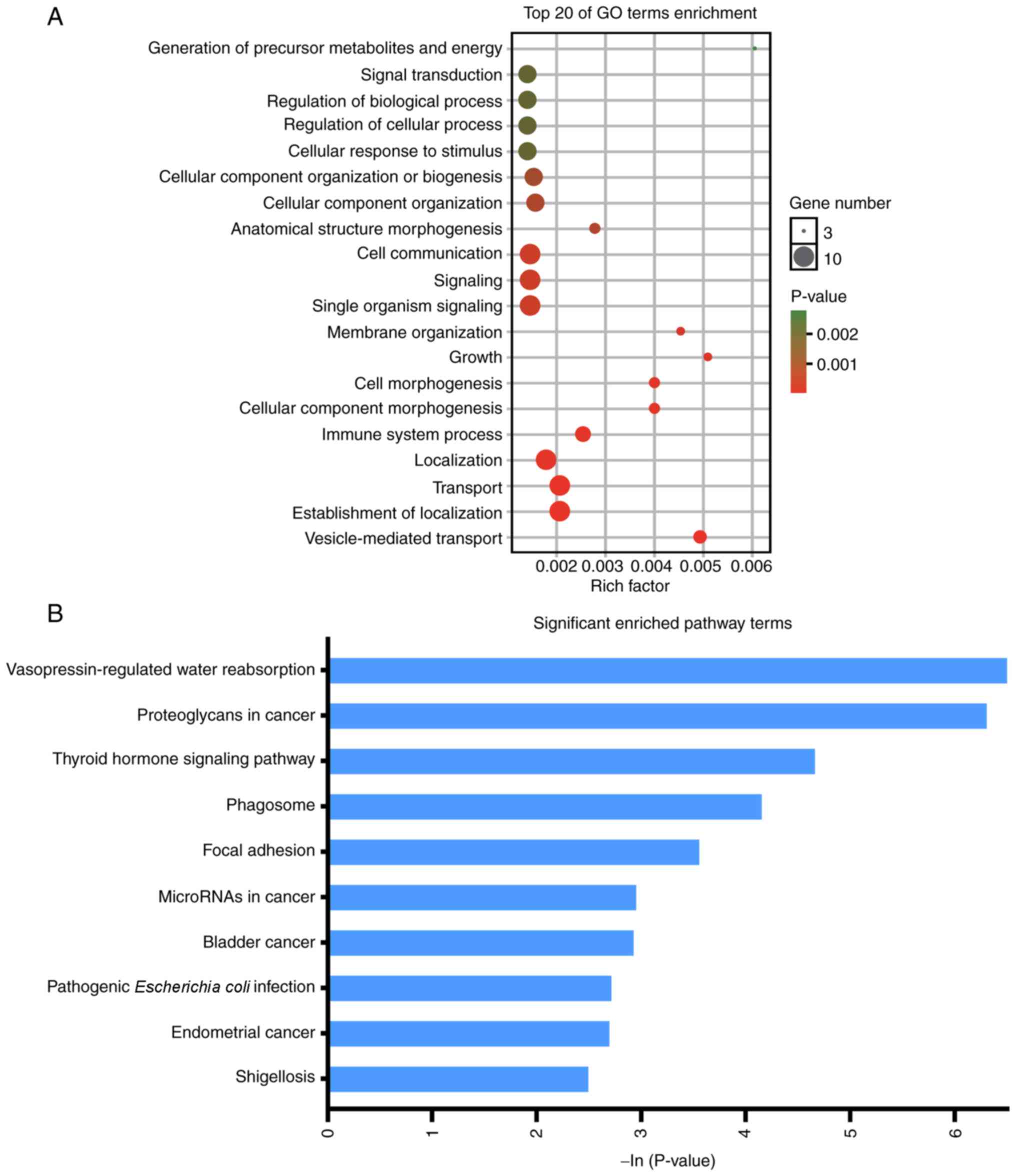
2). The enriched GO terms were ‘generation of precursor
metabolites and energy’, ‘signal transduction’, ‘regulation of
biological process’, ‘regulation of cellular process’, ‘cellular
response to stimulus’, ‘cellular component organization or
biogenesis’, ‘cellular component organization’, ‘anatomical
structure morphogenesis’, ‘cell communication’, ‘signaling’,
‘single organism signaling’, ‘membrane organization’, ‘growth’,
‘cell morphogenesis’, ‘cellular component morphogenesis’, ‘immune
system process’, ‘localization’, ‘transport’, ‘establishment of
localization’, and ‘vesicle-mediated transport’ (Fig. 3A). KEGG analysis revealed that DEGs
were enriched in ‘ErbB signaling pathway’, ‘microRNAs in cancer’,
‘endometrial cancer’, ‘Jak-STAT signaling pathway’, ‘non-small cell
lung cancer’, ‘chronic myeloid leukemia’, ‘hypertrophic
cardiomyopathy’ and ‘Hippo signaling pathway’ (Fig. 3B).
Construction of the PPI network and
identification of the significant module
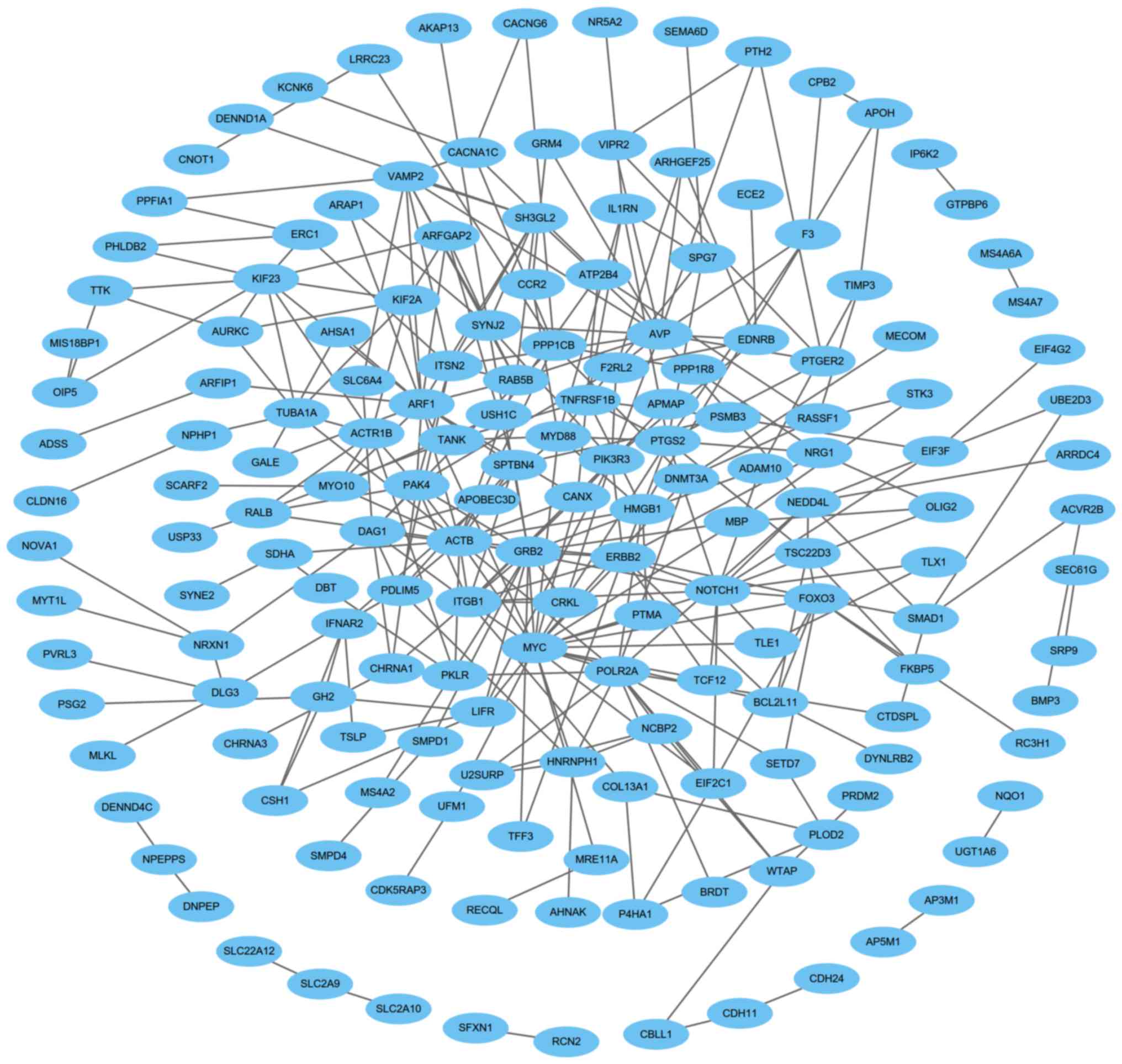
Construction of the PPI network and identification
of the significant module were performed, and there were 315 edges
and 167 nodes in the PPI network (Fig.
4). Furthermore, there were 26 edges and 11 nodes in the
significant module (Fig. 5). Using
DAVID, KEGG and GO analyses of DEGs involved in the significant
module were performed. The results demonstrated that genes in the
significant module were enriched in the following categories:
‘Cellular process’, ‘localization’, ‘signaling’, ‘cell’,
‘organelle’, ‘extracellular region’, ‘membrane’, ‘binding’,
‘molecular function regulator’ and ‘molecular transducer activity’
(Fig. 6). The enriched GO terms were
‘generation of precursor metabolites and energy’, ‘signal
transduction’, ‘regulation of biological processes’, ‘regulation of
cellular processes’, ‘cellular response to stimulus’, ‘cellular
component organization or biogenesis’, ‘cellular component
organization’, ‘anatomical structure morphogenesis’, and so on
(Fig. 7A). The KEGG pathway analysis
revealed that genes in the significant module were mainly enriched
in ‘vasopressin-regulated water reabsorption’, ‘proteoglycans in
cancer’, ‘thyroid hormone signaling pathway’, ‘phagosome’, ‘focal
adhesion’, ‘microRNAs in cancer’, ‘bladder cancer’, ‘pathogenic
Escherichia coli infection’, ‘endometrial cancer’ and
‘shigellosis’ (Fig. 7B).
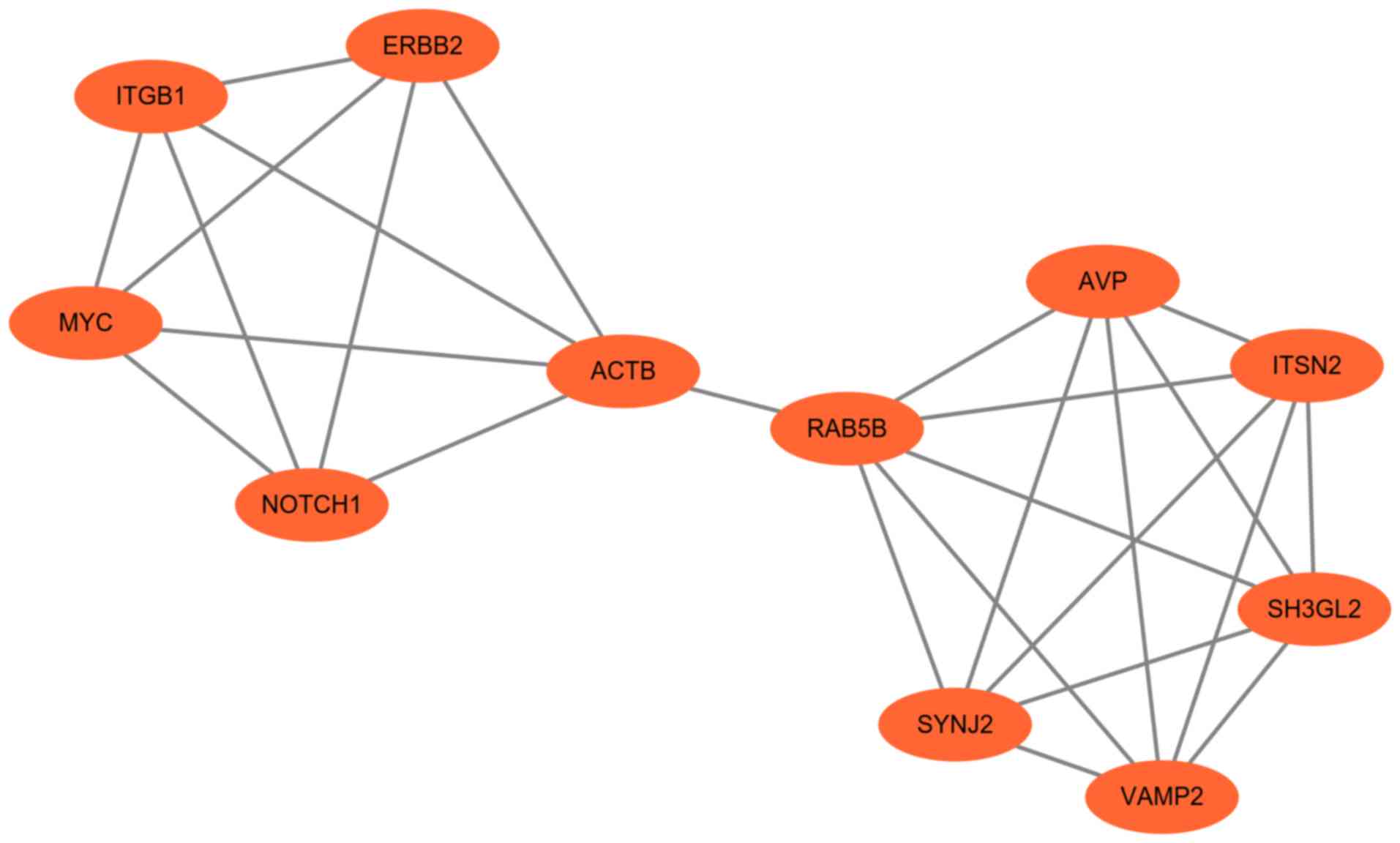 | Figure 5.Significant module obtained from the
protein-protein interaction network of differentially expressed
genes using Molecular Complex Detection. The significant module
included 11 nodes and 26 edges. ACTB, actin β; AVP, arginine
vasopressin; ERBB2, ERb-b2 receptor tyrosine kinase 2; ITGB1,
integrin subunit β1; ITSN2, intersectin 2; RAB5B, RAB5B, member RAS
oncogene family; SH3GL2, SH3 domain containing GRB2 like 2,
endophilin A1; SYNJ2, synaptojanin 2; VAMP2, vesicle associated
membrane protein 2. |
Hub gene selection and analysis
Degrees ≥10 was considered as the criterion of
judgment. A total of 11 genes were identified as hub genes using
Cytoscape: ITGB1, MYC, ERBB2, NOTCH1, ACTB, RAB5B, arginine
vasopressin (AVP), synaptojanin 2 (SYNJ2), ITSN2, SH3 domain
containing GRB2 like 2 endophilin A1 and vesicle associated
membrane protein 2 (VAMP2; Table I).
A co-expression network of these significant genes was obtained
using cBioPortal (Fig. 8). The BP,
CC and MF analyses by BiNGO for these genes supported the results
of the GO analysis (Figs.
S1–S3). The results of the
BiNGO analysis demonstrated that variations in the BP were also
mainly enriched for the ‘cellular process’ term (Fig. S1). Changes in CC were also enriched
in cell, membrane (‘plasma membrane’ and ‘plasma membrane part’)
and organelle terms (Fig. S2).
Additionally, the variations in MF were enriched in binding
(‘kinesin binding’, ‘nitric-oxide synthase binding’ and ‘Hsp90
protein binding’), ‘molecular transducer activity’ (Fig. S3).
 | Table I.Summary of the functions of the 11
hub genes. |
Table I.
Summary of the functions of the 11
hub genes.
| No. | Gene symbol | Full name | Function | (Refs.) |
|---|
| 1 | ITGB1 | Integrin subunit
β1 | Diseases associated
with ITGB1 include gallbladder cancer and breast cancer. Among its
related pathways are ERK signaling and focal adhesion. Gene
Ontology annotations related to this gene include protein
heterodimerization activity and signaling receptor binding. | (49) |
| 2 | MYC | V-myc avian
myelocytomatosis viral oncogene homolog | Activates the
transcription of growth-related genes. Binds to the VEGFA promoter,
promoting VEGFA production and subsequent sprouting
angiogenesis. | (52) |
| 3 | ERBB2 | Erb-b2 receptor
tyrosine kinase 2 | Regulates outgrowth
and stabilization of peripheral microtubules. Involved in the
transcription of ribosomal RNA genes by RNA Pol I and enhances
protein synthesis and cell growth. | (53) |
| 4 | NOTCH1 | Notch 1 | Affects the
implementation of differentiation, proliferation and apoptotic
programs. Involved in angiogenesis; negatively regulates
endothelial cell proliferation and migration and angiogenic
sprouting. | (54) |
| 5 | ACTB | Actin β | Among its related
pathways are ERK signaling and cytoskeleton remodeling regulation
of actin cytoskeleton by ρ GTPases. | (55) |
| 6 | RAB5B | RAB5B, member RAS
oncogene family | Protein transport.
Probably involved in vesicular traffic. | (56) |
| 7 | AVP | Arginine
vasopressin | Vasopressin has a
direct antidiuretic action on the kidney, it also causes
vasoconstriction of the peripheral vessels. | (57) |
| 8 | SYNJ2 | Synaptojanin 2 | Inositol
5-phosphatase which may be involved in distinct membrane
trafficking and signal transduction pathways. May mediate the
inhibitory effect of Rac1 on endocytosis. | (58) |
| 9 | ITSN2 | Intersectin 2 | May regulate the
formation of CCPs. Seems to be involved in CCPs maturation
including invagination or budding. | (59) |
| 10 | SH3GL2 | SH3 domain
containing GRB2 like 2, endophilin A1 | Implicated in
synaptic vesicle endocytosis. Cooperates with SH3GL2 to mediate
brain derived neurotrophic factor-neurotrophic receptor tyrosine
kinase 2 early endocytic trafficking and signaling from early
endosomes. | (60) |
| 11 | VAMP2 | Vesicle associated
membrane protein 2 | Involved in the
targeting and/or fusion of transport vesicles to their target
membrane. Modulates the gating characteristics of the delayed
rectifier voltage-dependent potassium channel potassium
voltage-gated channel subfamily B member 1. | (61) |
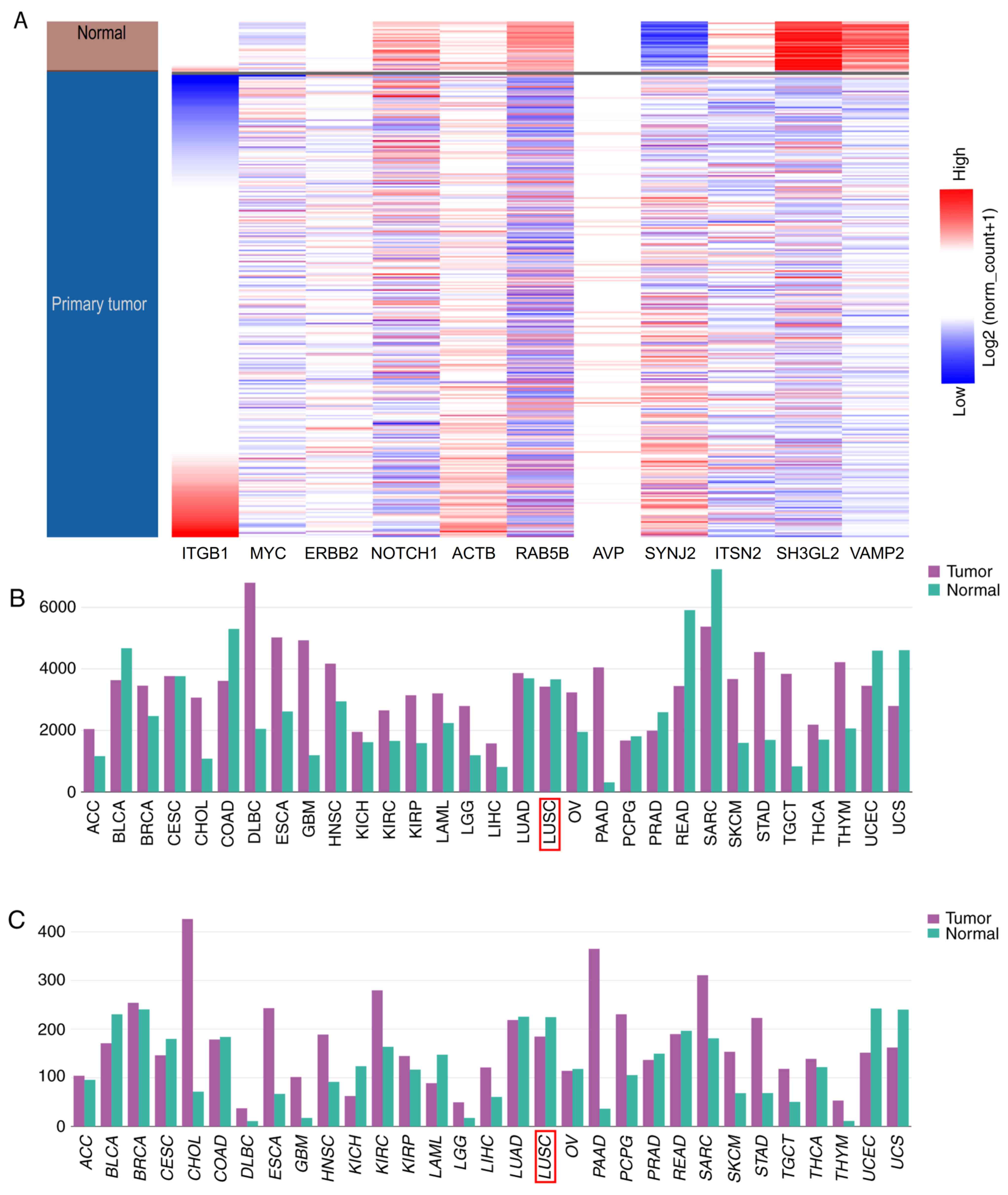
Hierarchical clustering revealed that the hub genes
could differentiate the MM LC samples from the non-MM LC samples
(Fig. 9). Subsequently, a
Kaplan-Meier plotter was used to perform overall survival (OS)
analysis (Figs. 10 and 11). The samples for OS analysis, derived
from the Kaplan-Meier plotter, were different from those used in
the analysis of DEGs. Patients with LC with genomic alterations in
high expression of ITGB1 (Fig.
10A), high expression of MYC (Fig.
10B), low expression of NOTCH1 (Fig. 10D), high expression of ACTB
(Fig. 10E), high expression of AVP
(Fig. 11B), low expression of SYNJ2
(Fig. 11C), low expression of ITSN2
(Fig. 11D) and high expression of
VAMP2, exhibited poorer OS. However, via the GEPIA, the expression
of ACTB (Fig. 10F), ERBB2 (Fig. 10C) RAB5B (Fig. 11A) and SH3GL2 (Fig. 11E), were not associated with OS. In
the UCSC Xena analysis, hierarchical clustering revealed that these
hub genes could differentiate the patients with LC from the normal
patients (Fig. 12A). Among the hub
genes, ACTB and ITGB1 had the highest score of 6.415, suggesting
that they may serve important roles in the occurrence or
development of MM LC. Using the database of the Kaplan-Meier
plotter, the present study identified that high expression of ACTB
was associated with poor OS in patients with LE (P<0.001).
Additionally, high expression of ITGB1 was associated with worse OS
(P=0.024). The expression profiles of ACTB and ITGB1 in human
tissues were visualized using GEPIA. The present study revealed
that ACTB and ITGB1 exhibited lower expression levels in lung
squamous cell carcinoma compared with the matched normal samples,
but no statistically significant difference was identified
(P>0.05; Fig. 12B and C).
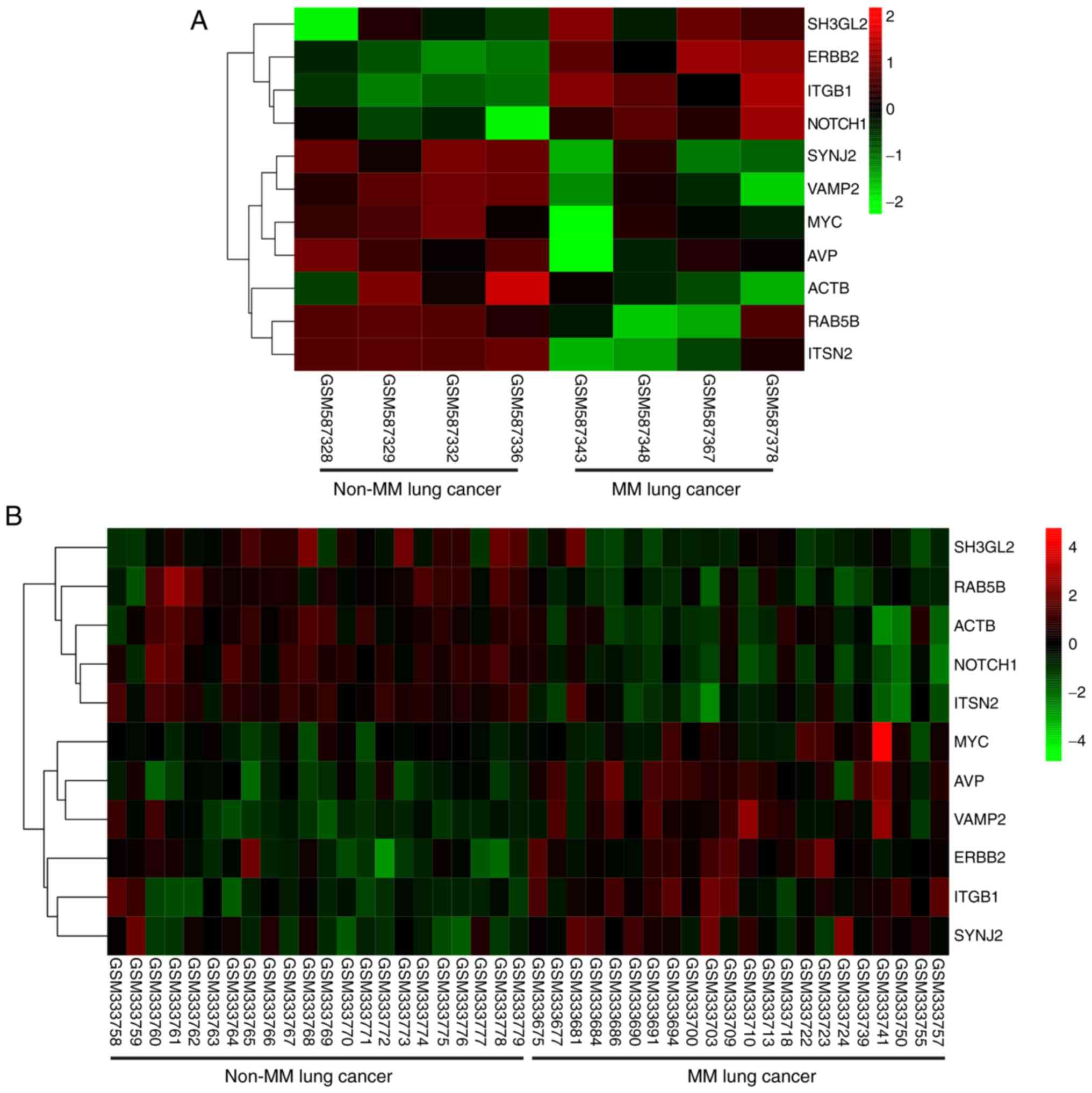 | Figure 9.Hierarchical clustering reveals that
the hub genes may differentiate the MM lung cancer samples from the
non-MM lung cancer samples. This was conducted in the (A) GSE23822
and (B) GSE13213 datasets. Upregulation of genes is marked in red
and downregulation of genes is marked in green. ACTB, actin β; AVP,
arginine vasopressin; ERBB2, ERb-b2 receptor tyrosine kinase 2;
ITGB1, integrin subunit β1; ITSN2, intersectin 2; MM lung cancer,
lung cancer with mediastinal lymph node metastasis; non-MM lung
cancer, lung cancer without mediastinal lymph node metastasis;
RAB5B, RAB5B, member RAS oncogene family; SH3GL2, SH3 domain
containing GRB2 like 2, endophilin A1; SYNJ2, synaptojanin 2;
VAMP2, vesicle associated membrane protein 2. |
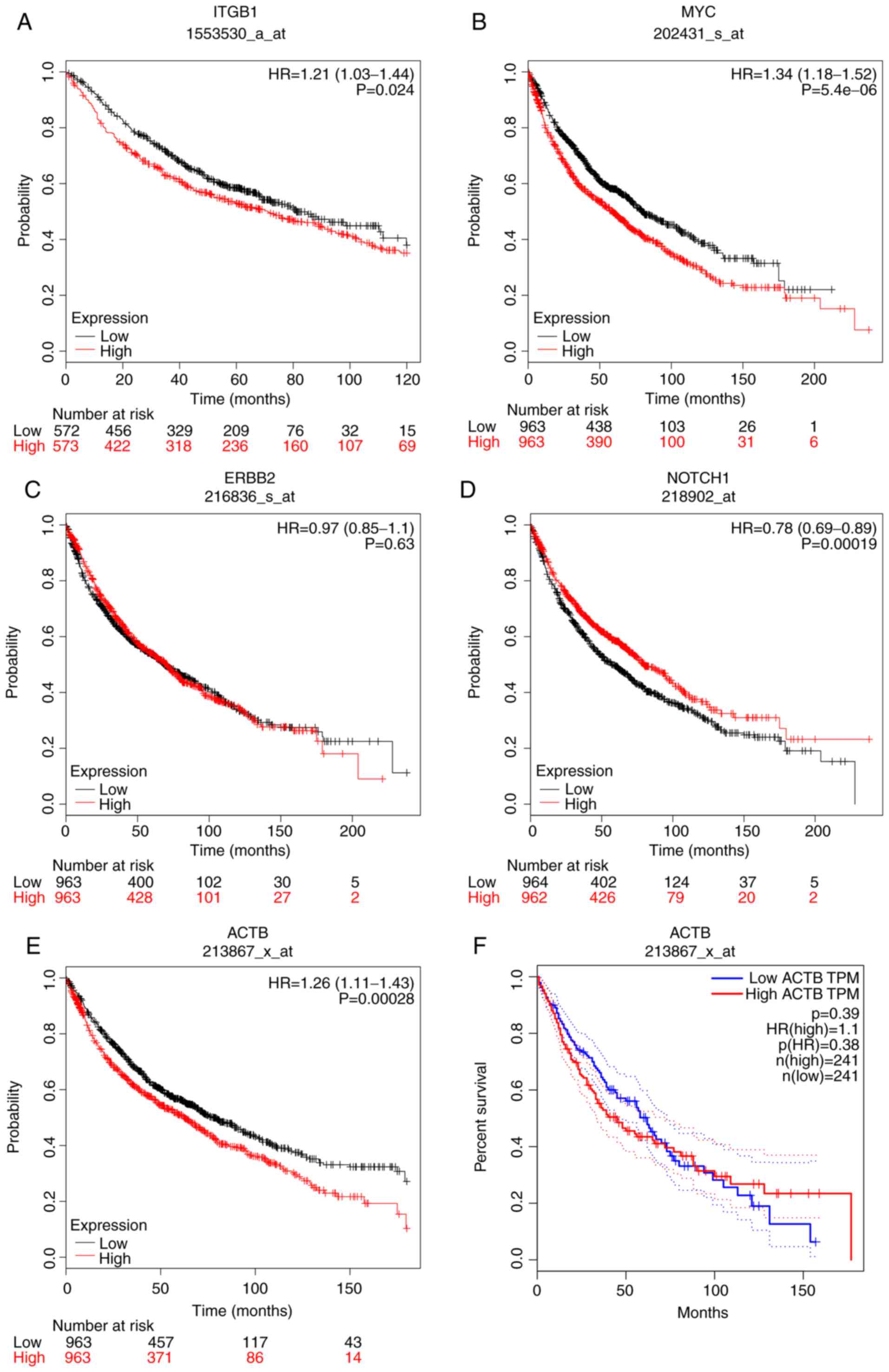 | Figure 10.Overall survival analysis of five hub
genes (A) Overall survival analysis of ITGB1, (B) MYC, (C) ERBB2,
(D) NOTCH1, (E) ACTB, using a Kaplan-Meier plotter online platform.
(F) Overall survival analysis of ACTB using GEPIA. P<0.05 was
considered to indicate a statistically significant difference.
ACTB, actin β; ERBB2, ERb-b2 receptor tyrosine kinase 2; HR, hazard
ratio; ITGB1, integrin subunit β1; TPM, transcripts per
million. |
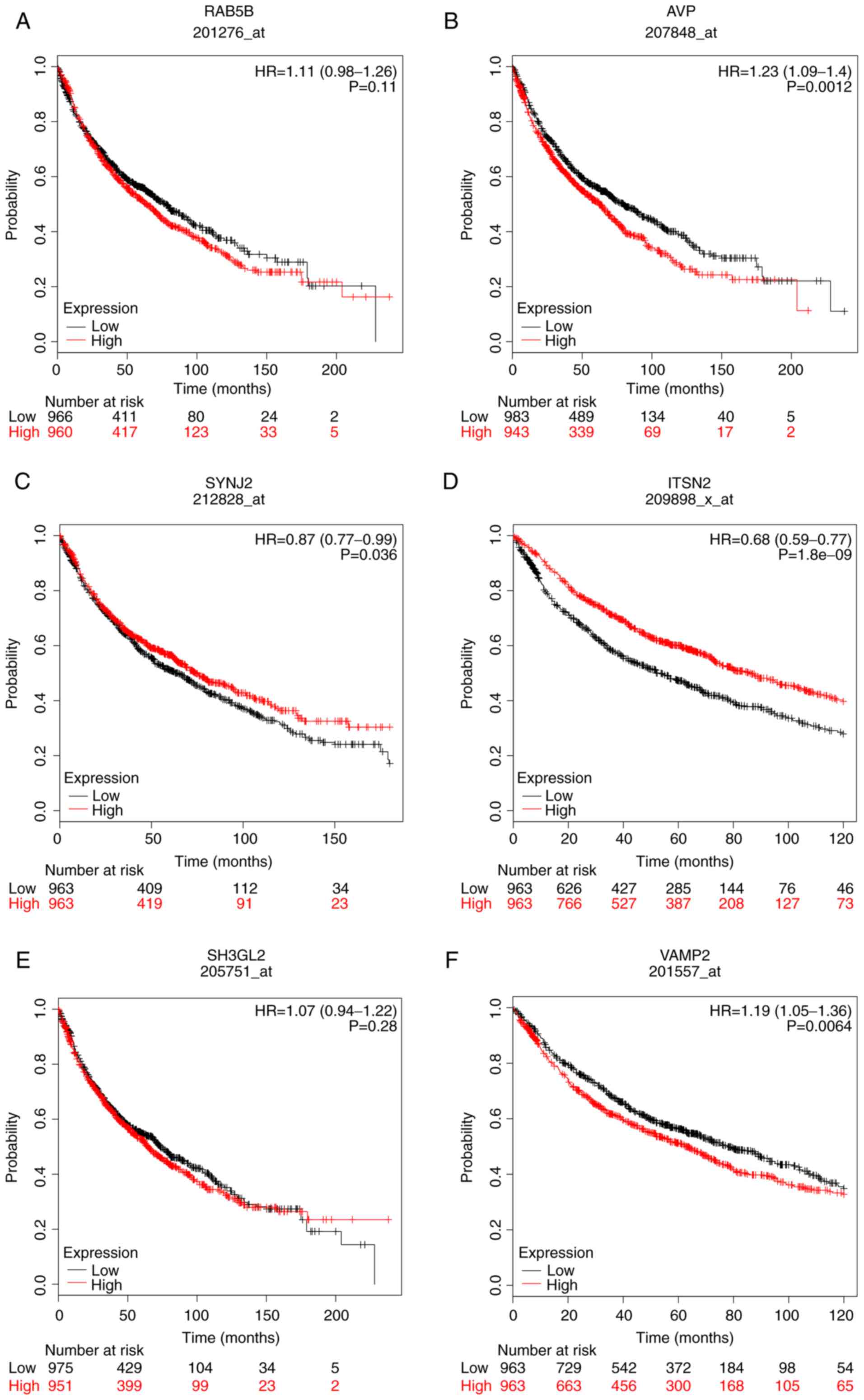 | Figure 11.Overall survival analysis of the
other six hub genes using a Kaplan-Meier plotter online platform.
Overall survival analysis of (A) RAB5B, (B) AVP, (C) SYNJ2, (D)
ITSN2, (E) SH3GL2 and (F) VAMP2. P<0.05 was considered to
indicate a statistically significant difference. AVP, arginine
vasopressin; HR, hazard ratio; ITSN2, intersectin 2; RAB5B, RAB5B,
member RAS oncogene family; SH3GL2, SH3 domain containing GRB2 like
2, endophilin A1; SYNJ2, synaptojanin 2; VAMP2, vesicle associated
membrane protein 2. |
Discussion
LC causes the highest cancer-associated mortality
rate in China and the majority of countries worldwide. The most
common pathological type of LC is non-small-cell LC, which accounts
for 80–85% of LC cases (33).
Optimization of LC treatment strategies depends on accurate
pathological staging and International Association for the Study of
Lung Cancer pathological TNM staging (34). In clinical practice, patients with
stages 0, I, II and IIIa may benefit from surgery. For patients
with MLNM, neoadjuvant chemotherapy could prolong their
postoperative survival (35). For
patients without surgical indications, the definite diagnosis of
mediastinal lymph node staging could effectively reduce the
irradiation area of the radiation target area, which may reduce the
incidence of radioactive lung injury. Therefore, if MLNM can
clearly defined prior to treatment, it is of great significance to
develop appropriate treatment schemes to improve the prognosis of
patients.
Currently, there are numerous clinical diagnostic
methods for MLNM of LC, including computed tomography (CT),
positron emission tomography (PET) and PET-CT. CT is the most
widely used diagnostic method for MLNM. A previous study
demonstrated that imaging methods continue to have limitations in
the evaluation of MLNM, and some patients cannot be clearly
diagnosed or are misdiagnosed (36).
Diagnosis of MLNM using CT is mainly based on the size of lymph
nodes. The larger the lymph nodes, the higher the metastasis rate.
The average size of the short diameter of normal lymph nodes
depends on the region in which they are located. McLoud et
al (37) evaluated 443 lymph
nodes in 143 patients using CT. The sensitivity to lymph nodes of
various regions was 17–78%, and the specificity was 72–94%. The
previous study demonstrated that there were some differences in the
sensitivity and specificity of diagnosis using CT to evaluate
mediastinal lymph nodes in different subgroups (31). In addition, PET is superior to CT in
the diagnosis of MLNM, since PET considers not only lymph node
size, but also lymph node metabolism information. A meta-analysis
by Toloza et al (38) of PET
examination of 1,045 patients in 18 studies manifested that PET was
more accurate than CT, and the total sensitivity and specificity
were 0.84 (95% CI, 0.78–0.89) and 0.89 (95% CI, 0.83–0.93),
respectively. However, the diagnostic method of PET has relatively
high rates of false negatives and false positives (39). Furthermore, PET-CT effectively
combines the technical advantages of CT and PET, which could
significantly improve the accuracy of preoperative diagnosis of
MLNM in LC. However, several previous studies have reported that
PET-CT has high specificity and low sensitivity (40,41). The
sensitivity of PET-CT in preoperative assessment of MLNM of lung
adenocarcinoma is too low, and further surgical staging is required
for patients with LC without MLNM detected by PET-CT.
To overcome the limitations of imaging evaluation of
MLNM, previous studies have attempted to identify molecular
biomarkers of MM LC (7–10). During the past decades,
bioinformatics technology has been generally used to screen
potential genetic targets of diseases, which assisted the
authentication of DEGs and underlying pathways associated with the
occurrence and recurrence of diseases.
Following analysis of the two microarray datasets in
the present study, DEGs between non-MM LC and MM LC were
identified. A total of 308 DEGs were contained in the two datasets
simultaneously. From the KEGG and GO analyses, the interactions of
the DEGs were explored. The DEGs were mainly enriched in the GO
terms ‘cellular process’, ‘signaling’, ‘cell’, ‘organelle’,
‘binding’, ‘molecular transducer activity’ and ‘molecular function
regulator’. Among the hub genes, ACTB and ITGB1 exhibited the
highest score of 6.415 via the MCODE analysis, suggesting that they
may serve important roles in the occurrence or development of MM
LC.
Actin β, encoded by the ACTB gene, is widely present
in non-muscle cells in the form of a ball or fiber, and
participates in the construction of the cytoskeleton and cell
movement. As a downstream regulatory protein, actin β has the
function of maintaining normal cell migration, growth,
differentiation and signal transduction (42). Therefore, it may also be involved in
the occurrence mechanism of vascular remodeling. Numerous previous
studies (43–45) have demonstrated a close association
between ACTB and the occurrence of tumors. Lim et al
(43) reported that the mutation of
ACTB may cause pilocytic astrocytoma in their clinical experience.
Furthermore, the fusions of ACTB and glioma-associated oncogene
homolog 1 (GLI1) were regarded as a specific genetic abnormality,
which could result in a distinctive type of actin-positive,
perivascular myoid tumors, known as ‘pericytoma with the t (7;12)
translocation’ (44). Furthermore,
Castro et al (45) reported
that the extremely unusual translocation t (7;12) may lead to the
gene fusion of ACTB and GLI1, which may induce an infrequent
gastric tumor derived from the pyloric wall of the stomach. The
results of the present study revealed that the expression levels of
ACTB in MM LC were downregulated; therefore, the production of
actin β was reduced, which may lead to abnormal growth,
differentiation and exfoliation of LC cells. The detached cancer
cells first enter the mediastinal lymph nodes to form a cancer
embolus. According to the Kaplan-Meier survival analysis, patients
with low expression levels of ACTB had a good prognosis
(P<0.05). However, the expression levels of ACTB had no
significant effects on the prognosis based on the survival analysis
of GEPIA (P>0.05). The influence of ACTB expression on the
prognosis was undefined; therefore, more data are required to
verify the suggested effect.
ITGB1 is a member of the integrin family of
proteins. Integrin family proteins are involved in the regulation
of cell adhesion and recognition processes, including hemostasis,
embryogenesis, immune response, tissue repair and tumor cell
metastasis (46). Yan et al
(47) reported that the expression
levels of ITGB1 were associated with OS and metastasis in patients
with aggressive breast cancer. Wang et al (48) reported that linc-ITGB1 promoted the
invasion and migration of gallbladder cancer cells by activating
epithelial-mesenchymal transition, and knockout of ITGB1
significantly inhibited the metastasis and invasion of gallbladder
cancer cells. Klahan et al (49) knocked out ITGB1 in breast cancer
cells and revealed that calcium influx decreased, resulting in a
significant decrease in the invasion and metastasis of
triple-negative breast cancer cells. Wang et al (50) reported that ITGB1 serves important
roles in the occurrence and metastasis of LC. The findings of Qin
et al (51) suggested that
microRNA-134 suppresses migration and invasion of non-small cell LC
by targeting ITGB1. The present study demonstrated that the
expression levels of ITGB1 were upregulated in MM LC. According to
the OS analysis, patients with high expression levels of ITGB1 had
a poor prognosis. The reason for this may be that high ITGB1
expression induces the occurrence of MLNM, which could invade the
systemic organs along the lymphatic duct, causing systemic organ
failure and a shorter lifespan. Based on the aforementioned
results, the alterations in the expression levels of ITGB1 may be a
molecular mechanism for stimulating the metastasis and invasion of
LC cells to the mediastinal lymph node. However, currently, studies
regarding IGTB1 are rare, so more efforts should be made in the
future.
There are some limitations of the present study.
First, the results of the present study are based on bioinformatics
analysis only. Therefore, they require laboratory work to be
verified using a large set of samples of patients with LC.
Currently, it is difficult to obtain the ethical approval documents
and informed consent. In the next stage of research, ethical
approval and informed consent will be obtained to perform
verification of the results of the present study in humans and
animals.
In conclusion, the present study aimed to identify
DEGs which may be involved in the occurrence or development of LC.
Finally, 308 DEGs and 11 hub genes were identified by comparisons
between MM LC and non-MM LC samples, which could be used as
diagnostic and therapeutic biomarkers for MM LC. The present study
provided novel insight for the diagnosis and treatment of MM LC.
The results suggested that data mining and integration could be a
promising tool to predict biomarkers of malignant tumors. However,
the present study is only a preliminary report, and the number of
samples in the present study was limited. Since cancer biomarkers
only have meaning if they are integrated with clinical data,
further experiments should be conducted to confirm the conclusions
of the present study.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
NZ performed the experiment, and was a major
contributor in writing and submitting the manuscript. SWZ made
substantial contributions to the conception and design of the
study, as well as the acquisition, analysis and interpretation of
the data, and also designed the draft of the research process. NZ
was involved in critically revising the manuscript for intellectual
content. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ACTB
|
actin β
|
|
ADAM
|
ADAM metallopeptidase domain
|
|
AVP
|
arginine vasopressin
|
|
BiNGO
|
Biological Networks Gene Oncology
tool
|
|
BP
|
biological processes
|
|
CC
|
cellular components
|
|
CXCR4
|
C-X-C motif chemokine receptor 4
|
|
DEGs
|
differentially expressed genes
|
|
ERBB2
|
Erb-b2 receptor tyrosine kinase 2
|
|
GEO
|
Gene Expression Omnibus
|
|
GEPIA
|
Gene Expression Profiling Interactive
Analysis
|
|
GO
|
Gene Ontology
|
|
ITGB1
|
integrin subunit β1
|
|
ITSN2
|
intersectin 2
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
LC
|
lung cancer
|
|
MCODE
|
Molecular Complex Detection
|
|
MF
|
molecular function
|
|
MLNM
|
mediastinal lymph node metastasis
|
|
MM LC
|
lung cancer with mediastinal lymph
node metastasis
|
|
non-MM LC
|
lung cancer samples without
mediastinal lymph node metastasis
|
|
OS
|
overall survival
|
|
PET
|
positron emission tomography
|
|
PPI
|
protein-protein interaction
|
|
RAB5B
|
RAB5B, member RAS oncogene family
|
|
SYNJ2
|
synaptojanin 2
|
|
VEGF-C
|
vascular endothelial growth
factor-C
|
|
VEGF-D
|
vascular endothelial growth
factor-D
|
|
VEGFR-3
|
vascular endothelial growth factor
receptor-3
|
|
VAMP2
|
vesicle associated membrane protein
2
|
References
|
1
|
Lopez-Pastorini A, Riedel R, Koryllos A,
Beckers F, Ludwig C and Stoelben E: The impact of preoperative
elevated serum C-reactive protein on postoperative morbidity and
mortality after anatomic resection for lung cancer. Lung Cancer.
109:68–73. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Isaka M, Kojima H, Takahashi S, Omae K and
Ohde Y: Risk factors for local recurrence after lobectomy and lymph
node dissection in patients with non-small cell lung cancer:
Implications for adjuvant therapy. Lung Cancer. 115:28–33. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yeh YC, Kadota K, Nitadori J, Sima CS,
Rizk NP, Jones DR, Travis WD and Adusumilli PS: International
association for the study of lung cancer/American thoracic
society/European respiratory society classification predicts occult
lymph node metastasis in clinically mediastinal node-negative lung
adenocarcinoma. Eur J Cardiothorac Surg. 49:e9–e15. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang S, Zhou W, Zhang H, Zhao M and Chen
X: Analysis of predictive factors for postoperative survival for
non small cell lung carcinoma patients with unexpected mediastinal
lymph nodes metastasis. Thorac Cardiovasc Surg. 62:126–132. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ma K, Chang D, He B, Gong M, Tian F, Hu X,
Ji Z and Wang T: Radical systematic mediastinal lymphadenectomy
versus mediastinal lymph node sampling in patients with clinical
stage IA and pathological stage T1 non-small cell lung cancer. J
Cancer Res Clin Oncol. 134:1289–1295. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Saintigny P, Kambouchner M, Ly M, Gomes N,
Sainte-Catherine O, Vassy R, Czernichow S, Letoumelin P, Breau JL,
Bernaudin JF and Kraemer M: Vascular endothelial growth factor-C
and its receptor VEGFR-3 in non-small-cell lung cancer: Concurrent
expression in cancer cells from primary tumour and metastatic lymph
node. Lung Cancer. 58:205–213. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ohtsuka T, Shiomi T, Shimoda M, Kodama T,
Amour A, Murphy G, Ohuchi E, Kobayashi K and Okada Y: ADAM28 is
overexpressed in human non-small cell lung carcinomas and
correlates with cell proliferation and lymph node metastasis. Int J
Cancer. 118:263–273. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Na IK, Scheibenbogen C, Adam C, Stroux A,
Ghadjar P, Thiel E, Keilholz U and Coupland SE: Nuclear expression
of CXCR4 in tumor cells of non-small cell lung cancer is correlated
with lymph node metastasis. Hum Pathol. 39:1751–1755. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Maekawa S, Iwasaki A, Shirakusa T, Enatsu
S, Kawakami T and Kuroki M and Kuroki M: Correlation between lymph
node metastasis and the expression of VEGF-C, VEGF-D and VEGFR-3 in
T1 lung adenocarcinoma. Anticancer Res. 27:3735–3741.
2007.PubMed/NCBI
|
|
11
|
Li J, Li BL, Zhang HQ, Xu SF, Liu ZD, Yue
WT and Han Y: Relationship between vascular endothelial growth
factor C expression level and lymph node metastasis in non small
cell lung cancer. Zhonghua Yi Xue Za Zhi. 88:2982–2985.
2008.PubMed/NCBI
|
|
12
|
Kikuchi T, Daigo Y, Katagiri T, Tsunoda T,
Okada K, Kakiuchi S, Zembutsu H, Furukawa Y, Kawamura M, Kobayashi
K, et al: Expression profiles of non-small cell lung cancers on
cDNA microarrays: Identification of genes for prediction of
lymph-node metastasis and sensitivity to anti-cancer drugs.
Oncogene. 22:2192–2205. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li L, Lei Q, Zhang S, Kong L and Qin B:
Screening and identification of key biomarkers in hepatocellular
carcinoma: Evidence from bioinformatic analysis. Oncol Rep.
38:2607–2618. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kan T, Shimada Y, Sato F, Ito T, Kondo K,
Watanabe G, Maeda M, Yamasaki S, Meltzer SJ and Imamura M:
Prediction of lymph node metastasis with use of artificial neural
networks based on gene expression profiles in esophageal squamous
cell carcinoma. Ann Surg Oncol. 11:1070–1078. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
O'Donnell RK, Kupferman M, Wei SJ, Singhal
S, Weber R, O'Malley B, Cheng Y, Putt M, Feldman M, Ziober B and
Muschel RJ: Gene expression signature predicts lymphatic metastasis
in squamous cell carcinoma of the oral cavity. Oncogene.
24:1244–1251. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nguyen ST, Hasegawa S, Tsuda H, Tomioka H,
Ushijima M, Noda M, Omura K and Miki Y: Identification of a
predictive gene expression signature of cervical lymph node
metastasis in oral squamous cell carcinoma. Cancer Sci. 98:740–746.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim TJ, Choi JJ, Kim WY, Choi CH, Lee JW,
Bae DS, Son DS, Kim J, Park BK, Ahn G, et al: Gene expression
profiling for the prediction of lymph node metastasis in patients
with cervical cancer. Cancer Sci. 99:31–38. 2008.PubMed/NCBI
|
|
18
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wright CM, Savarimuthu Francis SM, Tan ME,
Martins MU, Winterford C, Davidson MR, Duhig EE, Clarke BE, Hayward
NK, Yang IA, et al: MS4A1 dysregulation in asbestos-related lung
squamous cell carcinoma is due to CD20 stromal lymphocyte
expression. PLoS One. 7:e349432012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tomida S, Takeuchi T, Shimada Y, Arima C,
Matsuo K, Mitsudomi T, Yatabe Y and Takahashi T: Relapse-related
molecular signature in lung adenocarcinomas identifies patients
with dismal prognosis. J Clin Oncol. 27:2793–2799. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets - update. Nucleic Acids Res. 41(Database issue):
D991–D995. 2013.PubMed/NCBI
|
|
22
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kanehisa M: The KEGG database. Novartis
Found Symp. 247:91–101. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ni M, Liu X, Wu J, Zhang D, Tian J, Wang
T, Liu S, Meng Z, Wang K, Duan X, et al: Identification of
candidate biomarkers correlated with the pathogenesis and prognosis
of non-small cell lung cancer via integrated bioinformatics
analysis. Front Genet. 9:4692018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43(Database issue): D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang HM, Jiang X, Hao ML, Shan MJ, Qiu Y,
Hu GF, Wang Q, Yu ZQ, Meng LB and Zou YY: Identification of
biomarkers in macrophages of atherosclerosis by microarray
analysis. Lipids Health Dis. 18:1072019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Maere S, Heymans K and Kuiper M: BiNGO: A
cytoscape plugin to assess overrepresentation of gene ontology
categories in biological networks. Bioinformatics. 21:3448–3449.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nagy Á, Lánczky A, Menyhárt O and Győrffy
B: Validation of miRNA prognostic power in hepatocellular carcinoma
using expression data of independent datasets. Sci Rep. 8:92272018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Detterbeck FC, Boffa DJ and Tanoue LT: The
new lung cancer staging system. Chest. 136:260–271. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Suntharalingam M, Paulus R, Edelman MJ,
Krasna M, Burrows W, Gore E, Wilson LD and Choy H: Radiation
therapy oncology group protocol 02-29: A phase II trial of
neoadjuvant therapy with concurrent chemotherapy and full-dose
radiation therapy followed by surgical resection and consolidative
therapy for locally advanced non-small cell carcinoma of the lung.
Int J Radiat Oncol Biol Phys. 84:456–463. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shim SS, Lee KS, Kim BT, Chung MJ, Lee EJ,
Han J, Choi JY, Kwon OJ, Shim YM and Kim S: Non-small cell lung
cancer: Prospective comparison of integrated FDG PET/CT and CT
alone for preoperative staging. Radiology. 236:1011–1019. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
McLoud TC, Bourgouin PM, Greenberg RW,
Kosiuk JP, Templeton PA, Shepard JA, Moore EH, Wain JC, Mathisen DJ
and Grillo HC: Bronchogenic carcinoma: Analysis of staging in the
mediastinum with CT by correlative lymph node mapping and sampling.
Radiology. 182:319–323. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Toloza EM, Harpole L and McCrory DC:
Noninvasive staging of non-small cell lung cancer: A review of the
current evidence. Chest 123(1 Suppl). 137S–146S. 2003. View Article : Google Scholar
|
|
39
|
Takamochi K, Yoshida J, Murakami K, Niho
S, Ishii G, Nishimura M, Nishiwaki Y, Suzuki K and Nagai K:
Pitfalls in lymph node staging with positron emission tomography in
non-small cell lung cancer patients. Lung Cancer. 47:235–242. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Billé A, Pelosi E, Skanjeti A, Arena V,
Errico L, Borasio P, Mancini M and Ardissone F: Preoperative
intrathoracic lymph node staging in patients with non-small-cell
lung cancer: Accuracy of integrated positron emission tomography
and computed tomography. Eur J Cardiothorac Surg. 36:440–445. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Billè A, Okiror L, Skanjeti A, Errico L,
Arena V, Penna D, Ardissone F and Pelosi E: Evaluation of
integrated positron emission tomography and computed tomography
accuracy in detecting lymph node metastasis in patients with
adenocarcinoma vs. squamous cell carcinoma. Eur J Cardiothorac
Surg. 43:574–579. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pavlyk I, Leu NA, Vedula P, Kurosaka S and
Kashina A: Rapid and dynamic arginylation of the leading edge
β-actin is required for cell migration. Traffic. 19:263–272. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lim YH, Burke AB, Roberts MS, Collins MT
and Choate KA: Multilineage ACTB mutation in a patient with
fibro-osseous maxillary lesion and pilocytic astrocytoma. Am J Med
Genet A. 176:2037–2040. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Antonescu CR, Agaram NP, Sung YS, Zhang L,
Swanson D and Dickson BC: A distinct malignant epithelioid neoplasm
with GLI1 gene rearrangements, frequent S100 protein expression,
and metastatic potential: Expanding the spectrum of pathologic
entities with ACTB/MALAT1/PTCH1-GLI1 fusions. Am J Surg Pathol.
42:553–560. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Castro E, Cortes-Santiago N, Ferguson LM,
Rao PH, Venkatramani R and López-Terrada D: Translocation t(7;12)
as the sole chromosomal abnormality resulting in ACTB-GLI1 fusion
in pediatric gastric pericytoma. Hum Pathol. 53:137–141. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Huang R and Rofstad EK: Integrins as
therapeutic targets in the organ-specific metastasis of human
malignant melanoma. J Exp Clin Cancer Res. 37:922018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yan M, Zhang L, Li G, Xiao S, Dai J and
Cen X: Long noncoding RNA linc-ITGB1 promotes cell migration and
invasion in human breast cancer. Biotechnol Appl Biochem. 64:5–13.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang L, Zhang Y, Lv W, Lu J, Mu J, Liu Y
and Dong P: Long non-coding RNA Linc-ITGB1 knockdown inhibits cell
migration and invasion in GBC-SD/M and GBC-SD gallbladder cancer
cell lines. Chem Biol Drug Des. 86:1064–1071. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Klahan S, Huang WC, Chang CM, Wong HS,
Huang CC, Wu MS, Lin YC, Lu HF, Hou MF and Chang WC: Gene
expression profiling combined with functional analysis identify
integrin beta1 (ITGB1) as a potential prognosis biomarker in triple
negative breast cancer. Pharmacol Res. 104:31–37. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang XM, Li J, Yan MX, Liu L, Jia DS, Geng
Q, Lin HC, He XH, Li JJ and Yao M: Integrative analyses identify
osteopontin, LAMB3 and ITGB1 as critical pro-metastatic genes for
lung cancer. PLoS One. 8:e557142013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Qin Q, Wei F, Zhang J and Li B: miR-134
suppresses the migration and invasion of nonsmall cell lung cancer
by targeting ITGB1. Oncol Rep. 37:823–830. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Demin DE, Bogolyubova AV, Zlenko DV,
Uvarova AN, Deikin AV, Putlyaeva LV, Belousov PV, Mitkin NA,
Korneev KV, Sviryaeva EN, et al: The novel short isoform of securin
stimulates the expression of cyclin D3 and angiogenesis factors
VEGFA and FGF2, but does not affect the expression of MYC
transcription factor. Mol Biol (Mosk). 52:508–518. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zaoui K, Benseddik K, Daou P, Salaün D and
Badache A: ErbB2 receptor controls microtubule capture by
recruiting ACF7 to the plasma membrane of migrating cells. Proc
Natl Acad Sci USA. 107:18517–18522. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Huang CC, Kuo HM, Wu PC, Cheng SH, Chang
TT, Chang YC, Kung ML, Wu DC, Chuang JH and Tai MH: Soluble
delta-like 1 homolog (DLK1) stimulates angiogenesis through
Notch1/Akt/eNOS signaling in endothelial cells. Angiogenesis.
21:299–312. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Joassard OR, Amirouche A, Gallot YS,
Desgeorges MM, Castells J, Durieux AC, Berthon P and Freyssenet DG:
Regulation of Akt-mTOR, ubiquitin-proteasome and autophagy-lysosome
pathways in response to formoterol administration in rat skeletal
muscle. Int J Biochem Cell Biol. 45:2444–2455. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Inoue J, Ninomiya M, Umetsu T, Nakamura T,
Kogure T, Kakazu E, Iwata T, Takai S, Sano A, Fukuda M, et al:
Small interfering RNA screening for the small GTPase rab proteins
identifies Rab5B as a major regulator of hepatitis B virus
production. J Virol. 93:e006212019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yamada K, Nakayama M, Miura Y, Nakano H,
Mimura N and Yoshida S: Role of AVP in the regulation of vascular
tonus and blood pressure in patients with chronic renal failure.
Regul Pept. 45:91–95. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Du Q, Guo X, Zhang X, Zhou W, Liu Z, Wang
J, Zhang T, Mao Z, Luo J, Jin T and Liu C: SYNJ2 variant rs9365723
is associated with colorectal cancer risk in Chinese Han
population. Int J Biol Markers. 31:e138–e143. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Nakatsu F, Perera RM, Lucast L, Zoncu R,
Domin J, Gertler FB, Toomre D and De Camilli P: The inositol
5-phosphatase SHIP2 regulates endocytic clathrin-coated pit
dynamics. J Cell Biol. 190:307–315. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Dasgupta S, Jang JS, Shao C, Mukhopadhyay
ND, Sokhi UK, Das SK, Brait M, Talbot C, Yung RC, Begum S, et al:
SH3GL2 is frequently deleted in non-small cell lung cancer and
downregulates tumor growth by modulating EGFR signaling. J Mol Med
(Berl). 91:381–393. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Caceres PS, Mendez M and Ortiz PA:
Vesicle-associated membrane protein 2 (VAMP2) but not VAMP3
mediates cAMP-stimulated trafficking of the renal Na+-K+-2Cl-
co-transporter NKCC2 in thick ascending limbs. J Biol Chem.
289:23951–23962. 2014. View Article : Google Scholar : PubMed/NCBI
|