Introduction
Colorectal cancer (CRC) is a common malignant
disease that occurs worldwide and is one of the leading causes of
cancer-associated mortality in humans (1,2).
Although chemotherapeutic and surgical treatments have improved the
5-year survival rate, >50% of patients present with metastasis
at the time of diagnosis, a primary explanation for the high 5-year
mortality (3). It is difficult to
cure patients with distant metastasis with CRC; therefore, it
imperative to identify novel targeted therapy genes (3).
Neural precursor cell-expressed,
developmentally-downregulated 9 (NEDD9), also called HEF1 and Cas-L
(4), is a multidomain scaffolding
protein, belonging to the crk-associated substrate family (5). The most thorough study of NEDD9
suggested that it coordinates adhesion, migration, invasion and
cascade reactions of Src and FAK signals (4–7).
Numerous studies have reported that NEDD9 modulates invasion and
metastasis of gastric cancer (8),
breast cancer (9), cervical cancer
(6), melanoma (5) and lung cancer (10). We hypothesized that NEDD9 was a
biomarker of tumor invasion and metastasis. Previous studies have
identified that NEDD9 is expressed in CRC and is closely associated
with invasion, metastasis and poor prognosis (11–13).
Nevertheless, the specific mechanisms of the effect of NEDD9 on CRC
have yet to be completely elucidated.
Epithelial-mesenchymal transition (EMT) is a
reversible process of differentiation that causes polarized
epithelial cells to lose epithelial characteristics and obtain
typical mesenchymal properties, and it has been reported that EMT
is closely associated with the progression of malignant tumors
(14–16). Studies have demonstrated that NEDD9
promotes tumor invasion and metastasis by activating EMT (9,17,18). The
c-Jun NH-terminal kinase (JNK) is a member of the family of
mitogen-activated protein (MAP) kinases (19), and is primarily associated with
proliferation, differentiation, apoptosis and migration (20,21).
Previous studies have reported that activated JNK promotes the
invasion and metastasis of tumors by promoting the development of
EMT (22–24).
The aim of the present study was to evaluate whether
NEDD9 promoted cell invasion and migration by activating the
JNK/EMT signaling pathways in colorectal tumors.
Materials and methods
Cells and tissues
Colorectal cancer cells HCT116 (Jikai gene,
Shanghai, China) and normal colorectal tissue cells FHC (Suzhou
Jikai Gene Technology Co., Ltd. Shanghai, China) were cultured in
RPMI-1640 with 10% fetal bovine serum (ExCell Bio, Shanghai,
China). Trypsin-EDTA (Beijing Solarbio Science and Technology Co.,
Ltd., Beijing, China) was used to digest cells. Tissue samples were
from Qingdao Municipal Hospital between February 2016 and December
2017. Written informed consent was provided by patients. The
present study was approved by the Ethics Committee of the
Affiliated Hospital of Qingdao University (Qingdao, China). The
clinicopathological features including age, sex, stage of Tumor
Node Metastasis (25), data and
NEDD9 expression status. The inclusion criterion were the
following: <75 years; clinically proven colorectal cancer tissue
and no prior cancer chemotherapy. The exclusion criterion were the
following: ≥75 years and prior cancer chemotherapy.
Transfection
The lentiviral downregulation vector (lv-nedd9)
(Jikai Gene Chemical Technology Co., Ltd.) and the blank vector
(lv-nc) (Jikai Gene Chemical Technology Co., Ltd.) expression of
the target gene NEDD9 were constructed. The Lv-NEDD9 and Lv-NC two
groups of cells were seeded in 96-well plates at a density of
10,000 cells/well, and a Multiplicity of Infection=25 lentiviral
vector and a transfection enhancer Polybrene (Jikai Gene Chemical
Technology Co., Ltd.) were added simultaneously 24 h later. The
culture solution was changed after 10 h. The culture was kept for
10 days and then used for experimental research.
Transwell assays
Cells in each group (Lv-NC and Lv-NEDD9) were placed
in the Transwell chambers at a density of 30,000 cells/well (8 µm,
Corning, USA). Serum-free medium with 200 µl was added to the upper
compartment, and 500 µl fetal bovine serum was added to the lower
compartment. After 24 h of culture, the cells in the upper chamber
were wiped away. After being fixed with methanol for 10 min and
stained with 0.2% crystal violet (Solarbio, Beijing, China) for 10
min at 25°C, the cells were washed with PBS three times, then
images were obtained under an optical microscope (magnification,
×400) and counted.
Invasive ability test: Matrigel matrix (Corning
Inc.) was mixed with complete medium in a 1:9 ratio. The diluted
mixture was added to the Transwell chamber at 100 µl/well, in a
37°C incubator for 4 h. Cells in each group were placed into
Transwell chambers with 50,000 cells/well (8-µm, Corning Inc.), and
the other steps were the same as the migration experiment.
For HCT116 cell lines, the JNK inhibitor SP600125
(Cell Signaling Technology, Inc.) was dissolved in DMSO and added.
This was the experimental group (SP600125 group). By contast, DMSO
alone (Beijing Solarbio Science and Technology Co, Ltd.) was the
control group (DMSO group). The invasion and metastasis capacities
of cells in each were measured by Transwell experiments. The
experimental method was carried out as aforementioned.
Wound healing assay
Cells in each group (Lv-NC and Lv-NEDD9) were placed
in the six-well plates (Corning, Inc.) at a density of 500,000
cells/well. After 24 h, the cells were scratched with a 200 µl
sterile pipette tip and photographed at 0 and 24 h. DMSO and
SP600125 groups in the scratch test were the same as described
earlier.
cDNA synthesis and RT-qPCR
Tissue and cell total RNA (Takara, Japan) was
extracted using an ISO plus RNA extraction kit (Takara Bio, Inc.).
A PrimeScript RT reagent kit with gDNA Eraser (Perfect Real Time)
was used for reverse transcription (Takara Bio, Inc., Otsu, Japan).
SYBR Premix Ex Taq RR420A (Takara Bio, Inc.) was used for
quantitative-PCR (RT-qPCR) (Bio-Rad Laboratories, Inc. CFX96) and
the thermocycling conditions were as follows: 30 sec, 1 cycle, 95°C
for 5 sec, 60°C for 30 sec, 40 cycles. The expression levels were
analysed by 2−ΔΔCq method (26). Primers used in the experiment were
purchased from Shanghai Sangon Biotech. NEDD9: Forward:
5′-GAGCTGGATGGATGACTACGA-3′; Reverse: 5′-AGCTCTTTCTGTTGCCTCTCA-3′.
E-cadherin: Forward: 5′-CTGATTCTGCTGCTCTTGCTG-3′; Reverse:
5′-CTTCTCCGCCTCCTTCTTCA-3′. Vimentin: Forward:
5′-GAAATTGCAGGAGGAGATGC-3′; Reverse: 5′-ATTCCACTTTGCGTTCAAGG-3′.
β-actin: Forward: 5′-CACCATGAAGATCAAGATCATTGC-3′; Reverse:
5′-GGCCGGACTCATCGTACTCCTGC-3′.
Western blot analysis
The four groups cells, Lv-NEDD9, Lv-NC, DMSO and
SP600125 (3 ml 50 µM SP600125 for 5 h at 37°C), were washed twice
with PBS and 1 ml PBS was added. The cells in the culture flask
were scraped off with a cell scalpel and centrifuged for 5 min at
4°C at 5,000 × g. Following removal of the supernatant, RIPA lysis
buffer (Beijing Kangwei Century Biotechnology Co., Ltd.) was used
to extract whole protein. After adding protease and phosphatase
inhibitors, proteins were placed on ice for 15 min. After
vortexing, proteins were again placed on ice for 15 min. The
lysates were centrifuged at 12,000 × g for 10 min. The supernatants
contained total protein, the protein determination (Thermo Fisher
Scientific, Inc.) method was the BCA method. The extracted protein
was mixed with the sample buffer SDS-loading buffer, and boiled for
5 min. A total of 15 µl of protein was loaded per lane. Samples
were separated by 10% gel electrophoresis. Proteins were then
transferred to PVDF membranes (EMD Millipore). The membranes were
blocked in 5% skim milk for 1 h at room temperature. NEDD9, JNK,
β-actin, E-cadherin and vimentin antibodies were incubated
overnight at 4°C. The enhanced chemiluminescent substrate reagent
(ECL) was applied to the film and was analyzed on a Quant LAS 4010
imaging system (Ultra-Violet Products Ltd.).
The antibodies used in this experiment were as
follows: The primary antibodies were 1:1,000 mouse anti-NEDD9 (cat.
no., 4044, CST); 1:1,000 rabbit anti-JNK (cat. no., 9252, CST);
1:5,000 rabbit anti-β-actin (cat. no., bs-0061R, BIOSS, Beijing,
China); 1:1,000 rabbit anti-E-cadherin (cat. no., 3195, CST); and
1:1,000 rabbit anti-vimentin (cat. no., 5741, CST). All primary
antibodies were treated at 4°C for 12 h. The secondary antibodies
were 1:1,000 [cat. no., ab6708, goat anti-mouse IgG H&L (HRP),
Abcam] or 1:1,500 (cat. no., bs-0295G, goat anti-rabbit IgG,
BIOSS), and incubated for 2 h at 25°C.
Statistical analysis
GraphPad Prism 7 software (GraphPad Software, Inc.,
La Jolla, CA, USA) was used for statistical analysis. The results
of the experiments are presented as the means with standard
deviations. Data were compared using the paired t-test. P<0.05
was considered to indicate a statistically significant
difference.
Results
NEDD9 was highly expressed in
colorectal cancer
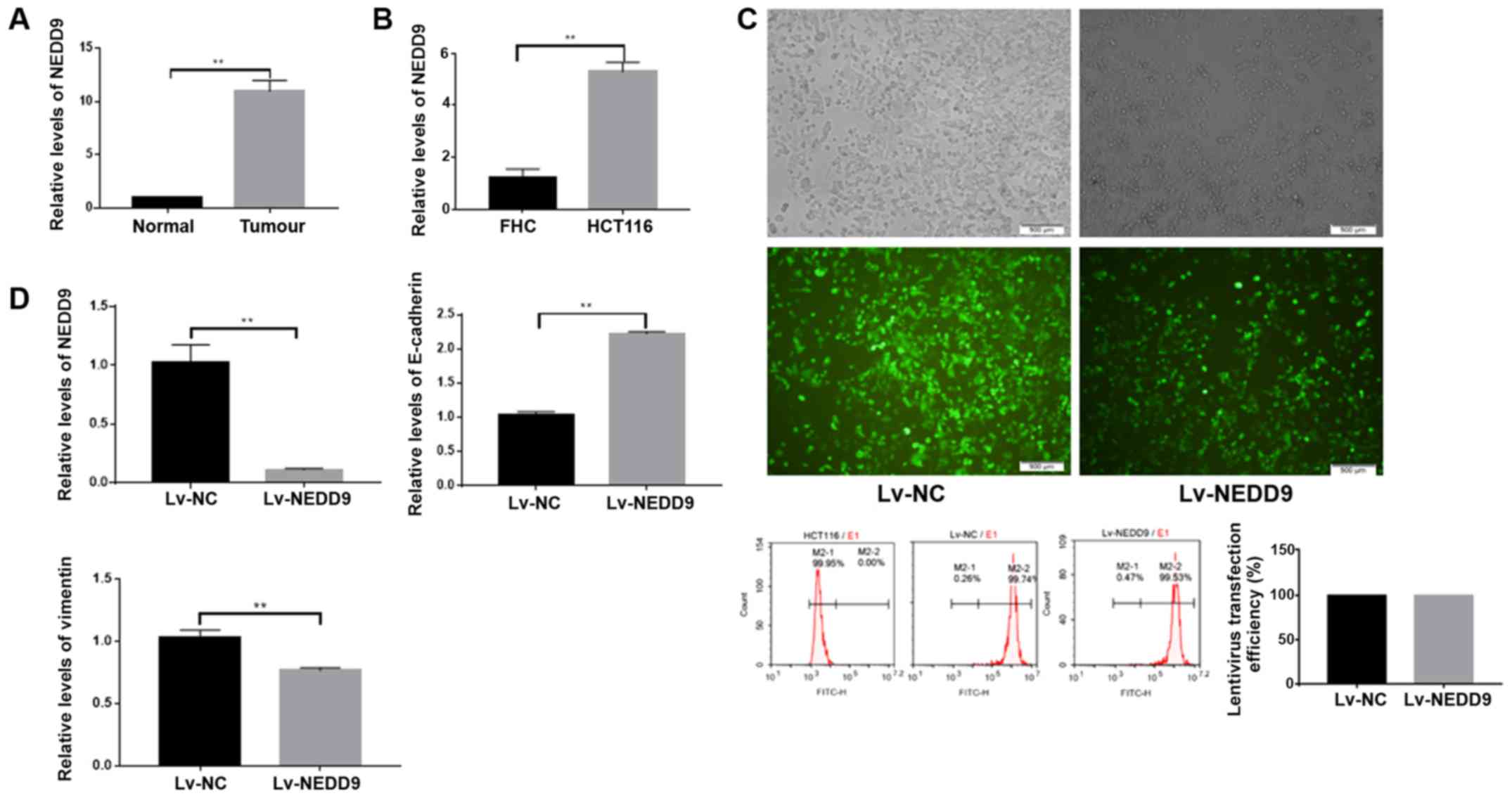
First, the present study measured expression levels
of NEDD9 in 40 normal colorectal tissues and colorectal cancer
tissues by RT-qPCR, and observed that NEDD9 was highly expressed
(Fig. 1A and Table I) in tumor tissues. Subsequently, the
expression levels of NEDD9 were measured in the normal colorectal
cell line FHC and CRC cell line HCT116, and identified that NEDD9
was also highly expressed in the CRC cell line (Fig. 1B).
 | Table I.Clinicopathological features. |
Table I.
Clinicopathological features.
| Features | Number | High expression of
NEDD9 |
|---|
| Age (years) |
|
|
|
≥50 | 35 | 30 |
|
<50 | 5 | 2 |
| Sex |
|
|
|
Female | 29 | 24 |
|
Male | 11 | 8 |
| Grade (TNM)
(25) |
|
|
| I | 5 | 2 |
| II | 11 | 8 |
|
III | 21 | 19 |
| IV | 3 | 3 |
| Stage (TNM)
(25) |
|
|
| pT |
|
|
| T1 | 2 | 1 |
| T2 | 5 | 3 |
| T3 | 17 | 13 |
| T4 | 16 | 15 |
| pN |
|
|
| N0 | 20 | 14 |
| N1 | 13 | 11 |
| N2 | 7 | 7 |
| pM |
|
|
| M0 | 37 | 29 |
| M1 | 3 | 3 |
| Date of tissue
collection |
|
|
|
02/2016-12/2016 | 12 | 10 |
|
01/2017-12/2017 | 28 | 23 |
Expression of EMT changed following
the downregulation of NEDD9
The present study downregulated NEDD9 in colorectal
cancer cells using lentiviral transfection, and observed that the
transfection efficacy was >90% (P<0.01; Fig. 1C). NEDD9 expression in the Lv-NEDD9
group was significantly lower than in the Lv-NC group as measured
by RT-qPCR (P<0.01; Fig. 1D). The
E-cadherin expression levels were significantly higher in the
Lv-NEDD9 group than that in the Lv-NC group, and the vimentin
expression levels were significantly lower in Lv-NEDD9 group when
compared with the Lv-NC group (P<0.01; Fig. 1D). These data suggest that the
expression of EMT markers was altered following NEDD9
downregulation.
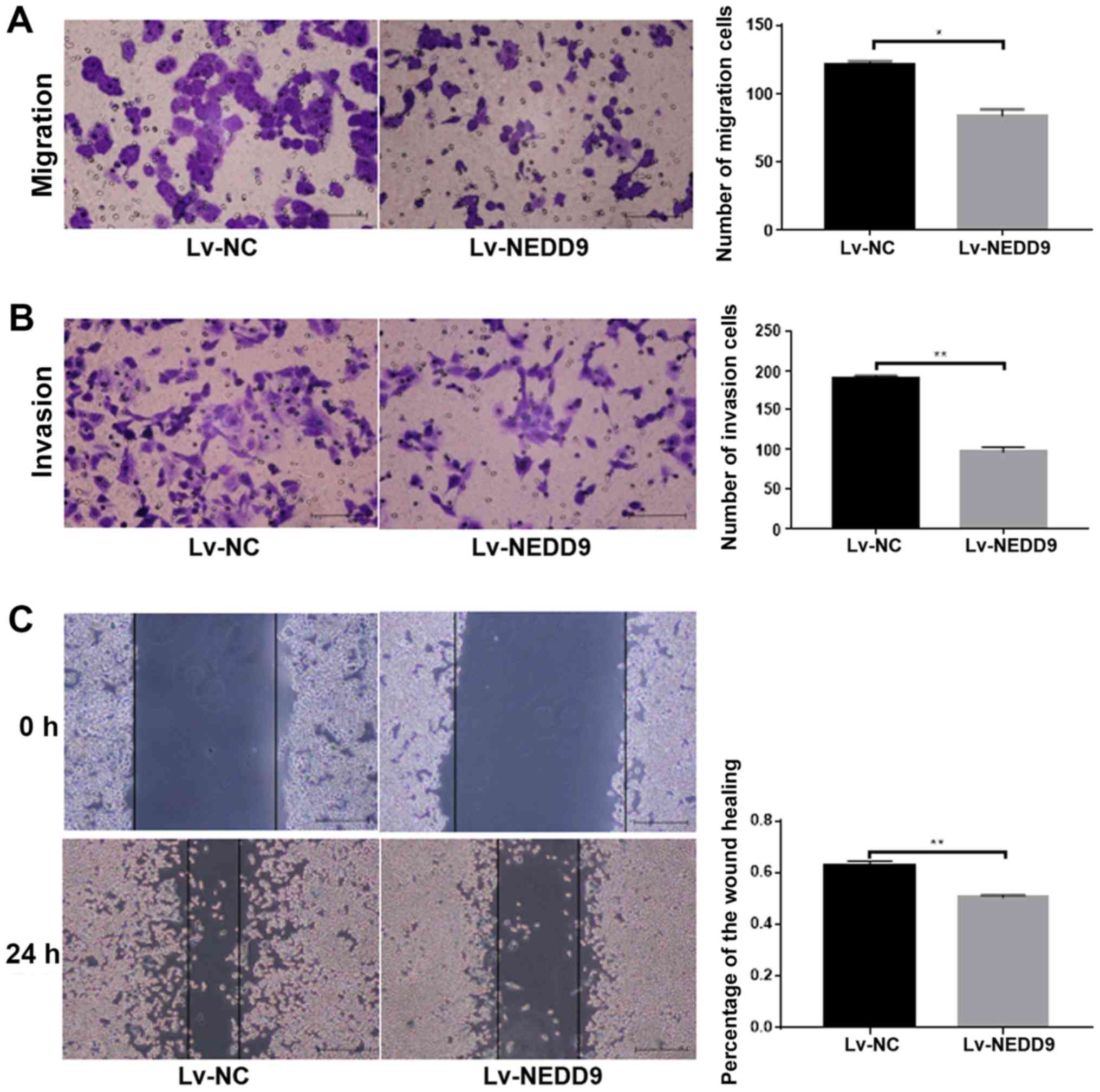
Attenuation of the ability of invasion
and migration ability of colorectal cancer cells following
downregulation of NEDD9
Cell migration ability (P<0.05; Fig. 2A) and invasion (P<0.01; Fig. 2B) was significantly lower in the
Lv-NEDD9 group than that in the Lv-NC group. The wound healing
assay results demonstrated that cell migration (P<0.01; Fig. 2C) was significantly lower in the
Lv-NEDD9 group than that in the Lv-NC group. These data suggested
that downregulation of NEDD9 inhibits the invasion and migration of
colorectal cancer.
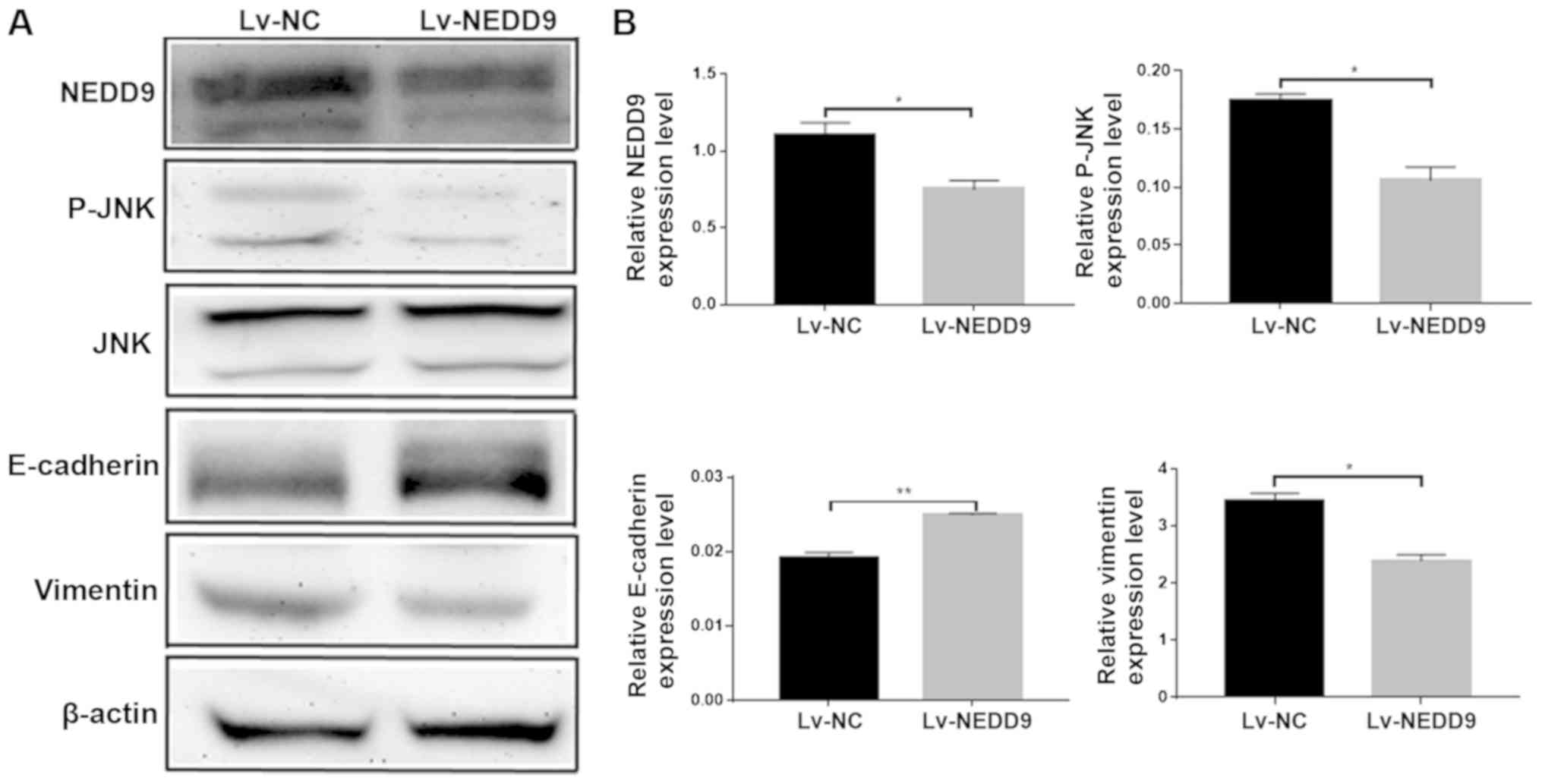
NEDD9 regulates the expression of
JNK/EMT in HCT116 cell lines
The present study performed western blot analysis to
measure the expressions of JNK, E-cadherin and vimentin following
the downregulation of NEDD9. The results revealed that the protein
expression levels of NEDD9 were decreased, the expression levels of
p-jnk were decreased, the expression levels of E-cadherin were
increased and the expression levels of vimentin were decreased
(Fig. 3B). E-cadherin and Vimentin
are EMT-related proteins (27), and
the changes of the expressions of JNK, E-cadherin and Vimentin
suggest that NEDD9 modulates invasion and migration of CRC by
regulating JNK and EMT.
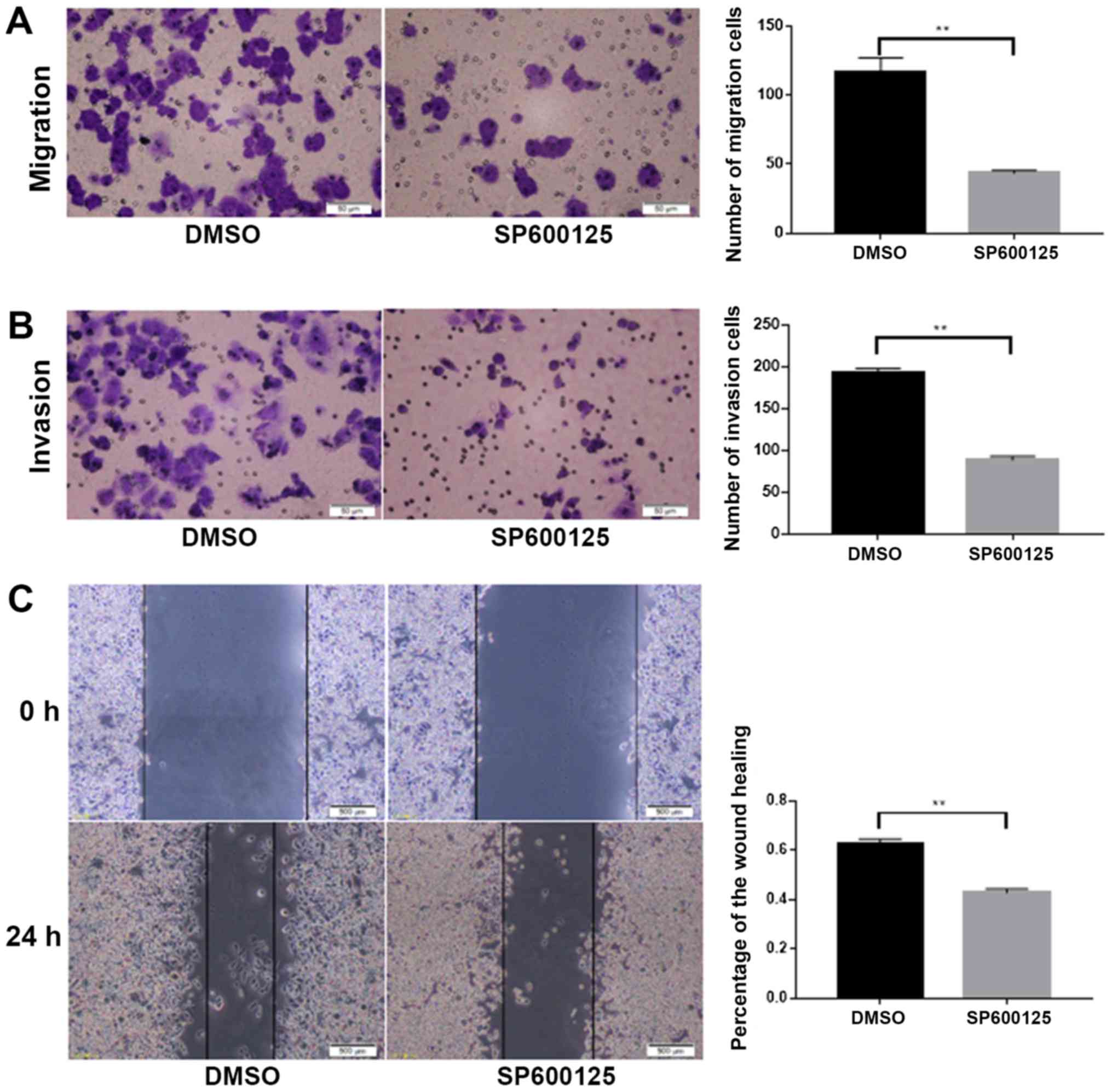
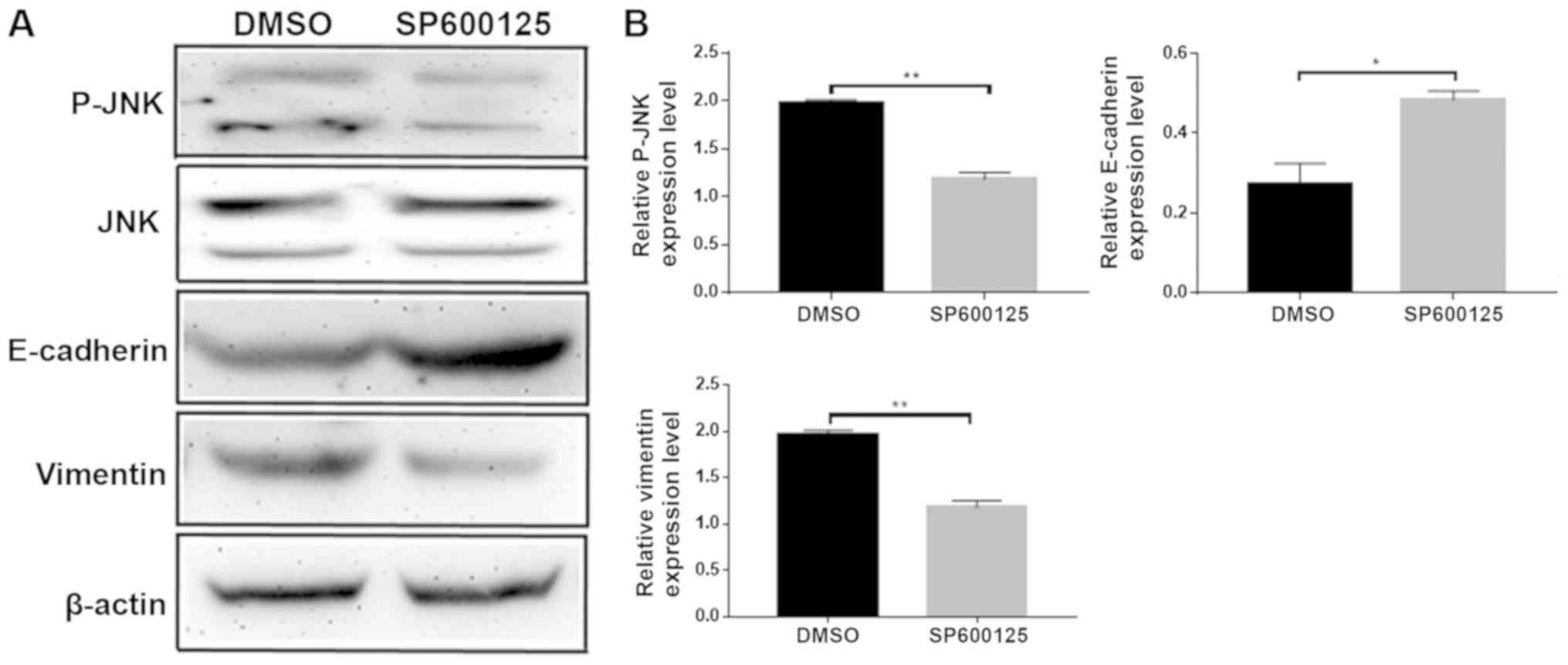
JNK inhibitor SP600125 suppressed
invasion and migration of colorectal cancer cells
To investigate whether NEDD9 affected the invasion
and migration of colorectal cancer through the JNK pathway, the JNK
inhibitor SP600125 was dissolved in DMSO and added to HCT116 cells.
Migration (P<0.01; Fig. 4A) and
invasion (P<0.01; Fig. 4B) were
significantly lower in the SP600125 group than in the DMSO group.
Wound healing assays demonstrated that migration was significantly
lower in the SP600125 group when compared with the DMSO group
(P<0.01; Fig. 4C). Taken
together, these data suggested that JNK promoted the invasion and
migration abilities of colorectal cancer.
JNK inhibitor SP600125 delayed the
progression of EMT
Following the addition of the JNK inhibitor
SP600125, changes in expression levels of the EMT-related proteins
E-cadherin and vimentin were examined by western blotting (Fig. 5A). E-cadherin expression levels
increased and the vimentin expression levels were decreased
(Fig. 5B). These data suggested that
JNK promoted EMT. Taken together, the results of the present study
demonstrated that NEDD9 regulates EMT by JNK to promote CRC
invasion and migration.
Discussion
Several studies have demonstrated that NEDD9 is
highly expressed in tumors and functions as an oncogene (4). For example, NEDD9 promotes invasion and
migration of melanoma (7) and
modulates the metastatic ability of gastric cancer (8), the invasion ability of breast (9) and the migration and invasion ability of
cervical cancer (6), which was
consistent with the findings of the present study. The present
study observed that NEDD9 was highly expressed in CRC and promoted
invasion and migration of colorectal cancer cells. The current
literature indicates that the majority of studies on NEDD9 focus on
its influence on tumor invasion and migration.
NEDD9 acts as a tumor-promoting factor, and its high
expression in tumors also promotes the development of EMT (9,28,29).
NEDD9 promoted EMT through the ERK pathway in breast cancer
(9), as well as promoting EMT by
inhibiting Smad7 in liver cancer (18), and promoting metastasis of gastric
cancer cells by regulating EMT in gastric cancer (30). The experimental data of the present
study demonstrated that the downregulation of NEDD9 in colorectal
cancer increased the expression of the EMT-related protein
E-cadherin and decreased the expression of vimentin, suggesting
that the downregulation of NEDD9 can attenuate EMT. For example,
TGF-β1 triggers EMT in lung cancer via the JNK pathway (31). Inhibition of JNK reversed EMT and
inhibited the migration of gastric cancer (32). JNK has also been reported to promote
EMT in renal cell carcinoma (33).
These data are consistent with our experimental hypothesis. The
present study downregulated NEDD9 and observed that the expression
of p-JNK was decreased and the EMT-related protein expression
changed. Therefore, it was hypothesized that NEDD9 may regulate the
development of EMT via the JNK signaling pathway in CRC. Therefore,
the JNK inhibitor SP600125 was added to colorectal cancer cells and
the expression of EMT-related proteins was measured. The results
revealed that the expression of EMT-related proteins changed. Taken
together the results of the present study demonstrated that NEDD9
promotes EMT via the JNK pathway in colorectal cancer.
NEDD9 mediated tumor invasion by promoting the
secretion of MMP9 (34), and the
negative regulation of tumor suppressor gene LKB1 modulated the
progression of lung cancer (35).
NEDD9 is also affected by miRNA regulation affecting tumor invasion
and metastasis (36). NEDD9 as a
tumor factor is closely associated with the occurrence and
development of tumors (37);
nevertheless, numerous mechanisms for NEDD9 are unknown and have
not been studied; therefore, the current state of knowledge of
NEDD9 remains scarce and requires further investigation.
Consult the relevant literature to know that NEDD9
can interact with FAK to affect tumors (38) and related literature has also found
that FAK can activate JNK (39), so
NEDD9 may activate JNK through FAK. In addition, NEDD9 may also
activate JNK via P38MAPK-Erk1/2 (40). NEDD9 may also activate JNK in other
ways, but it has not been discovered yet. A review of the
literature reveals the relevant situation downstream of JNK. For
example, IL-2/sorafenib can affect tumor survival, growth and
mobility through JNK-TAZ pathways (41), GPS1 can activate the c-Jun N-terminal
kinase (JNK)/Jun pathway to affect breast cancer growth and
migration (42), Saikosaponin D
induced apoptosis via the activation of TGFα-JNK-p53 (43), Thymoquinone inhibits the metastasis
through activation of JNK-p38 (44),
JNK-ELK1 The pathway can also have an effect on the tumor (45). These are the pathways already known
downstream of JNK, but there must be downstream pathways that have
not been studied.
Our experiment investigated the effect of NEDD9 on
colorectal cancer through JNK, which is currently not done in
colorectal cancer. It is innovative and meaningful. However, due to
the limitations of experimental conditions, we did not study the
specific mechanism of NEDD9 activation of JNK and the downstream
pathway of JNK in this experiment. In future experiments, we will
work to study the specific mechanism by which NEDD9 activates JNK
and the pathway downstream of JNK.
In conclusion, we found that NEDD9 was highly
expressed in colorectal cancer tissues and colorectal cancer cell
lines, and that NEDD9 promoted invasion and migration of colorectal
cancer. These data suggested that NEDD9 can be used as a new and
effective target for therapy of colorectal cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed are included in this
published article.
Authors' contributions
HM and RS were responsible for study design, the
acquisition and analysis of data. HM, JW performed the acquisition
of data, and wrote and revised the manuscript. JW, QH, XY, KY, YQ
and HQ aided with the acquisition of data. HM and JW aided with the
statistical analysis. RS and HQ participated in the design and
coordination of the study and helped modify the manuscript. JR
acquired the data, wrote and revised the manuscript, and helped to
perform the statistical analysis. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
the Affiliated Hospital of Qingdao University (Qingdao, China).
Witten informed consent was obtained from the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RGS, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mcquade RM, Stojanovska V, Bornstein JC
and Nurgali K: Colorectal cancer chemotherapy: The evolution of
treatment and new approaches. Curr Med Chem. 24:1537–1557. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Izumchenko E, Singh MK, Plotnikova OV,
Tikhmyanova N, Little JL, Serebriiskii IG, Seo S, Kurokawa M,
Egleston BL, Klein-Szanto A, et al: NEDD9 promotes oncogenic
signaling in mammary tumor development. Cancer Res. 69:7198–7206.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim M, Gans JD, Nogueira C, Wang A, Paik
JH, Feng B, Brennan C, Hahn WC, Cordon-Cardo C, Wagner SN, et al:
Comparative oncogenomics identifies NEDD9 as a melanoma metastasis
gene. Cell. 125:1269–1281. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sima N, Cheng X, Ye F, Ma D, Xie X and Lü
W: The overexpression of scaffolding protein NEDD9 promotes
migration and invasion in cervical cancer via tyrosine
phosphorylated FAK and SRC. PLoS One. 8:e745942013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
O'Neill G, Seo S, Serebriiskii IG, Lessin
SR and Golemis EA: A new central scaffold for metastasis: Parsing
HEF1/Cas-L/NEDD9. Cancer Res. 67:8975–8979. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang S, Wu L, Liu Q, Chen K and Zhang X:
Elevated expression of NEDD9 is associated with metastatic activity
in gastric cancer. Onco Targets Ther. 8:633–640. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kong C, Wang C, Wang L, Ma M, Niu C, Sun
X, Du J, Dong Z, Zhu S, Lu J and Huang B: NEDD9 is a positive
regulator of epithelial-mesenchymal transition and promotes
invasion in aggressive breast cancer. PLoS One. 6:e226662011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chang JX, Gao F, Zhao GQ and Zhang GJ:
Expression and clinical significance of NEDD9 in lung tissues. Med
Oncol. 29:2654–2660. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cui X, Shen K, Xie Z, Liu T and Zhang H:
Identification of key genes in colorectal cancer using random walk
with restart. Mol Med Rep. 15:867–872. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li P, Zhou H, Zhu X, Ma G, Liu C, Lin B
and Mao W: High expression of NEDD9 predicts adverse outcomes of
colorectal cancer patients. Int J Clin Exp Pathol. 7:2565–2570.
2014.PubMed/NCBI
|
|
13
|
Dai J, Van Wie PG, Fai LY, Kim D, Wang L,
Poyil P, Luo J and Zhang Z: Downregulation of NEDD9 by apigenin
suppresses migration, invasion, and metastasis of colorectal cancer
cells. Toxicol Appl Pharmacol. 311:106–112. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Thiery JP: Epithelial-mesenchymal
transitions in tumour progression. Nat Rev Cancer. 2:442–454. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Min J, Liu L, Li X, Jiang J, Wang J, Zhang
B, Cao D, Yu D, Tao D, Hu J, et al: Absence of DAB2IP promotes
cancer stem cell like signatures and indicates poor survival
outcome in colorectal cancer. Sci Rep. 5:165782015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guerrero MS, Parsons JT and Bouton AH: Cas
and NEDD9 contribute to tumor progression through dynamic
regulation of the cytoskeleton. Genes Cancer. 3:371–381. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Z, Shen M, Lu P, Li X, Zhu S and Yue
S: NEDD9 may regulate hepatocellular carcinoma cell metastasis by
promoting epithelial-mesenchymal-transition and stemness via
repressing Smad7. Oncotarget. 8:1714–1724. 2017.PubMed/NCBI
|
|
19
|
Weston CR and Davis RJ: The JNK signal
transduction pathway. Curr Opin Genet Dev. 12:14–21. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wagner EF and Nebreda AR: Signal
integration by JNK and p38 MAPK pathways in cancer development. Nat
Rev Cancer. 9:537–549. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bode AM and Dong Z: The functional
contrariety of JNK. Mol Carcinog. 46:591–598. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao X, Wu X, Qian M, Song Y, Wu D and
Zhang W: Knockdown of TGF-β1 expression in human umbilical cord
mesenchymal stem cells reverts their exosome-mediated EMT promoting
effect on lung cancer cells. Cancer Lett. 428:34–44. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Y, Xu Y, Yan W, Han P, Liu J, Gong J,
Li D, Ding X, Wang H, Lin Z, et al: CFIm25 inhibits hepatocellular
carcinoma metastasis by suppressing the p38 and JNK/c-Jun signaling
pathways. Oncotarget. 9:11783–11793. 2018.PubMed/NCBI
|
|
24
|
Dong Y, Wu Z, He M, Chen Y, Chen Y, Shen
X, Zhao X, Zhang L, Yuan B and Zeng Z: ADAM9 mediates the
interleukin-6-induced Epithelial-Mesenchymal transition and
metastasis through ROS production in hepatoma cells. Cancer Lett.
421:1–14. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Scanlon CS, Van Tubergen EA, Inglehart RC
and D'Silva NJ: Biomarkers of epithelial-mesenchymal transition in
squamous cell carcinoma. J Dent Res. 92:114–121. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Morimoto K, Tanaka T, Nitta Y, Ohnishi K,
Kawashima H and Nakatani T: NEDD9 crucially regulates
TGF-β-triggered epithelial-mesenchymal transition and cell invasion
in prostate cancer cells: Involvement in cancer progressiveness.
Prostate. 74:901–910. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Miao Y, Li AL, Wang L, Fan CF, Zhang XP,
Xu HT, Yang LH, Liu Y and Wang EH: Overexpression of NEDD9 is
associated with altered expression of E-cadherin, β-catenin and
N-cadherin and predictive of poor prognosis in non-small cell lung
cancer. Pathol Oncol Res. 19:281–286. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Feng J, Zhao J, Xie H, Yin Y, Luo G, Zhang
J, Feng Y and Li Z: Involvement of NEDD9 in the invasion and
migration of gastric cancer. Tumour Biol. 36:3621–3628. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Khan GJ, Gao Y, Gu M, Wang L, Khan S,
Naeem F, Semukunzi H, Roy D, Yuan S and Sun L: TGF-β1 Causes EMT by
regulating N-Acetyl glucosaminyl transferases via downregulation of
non muscle myosin II-A through JNK/P38/PI3K pathway in lung cancer.
Curr Cancer Drug Targets. 18:209–219. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Choi Y, Ko YS, Park J, Choi Y, Kim Y, Pyo
JS, Jang BG, Hwang DH, Kim WH and Lee BL: HER2-induced metastasis
is mediated by AKT/JNK/EMT signaling pathway in gastric cancer.
World J Gastroenterol. 22:9141–9153. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
An J, Guo Y, Wang T, Pantuck AJ and Rettig
MB: Pomegranate extract inhibits EMT in clear cell renal cell
carcinoma in a NF-κB and JNK dependent manner. Asian J Urol.
2:38–45. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Grauzam S, Brock AM, Holmes CO, Tiedeken
JA, Boniface SG, Pierson BN, Patterson DG, Coaxum SD, Neskey DM and
Rosenzweig SA: NEDD9 stimulated MMP9 secretion is required for
invadopodia formation in oral squamous cell carcinoma. Oncotarget.
9:25503–25516. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Feng Y, Wang Y, Wang Z, Fang Z, Li F, Gao
Y, Liu H, Xiao T, Li F, Zhou Y, et al: The CRTC1-NEDD9 signaling
axis mediates lung cancer progression caused by LKB1 loss. Cancer
Res. 72:6502–6511. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chu Q, Zhang J and Sun F: Study on
mechanism of mi R-203 inhibiting migration and invasion of breast
cancer cell MDA-MB-231 by regulating NEDD9 protein. J Diagnostics
Concepts Pract. 2014.
|
|
37
|
Shagisultanova E, Gaponova AV, Gabbasov R,
Nicolas E and Golemis EA: Preclinical and clinical studies of the
NEDD9 scaffold protein in cancer and other diseases. Gene.
567:1–11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Guerrero MS, Parsons JT and Bouton AH: Cas
and NEDD9 contribute to tumor progression through dynamic
regulation of the cytoskeleton. Genes Cancer. 3:371–381. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sulzmaier FJ, Jean C and Schlaepfer DD:
FAK in cancer: Mechanistic findings and clinical applications. Nat
Rev Cancer. 14:598–610. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tikhmyanova N, Little JL and Golemis EA:
CAS proteins in normal and pathological cell growth control. Cell
Mol Life Sci. 67:1025–1048. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ding X, Sun W and Chen J: IL-2 augments
the sorafenib-induced apoptosis in liver cancer by promoting
mitochondrial fission and activating the JNK/TAZ pathway. Cancer
Cell Int. 18:1762018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li Z, Lim SK, Liang X and Lim YP: The
transcriptional coactivator WBP2 primes triple-negative breast
cancer cells for responses to Wnt signaling via the JNK/Jun kinase
pathway. J Biol Chem. 293:20014–20028. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen X, Liu C, Zhao R, Zhao P, Wu J, Zhou
N and Ying M: Synergetic and antagonistic molecular effects
mediated by the feedback loop of p53 and JNK between Saikosaponin D
and SP600125 on lung cancer A549 cells. Mol Pharm. 15:4974–4984.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kubala MH, Punj V, Placencio-Hickok VR,
Fang H, Fernandez GE, Sposto R and DeClerck YA: Plasminogen
activator Inhibitor-1 promotes the recruitment and polarization of
macrophages in cancer. Cell Rep. 25:2177–2191.e7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hu Z, Tie Y, Lv G, Zhu J, Fu H and Zheng
X: Transcriptional activation of miR-320a by ATF2, ELK1 and YY1
induces cancer cell apoptosis under ionizing radiation conditions.
Int J Oncol. 53:1691–1702. 2018.PubMed/NCBI
|