Introduction
Gliomas are the most common primary brain tumors
with an incidence rate of ~5/100,000). Despite comprehensive
treatment strategies, including surgery, radiotherapy and
chemotherapy, the prognosis of patients remains unsatisfactory,
with a median survival time of 12–18 months (1–4). This
poor outcome is largely associated with the difficulty of curing
gliomas and the high relapse rates (5,6).
Therefore, the identification of novel therapeutic targets has
become a particular focus of research.
Ring finger protein 5 (RNF5) belongs to the
ring finger family of ubiquitin ligases (7,8), which
are anchored to the endoplasmic reticulum (ER) membrane and are
important components of the ER-associated degradation (ERAD)
mechanism. RNF5 serves a role in monitoring the folding of CF
transmembrane conductance regulator (CFTR), CFTRDF508 and nascent
CFTRΔF508 in the ER membrane (9,10).
Furthermore, a previous study reported that RNF5 regulates
cell movement by targeting paxillin ubiquitination and altering its
localization (11). RNF5
participates in the inflammatory response in viral infections by
ubiquitinating transmembrane protein 173 and inhibiting the
activation of virus-induced interferon regulatory factor 3,
expression of interferon β1 and the cellular antiviral response
(12). In breast cancer cells,
RNF5 ubiquitination degrades the L-glutamine carrier
proteins solute carrier family 1 member 5 and solute carrier family
38 member 2, which reduces glutamine uptake and levels of the
tricarboxylic acid cycle components, decreases mechanistic target
of rapamycin signaling and cell proliferation, and increases
autophagy and apoptosis (13).
RNF5 is highly expressed in breast cancer and related cell
lines, and inhibition of its expression decreases cell
proliferation (13). The present
study attempted to characterize the role of RNF5 in human
glioma and to determine its association with tumor grade and
survival time in patients with glioma.
The present study revealed that RNF5 was
differentially expressed in patients with different grades of
glioma and was closely associated with the prognosis of patients
with anaplastic glioma (AG) and glioblastoma multiforma (GBM).
Moreover, Gene Set Enrichment Analysis (GSEA) identified the Kyoto
Encyclopedia of Genes and Genomes (KEGG) signaling pathways that
were significantly associated with RNF5. Additionally, a
correlation analysis was used to predict the potential
ubiquitination substrates for RNF5 in human glioma.
Materials and methods
Patient samples
mRNA microarray expression for patients were
obtained from the Chinese Glioma Genome Atlas (CGGA; cgga.org.cn) and the Gene Expression Omnibus
(www.ncbi.nlm.nih.gov/geo). The CGGA
contains 301 glioma samples (including 84 astrocytoma, 89
oligodendroglioma and 128 glioblastoma samples). The grouping of
low-grade glioma (LGG), AG and GBM was performed as previously
described (14). The GSE16011
dataset (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE16011)
contains 276 glioma samples (including samples from 8 patients with
epilepsy and 24 astrocytoma, 85 oligodendroglioma and 159
glioblastoma samples) and 8 control samples, totaling 284 specimens
(15). The GSE4290 dataset
(www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE4290)
contains 23 samples from patients with epilepsy as non-cancerous
samples and 157 tumor samples, including 26 astrocytoma, 50
oligodendrogliomas and 81 glioblastoma samples (16). Gene mutation data were obtained from
The Cancer Genome Atlas (TCGA; cancergenome.nih.gov).
RNF5 expression and its association
with patient prognosis
The expression levels of RNF5 in the CGGA
database and GSE16011 and GSE4290 datasets were analyzed using
GraphPad Prism software version 6.01 (GraphPad Software, Inc.). In
addition, the association between the expression level of
RNF5 and the prognosis of patients was obtained using the
CGGA database. In order to analyze patient prognosis, patients were
equally divided into two groups according to RNF5 expression
levels.
Cell culture
The glioblastoma cell line U251 was obtained from
The Type Culture Collection of The Chinese Academy of Sciences.
U251 cells were cultured using DMEM (Invitrogen; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.) at 37°C and 5% CO2.
Plasmid and transfection
The RNF5 cDNA sequence was inserted between
the HindIII and XbaI restriction sites in the
p3XFLAG-CMV-14 vector (Shanghai GenePharma Co., Ltd.). The
p3XFLAG-CMV-14 plasmid was used as an empty vector. A total of 3 µg
plasmid was transfected into U251 cells using 9 µl Polyjet
transfection reagent (SignaGen Laboratories) according to the
manufacturer's protocol. The culture medium was changed after 12 h
and cells were transfected for 72 h.
Reverse transcription-quantitative PCR
(RT-qPCR)
RNF5 expression was assessed using RT-qPCR.
Total RNA was extracted from U251 cells using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and
reverse-transcribed into cDNA using the Quant One-Step RT-PCR kit
(Tiangen Biotech Co., Ltd.). qPCR was performed using FastStart
Universal SYBR Green Mix (Roche Diagnostics) and an ABI 7300
real-time PCR instrument (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The primers for RNF5 and β-actin were designed
as follows: RNF5, forward 5′-GTACCCATACGATGTTCCAGATTACGC-3′,
reverse 5′-CTGAGCAGCCAGAAAAAGAAAAAGATG-3′; and β-actin forward,
5′-CATGTACGTTGCTATCCAGGC-3′, and reverse,
5′-CGCTCGGTGAGGATCTTCATG-3′. Thermocycling conditions included
pre-denaturation at 95°C for 3 min, denaturation at 95°C for 15
sec, annealing at 60°C for 15 sec and extension at 72°C for 1 min
for 35 cycles. Expression level of RFN5 was calculated using
the 2−ΔΔCq method (17).
Cell colony formation assay
U251 cells (1×105) overexpressing
RNF5 were seeded into 6 cm dishes and cultured for 14 days
at 37°C. Cells were subsequently fixed in 4% paraformaldehyde for
30 min at room temperature and stained with 0.05% crystal violet
for 30 min at room temperature. A light Canon 70D camera (Canon,
Inc.) was used to capture images (magnification, ×1).
GSEA to evaluate RNF5-enriched KEGG
pathways
In order to elucidate the signaling pathways
associated with the possible actions of RNF5 in human
glioma, enrichment analysis was performed using GSEA software
version 6.2 (software.broadinstitute.org/gsea/login.jsp) in
the CGGA database. RNF5-enriched KEGG (www.genome.jp/kegg) signaling pathways were identified
through this analysis.
Identification of differentially
expressed genes (DEGs)
To predict the potential substrate(s) of
RNF5, CGGA, GSE16011 and GSE4290 data were sorted according
to RNF5 expression level from low to high. Data were
subsequently divided into four groups: A, B, C and D according to
the number of patients following RNF5 determination. To avoid data
with no significant differences from groups B and C, comparative
analysis was performed between groups A and D to identify the DEGs
from the three databases. To narrow the scope, the DEGs that
overlapped among the three databases were identified using limma
package of R software 3.4.4 (www.r-project.org) and selected for subsequent
analysis. A gene with |logFC|>1 was defined as DEG.
Correlation analysis between RNF5 and
overlapping genes
To demonstrate the correlation between RNF5
and the five overlapping genes, analysis using R and GraphPad Prism
software was performed. Correlation analysis was performed using
Pearson's correlation coefficient test.
Statistical analysis
GraphPad Prism software was used for statistical
analysis. Results are presented as the means ± standard error of
the mean. Statistical significance was analyzed using Student's
t-test (two groups) and one-way analysis of variance (multiple
groups) followed by Dunnett's post hoc test. Kaplan-Meier survival
analyses for overall survival were performed and compared with the
log-rank test. P<0.05 was considered to indicate a statistically
significant difference. For GSEA, a normalized enrichment score
>1, nominal P<0.05 and false discovery rate q-value <0.25
were considered to indicate a statistically significant
difference.
Results
Expression of RNF5 and its association
with prognosis
To characterize the expression of RNF5 and
its association with patient prognosis, the CGGA database and the
GSE16011 and GSE4290 datasets were used. RNF5 was
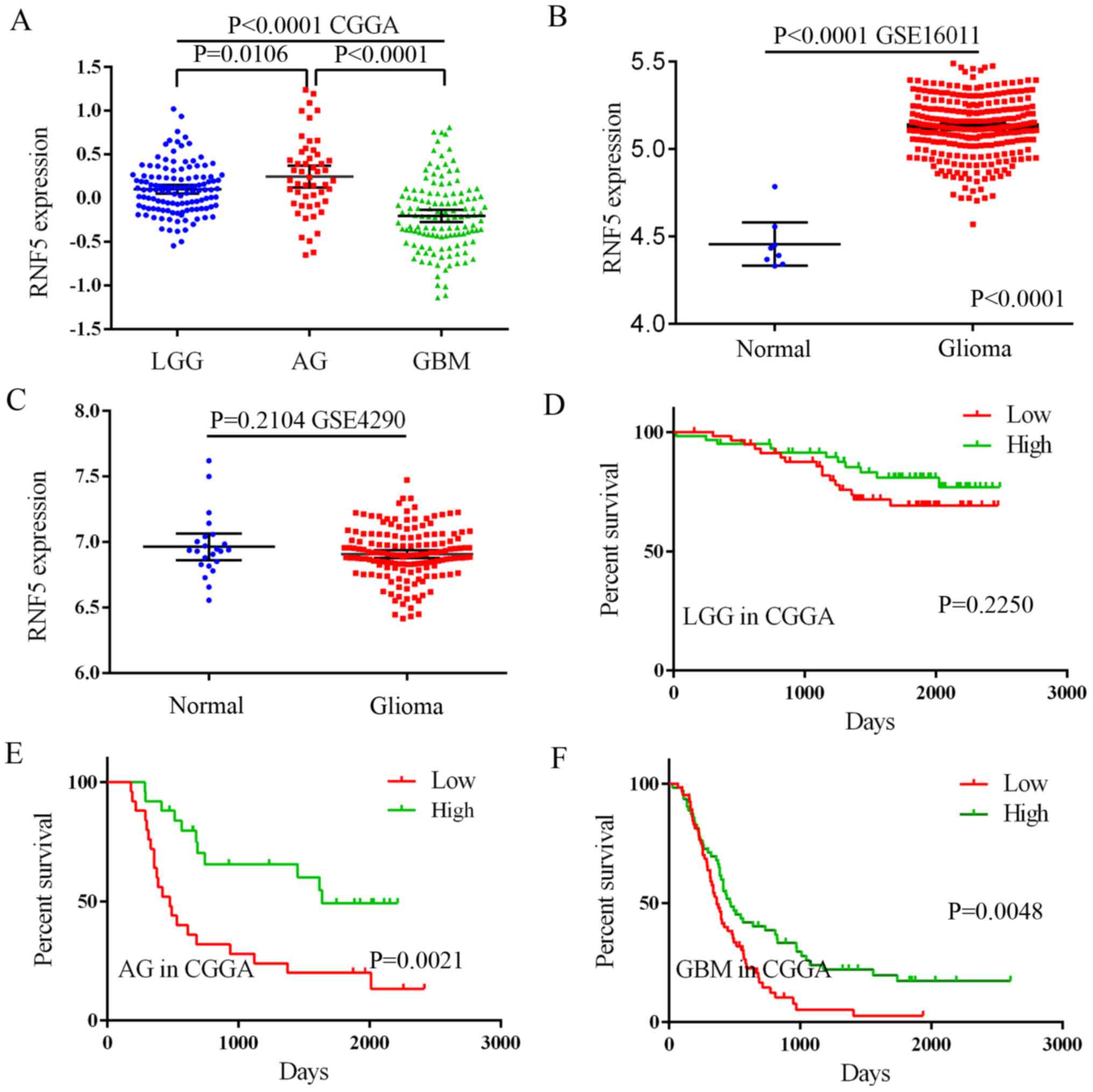
differentially expressed in LGG, AG and GBM. RNF5 expression
was significantly higher in LGG and AG compared with GBM (Fig. 1A). The GSE16011 and GSE4290 datasets
were selected to investigate the difference in RNF5
expression between non-cancerous brain tissue and glioma; however,
a consistent conclusion was not reached owing to the small number
of non-cancerous brain tissue samples in the datasets (Fig. 1B and C). However, an association
between RNF5 expression levels and patient prognosis was
determined using the CGGA database. In LGG, the expression level of
RNF5 and patient prognosis were not significantly
associated, while in AG and GBM, patients with high RNF5
expression had an improved prognosis compared with patients with
low expression (Fig. 1D-F).
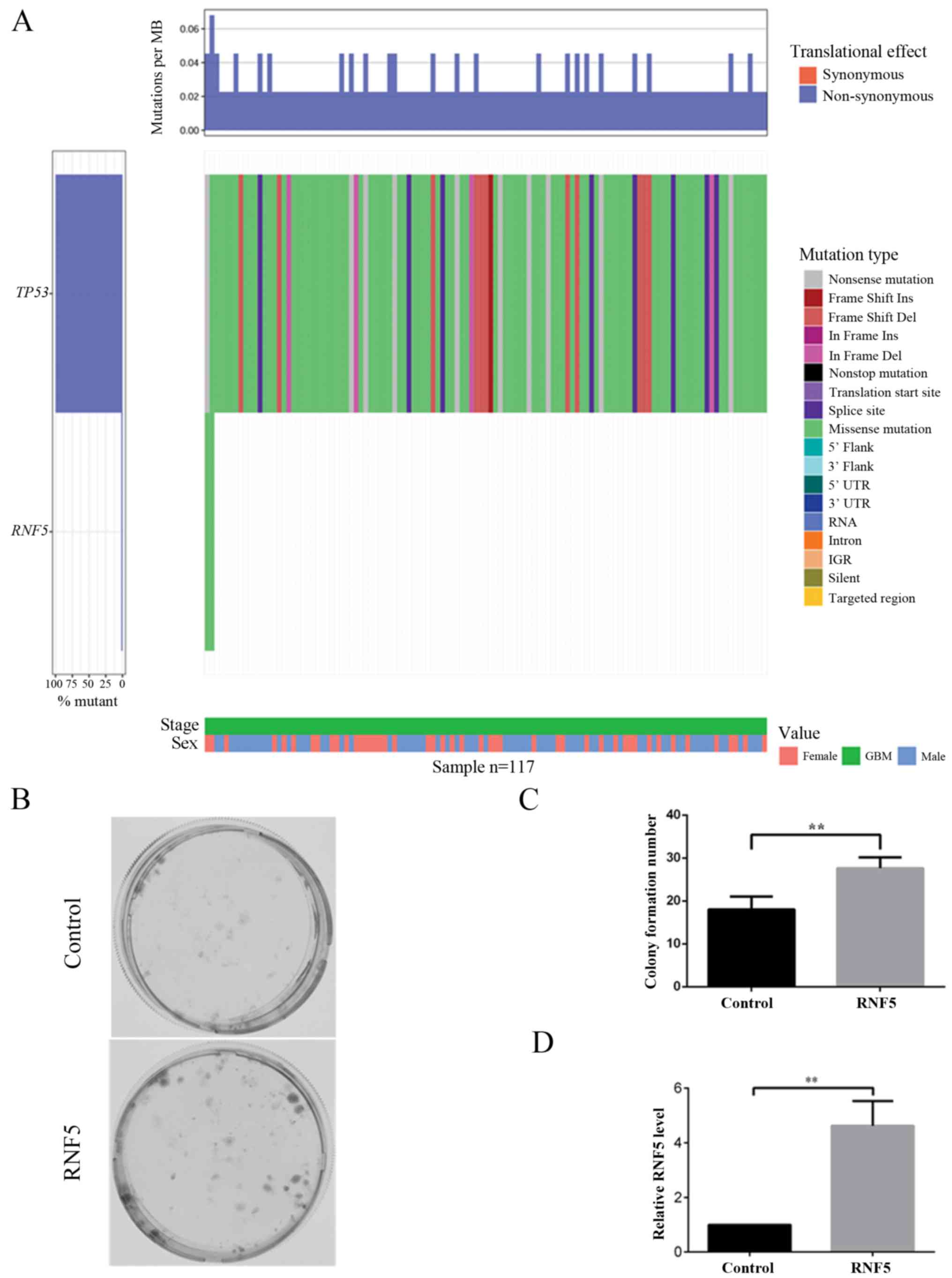
To further investigate the association between high
RNF5 expression and prognosis in patients with glioma, tumor
protein 53 (TP53) mutations were analyzed as indicated by
the literature (18–21). An analysis of TCGA database revealed
that TP53 has a high mutation rate in GBM, while only two
RNF5 mutations were identified (Fig. 2A). Additionally, in vitro
analysis revealed that cells overexpressing RNF5 exhibited
increased colony formation compared with control cells (Fig. 2B-D). Subsequently, the role of
RNF5 in glioma and its possible target proteins were further
analyzed using a bioinformatics approach.
GSEA of KEGG signaling pathways
associated with RNF5 in human glioma
To further analyze the role of RNF5 in human
glioma, GSEA of the CGGA database was performed. The expression
levels of RNF5 in the samples were sorted from low to high,
and the samples were divided into four groups: A, B, C and D. Group
A contained samples with low expression of RNF5, while Group
D contained samples with a high expression level of RNF5.
The newly grouped data were subsequently analyzed by GSEA. The GSEA
revealed that RNF5 was significantly associated with the
following KEGG signaling pathways: ‘Wnt signaling pathway’,
‘apoptosis’, ‘cell adhesion molecules CAMs’, ‘cytokine-cytokine
receptor interaction’, ‘focal adhesion’ and ‘ECM-receptor
interaction’ (Fig. 3). Therefore,
RNF5 may affect the development of glioma through these KEGG
signaling pathways.
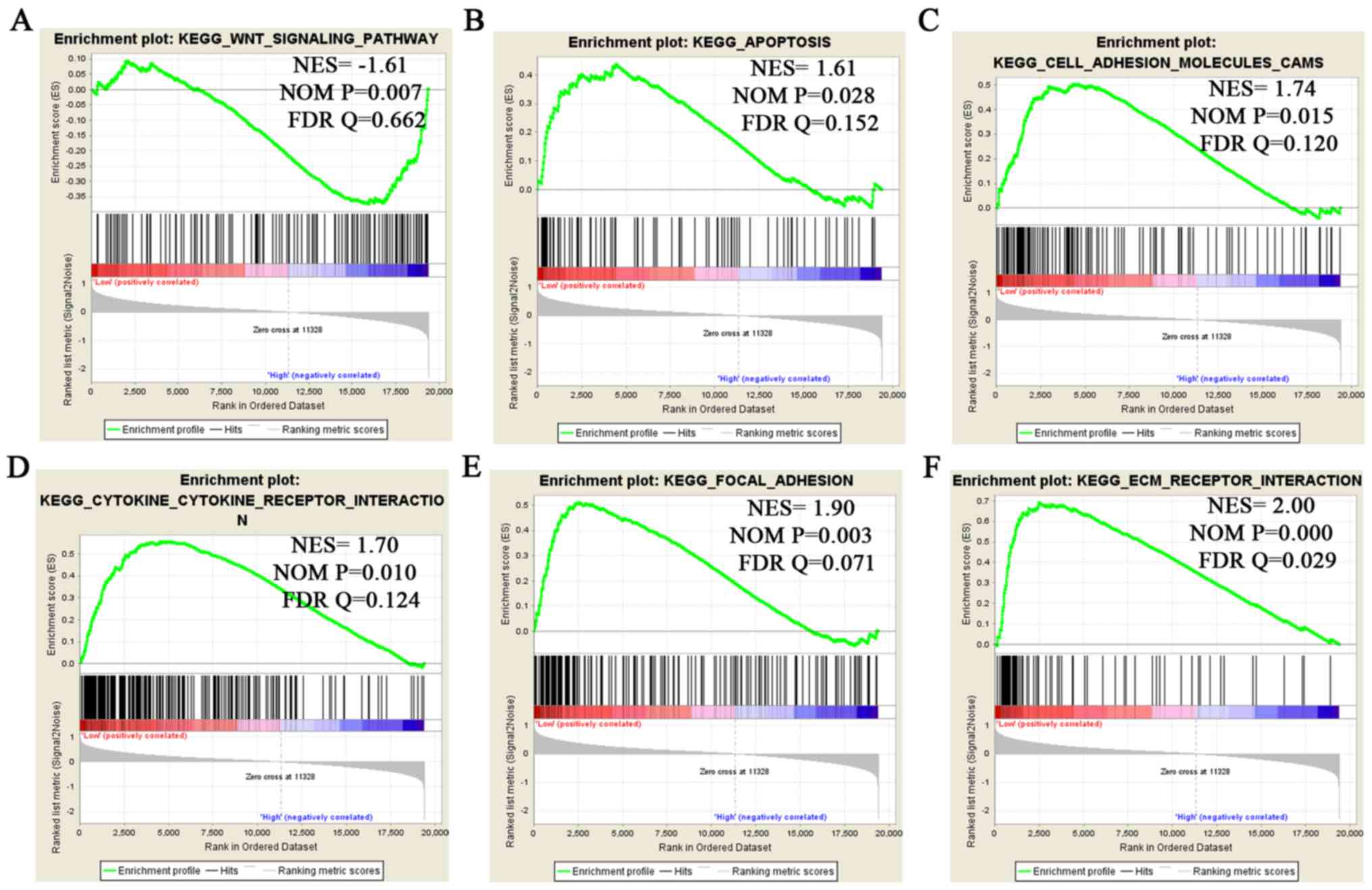 | Figure 3.GSEA to identify significant
RNF5-enriched KEGG signaling pathways. GSEA revealed that
RNF5 is enriched in the following pathways: (A) The ‘Wnt
signaling pathway’, (B) ‘apoptosis’, (C) ‘cell adhesion molecules
CAMs’, (D) ‘cytokine-cytokine receptor interaction’, (E) ‘focal
adhesion’ and (F) ‘ECM-receptor interaction’. GSEA, Gene Set
Enrichment Analysis; RNF5, ring finger protein 5; KEGG,
Kyoto Encyclopedia of Genes and Genomes; ECM, extracellular matrix;
NES, normalized enrichment score; NOM, nominal; FDR, false
discovery rate. |
Prediction of RNF5 ubiquitination
substrates using a bioinformatics approach
In order to identify the potential ubiquitination
substrates of RNF5 in human glioma, CGGA, GSE16011 and GSE4290 data
were divided into groups of low and high RNF5 expression.
Differential genetic analysis was then performed on these groups to
identify the DEGs. To further clarify the possible ubiquitination
substrates of RNF5, the overlapping genes between the CGGA
database and GSE16011 and GSE4290 datasets were identified
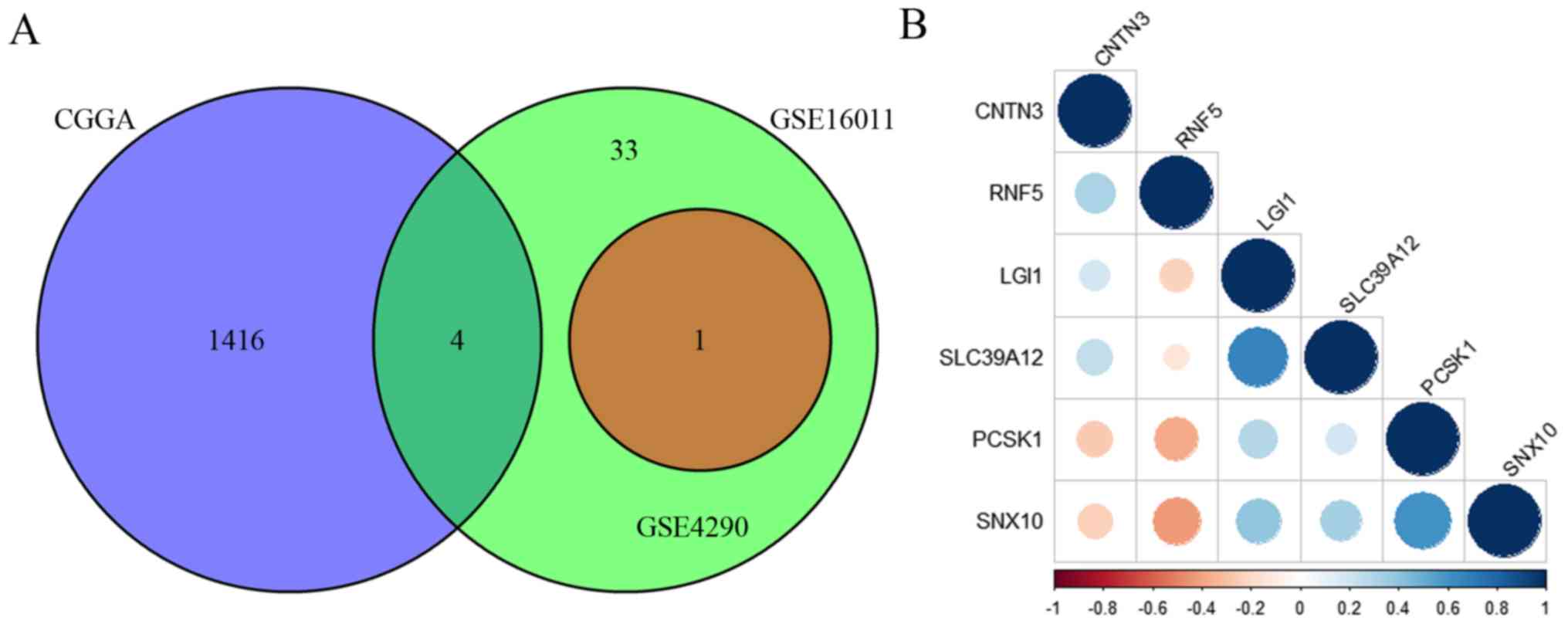
(Fig. 4A). A total of 4 overlapping
genes were identified between the CGGA database and the GSE16011
dataset, 1 overlapping gene was identified between the GSE16011 and
GSE4290 datasets, and no overlapping genes were identified between
the CGGA database and the GSE4290 dataset. The 5 overlapping genes
included contactin 3 (CNTN3), leucine rich glioma
inactivated 1 (LGI1), proprotein convertase subtilisin/kexin
type 1 (PCSK1), sorting nexin 10 (SNX10) and solute
carrier family 39 member 12 (SLC39A12; Fig. 4B). RNF5 expression was positively
associated with CNTN3, while a negative association was
demonstrated for the remaining four genes (SNX10, PCSK1, LGI1 and
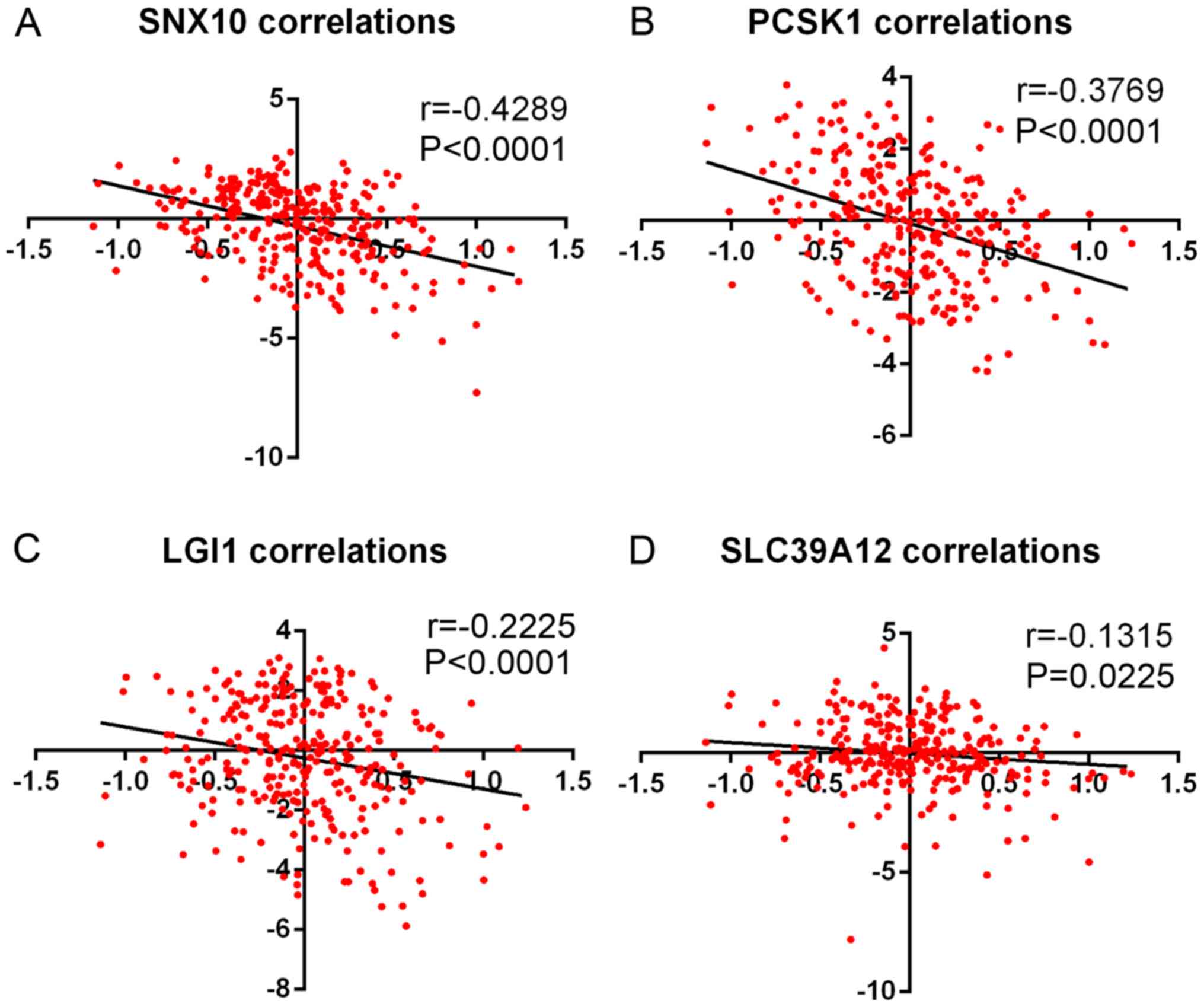
SLC39A12) (Fig. 4B). In addition,
the results from Fig. 5 demonstrated
that RNF5 was negatively correlated with SNX10, PCSK1, LGI1 and
SLC39A12.
Discussion
Previous studies have demonstrated that ubiquitin
ligase is closely associated with tumor development and metastasis
(13,22–25). The
present study used human glioma data to reveal that RNF5 was
differentially expressed in patients with different levels of
glioma and was correlated with prognosis in patients with AG and
GBM. Specifically, an improved prognosis was observed in patients
with AG and GBM with a high expression of RNF5 compared with
a low expression. Subsequently, RNF5 was overexpressed in
U251 cells in vitro, and it was revealed that colony forming
ability was enhanced in cells overexpressing RNF5 compared
with controls. The authors speculate that silencing RNF5
reduces the colony forming ability. Through GSEA enrichment
analysis, KEGG signaling pathways that were significantly
associated with RNF5 were identified. To further explore
possible ubiquitination substrates for RNF5 in human glioma,
correlation analysis were performed. A total of 4 genes were
negatively associated with RNF5 expression, and may serve as
potential RNF5 ubiquitination substrates.
Previous studies revealed that RNF5 was
highly expressed in breast cancer specimens and cell lines.
Additionally, tumor cell proliferation was inhibited after
silencing RNF5, and patients with breast cancer with high
RNF5 expression have a poor prognosis compared with patients
with low expression (13,21). Cell proliferation was inhibited after
silencing RNF5 expression in MCF-7 cells. However, cell
proliferation was not affected in MDA-MB-231, MDA-MB-435 and BT-474
cells following RNF5 silencing due to differences in
TP53 status, as TP53 is only functional in MCF-7
cells (21,26,27).
Furthermore, TP53 expression was increased following the
silencing of RNF5 in MCF-7 cells, suggesting that RNF5 may
be involved in the inhibition of TP53 by Rho GTPase, Src
networks or ERAD (21). The present
study demonstrated that higher RNF5 expression was
associated with an improved prognosis in patients with glioma,
which may be consistent with TP53 mutations in glioma,
particularly in GBM (28,29). Previous studies revealed a higher
rate of TP53 mutations in patients with glioma compared with
healthy subjects (30–33). Therefore, it is possible that in
patients with glioma, mutations in TP53 may reverse the
inhibition of proliferation induced by RNF5 silencing. The
apparently opposite prognostic effect of RNF5 expression
levels in patients with glioma necessitates further investigation
of the mechanism of action of RNF5. The present study
investigated cell colony formation of U251 cells overexpressing
RNF5. However, experiments on apoptosis, invasion and
migration should be performed in future studies. Furthermore, the
lack of validation on clinical samples is a limitation of the
present study.
In summary, the present study revealed that
RNF5 is differentially expressed in patients with glioma
with different disease grades and is associated with patient
prognosis. Moreover, GSEA revealed KEGG pathways that are
significantly associated with RNF5. Through correlation
analysis, possible ubiquitin substrates for RNF5 in patients
with glioma were predicted. These results provided a meaningful
insight into the treatment of glioma.
Acknowledgements
The authors would like to thank Mr. Xue Shengbai
(Clinical Department of Nanjing Medical University) for providing
technical support.
Funding
The present study was supported by the Foundation of
Jiangsu Provincial Health Department (grant no. YG201514), Xuzhou
Municipal Bureau on Science and Technology (grant no. XM12B055) and
Xuzhou Medical University (grant no. 2018KJ09).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YG, CX, MJ, QS and YS conceived and designed this
study. YG, CX, MJ, QA, BZ, XC, LW and YW performed the experiments.
YG, CX and LW conducted statistical analysis. YG, CX, QA, QS and YS
wrote the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study protocol was approved by the
Ethics Committee of Xuzhou Children's Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Stupp R and Roila F; ESMO Guidelines
Working Group, : Malignant glioma: ESMO clinical recommendations
for diagnosis, treatment and follow-up. Ann Oncol. 20 (Suppl
4):S126–S128. 2009. View Article : Google Scholar
|
|
2
|
Meyer MA: Malignant gliomas in adults. N
Engl J Med. 359:18502008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stupp R, Hegi ME, van den Bent MJ, Mason
WP, Weller M, Mirimanoff RO and Cairncross JG; European
Organisation for Research and Treatment of Cancer Brain Tumor and
Radiotherapy Groups; National Cancer Institute of Canada Clinical
Trials Group, : Changing paradigms-an update on the
multidisciplinary management of malignant glioma. Oncologist.
11:165–180. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stupp R, Hottinger AF, van den Bent MJ,
Dietrich PY and Brandes AA: Frequently asked questions in the
medical management of high-grade glioma: A short guide with
practical answers. Ann Oncol. 19 (Suppl 7):vii209–vii216. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Buonerba C, Di Lorenzo G, Marinelli A,
Federico P, Palmieri G, Imbimbo M, Conti P, Peluso G, De Placido S
and Sampson JH: A comprehensive outlook on intracerebral therapy of
malignant gliomas. Crit Rev Oncol Hematol. 80:54–68. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sherman JH, Hoes K, Marcus J, Komotar RJ,
Brennan CW and Gutin PH: Neurosurgery for brain tumors: Update on
recent technical advances. Curr Neurol Neurosci Rep. 11:313–319.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kyushiki H, Kuga Y, Suzuki M, Takahashi E
and Horie M: Cloning, expression and mapping of a novel RING-finger
gene (RNF5), a human homologue of a putative zinc-finger gene from
Caenorhabditis elegans. Cytogenet Cell Genet. 79:114–117. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Matsuda N, Suzuki T, Tanaka K and Nakano
A: Rma1, a novel type of RING finger protein conserved from
Arabidopsis to human, is a membrane-bound ubiquitin ligase.
J Cell Sci. 114:1949–1957. 2001.PubMed/NCBI
|
|
9
|
Younger JM, Chen L, Ren HY, Rosser MF,
Turnbull EL, Fan CY, Patterson C and Cyr DM: Sequential
quality-control checkpoints triage misfolded cystic fibrosis
transmembrane conductance regulator. Cell. 126:571–582. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Grove DE, Fan CY, Ren HY and Cyr DM: The
endoplasmic reticulum-associated Hsp40 DNAJB12 and Hsc70 cooperate
to facilitate RMA1 E3-dependent degradation of nascent
CFTRDeltaF508. Mol Biol Cell. 22:301–314. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Didier C, Broday L, Bhoumik A, Israeli S,
Takahashi S, Nakayama K, Thomas SM, Turner CE, Henderson S, Sabe H
and Ronai Z: RNF5, a RING finger protein that regulates cell
motility by targeting paxillin ubiquitination and altered
localization. Mol Cell Biol. 23:5331–5345. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhong B, Zhang L, Lei C, Li Y, Mao AP,
Yang Y, Wang YY, Zhang XL and Shu HB: The ubiquitin ligase RNF5
regulates antiviral responses by mediating degradation of the
adaptor protein MITA. Immunity. 30:397–407. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jeon YJ, Khelifa S, Ratnikov B, Scott DA,
Feng Y, Parisi F, Ruller C, Lau E, Kim H, Brill LM, et al:
Regulation of glutamine carrier proteins by RNF5 determines breast
cancer response to ER stress-inducing chemotherapies. Cancer Cell.
27:354–369. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gao Y, Han D, Sun L, Huang Q, Gai G, Wu Z,
Meng W and Chen X: PPARα regulates the proliferation of human
glioma cells through miR-214 and E2F2. Biomed Res Int.
2018:38427532018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gravendeel LA, Kouwenhoven MC, Gevaert O,
de Rooi JJ, Stubbs AP, Duijm JE, Daemen A, Bleeker FE, Bralten LB,
Kloosterhof NK, et al: Intrinsic gene expression profiles of
gliomas are a better predictor of survival than histology. Cancer
Res. 69:9065–9072. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sun L, Hui A, Su Q, Vortmeyer A, Kotliarov
Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey R, et al:
Neuronal and glioma-derived stem cell factor induces angiogenesis
within the brain. Cancer Cell. 9:287–300. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta DeltaC(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang X, Tian Q, Wang L, Liu Y, Li B,
Liang Z, Gao P, Zheng K, Zhao B and Lu H: Radiomics strategy for
molecular subtype stratification of lower-grade glioma: Detecting
IDH and TP53 mutations based on multimodal MRI. J Magn Reson
Imaging. 48:916–926. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lehrer S, Rheinstein PH, Green S and
Rosenzweig KE: von Willebrand factor gene expression in primary
lower grade glioma: Mutually Co-occurring mutations in von
Willebrand factor, ATRX, and TP53. Brain Tumor Res Treat. 7:33–38.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Malmer B, Gronberg H, Andersson U, Jonsson
BA and Henriksson R: Microsatellite instability, PTEN and p53
germline mutations in glioma families. Acta Oncol. 40:633–637.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bromberg KD, Kluger HM, Delaunay A, Abbas
S, DiVito KA, Krajewski S and Ronai Z: Increased expression of the
E3 ubiquitin ligase RNF5 is associated with decreased survival in
breast cancer. Cancer Res. 67:8172–8179. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shi H, Zheng B, Wu Y, Tang Y, Wang L, Gao
Y, Gong H, Du J and Yu R: Ubiquitin ligase Siah1 promotes the
migration and invasion of human glioma cells by regulating HIF-1α
signaling under hypoxia. Oncol Rep. 33:1185–1190. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu Y, Wang L, Bao H, Zou S, Fu C, Gong H,
Gao Y, Tang Y, Yu R and Shi H: Nrdp1S, short variant of Nrdp1,
inhibits human glioma progression by increasing Nrdp1-mediated
ErbB3 ubiquitination and degradation. J Cell Mol Med. 20:422–429.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shi H, Du J, Wang L, Zheng B, Gong H, Wu
Y, Tang Y, Gao Y and Yu R: Lower expression of Nrdp1 in human
glioma contributes tumor progression by reducing apoptosis. IUBMB
Life. 66:704–710. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shi H, Gong H, Cao K, Zou S, Zhu B, Bao H,
Wu Y, Gao Y, Tang Y and Yu R: Nrdp1-mediated ErbB3 degradation
inhibits glioma cell migration and invasion by reducing cytoplasmic
localization of p27(Kip1). J Neurooncol. 124:357–364. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Runnebaum IB, Nagarajan M, Bowman M, Soto
D and Sukumar S: Mutations in p53 as potential molecular markers
for human breast cancer. Proc Natl Acad Sci USA. 88:10657–10661.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nieves-Neira W and Pommier Y: Apoptotic
response to camptothecin and 7-hydroxystaurosporine (UCN-01) in the
8 human breast cancer cell lines of the NCI Anticancer Drug Screen:
Multifactorial relationships with topoisomerase I, protein kinase
C, Bcl-2, p53, MDM-2 and caspase pathways. Int J Cancer.
82:396–404. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kawasoe T, Takeshima H, Yamashita S,
Mizuguchi S, Fukushima T, Yokogami K and Yamasaki K: Detection of
p53 mutations in proliferating vascular cells in glioblastoma
multiforme. J Neurosurg. 122:317–323. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Marutani M, Tonoki H, Tada M, Takahashi M,
Kashiwazaki H, Hida Y, Hamada J, Asaka M and Moriuchi T:
Dominant-negative mutations of the tumor suppressor p53 relating to
early onset of glioblastoma multiforme. Cancer Res. 59:4765–4769.
1999.PubMed/NCBI
|
|
30
|
Lin C, Liang Y, Zhu H, Zhang J and Zhong
X: R280T mutation of p53 gene promotes proliferation of human
glioma cells through GSK-3β/PTEN pathway. Neurosci Lett. 529:60–65.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Robertson LB, Armstrong GN, Olver BD,
Lloyd AL, Shete S, Lau C, Claus EB, Barnholtz-Sloan J, Lai R,
Il'yasova D, et al: Survey of familial glioma and role of germline
p16INK4A/p14ARF and p53 mutation. Fam Cancer. 9:413–421. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cui W, Wu R, Cao H, Gao J, Wang X and Ren
Q: P53 gene mutation and expression of MDM2, P53, P16 protein and
their relationship in human glioma. J Huazhong Univ Sci Technolog
Med Sci. 25:622–624, 635. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hayashi Y, Yamashita J and Yamaguchi K:
Timing and role of p53 gene mutation in the recurrence of glioma.
Biochem Biophys Res Commun. 180:1145–1150. 1991. View Article : Google Scholar : PubMed/NCBI
|