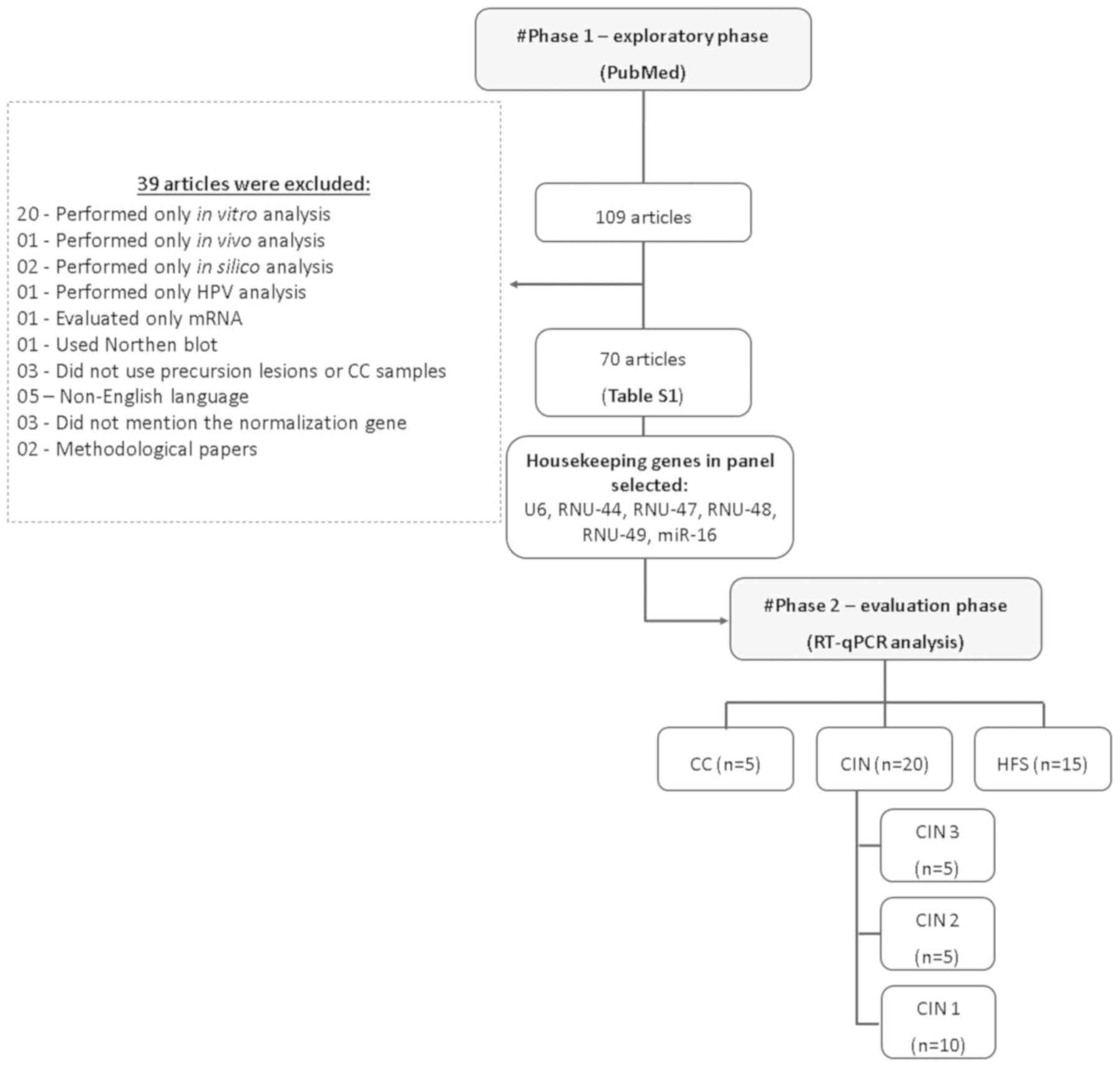
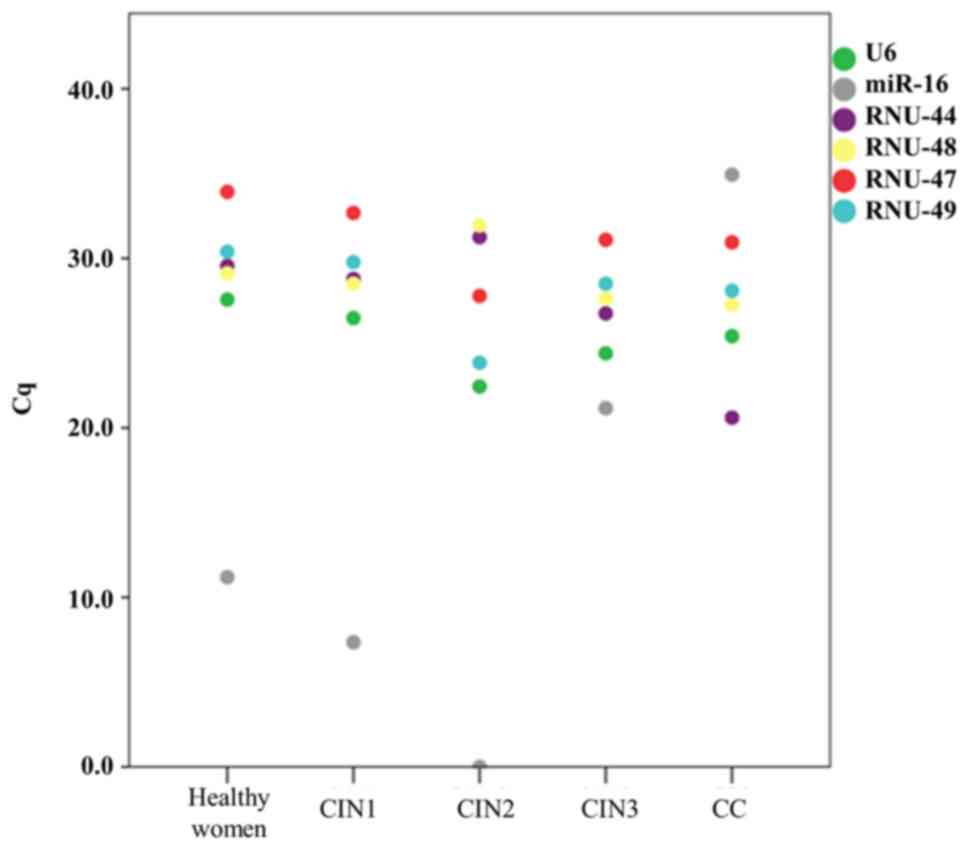
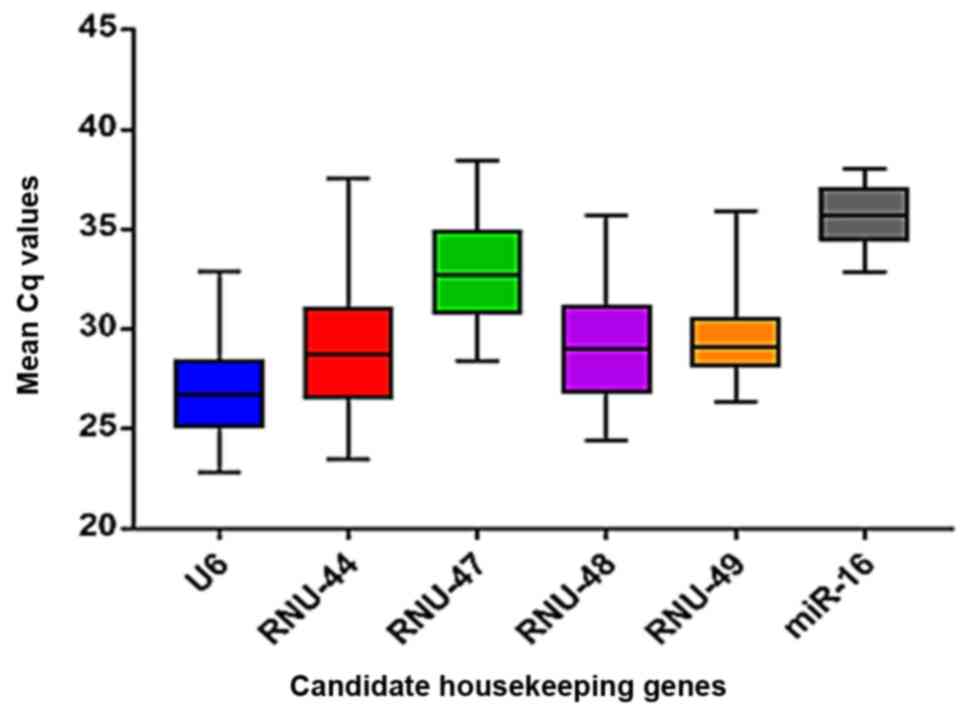
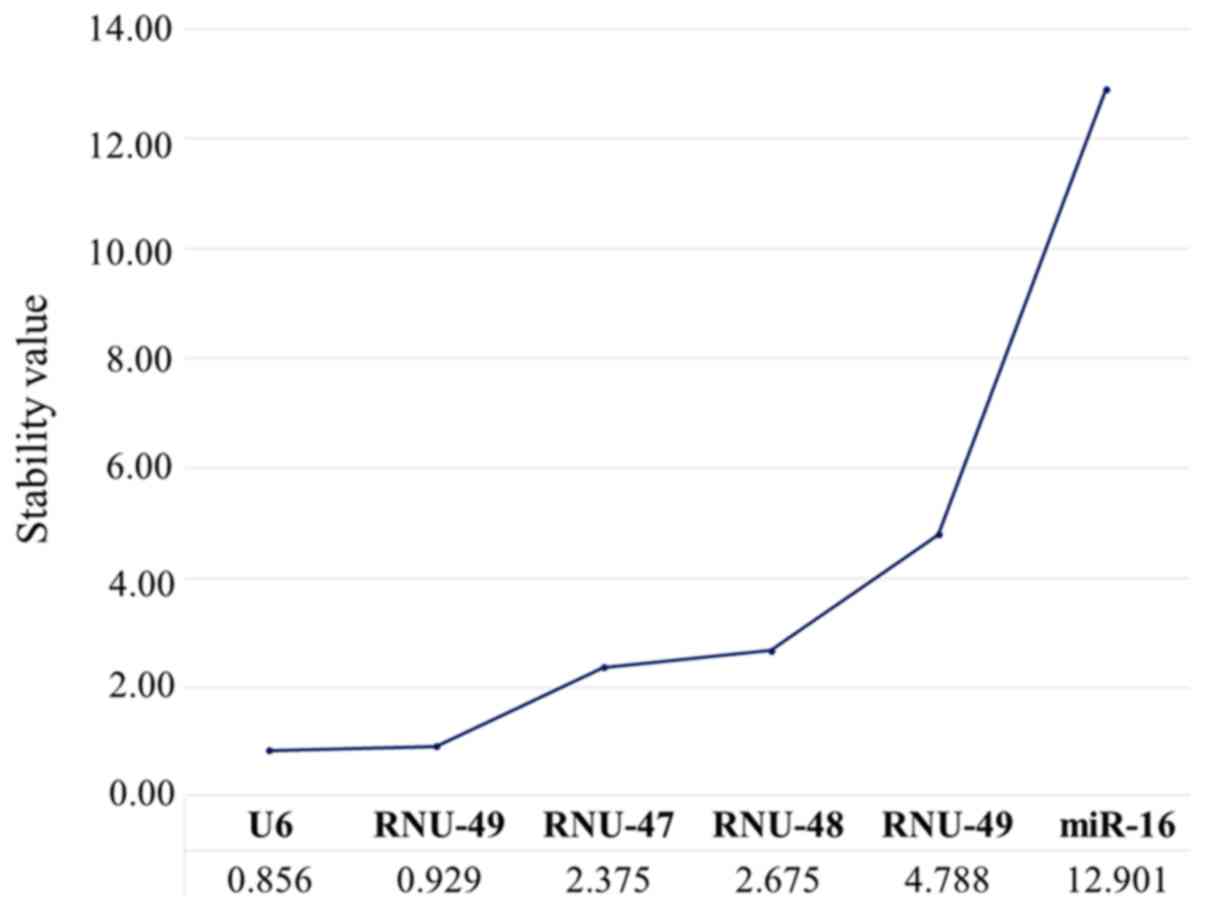
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parkin DM and Bray F: The burden of
HPV-related cancers. Vaccine. 24 (Suppl 3):S11–S25. 2006.
View Article : Google Scholar
|
|
3
|
Iorio MV and Croce CM: Causes and
consequences of microRNA dysregulation. Cancer J. 18:215–222. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
He Y, Lin J, Ding Y, Liu G, Luo Y, Huang
M, Xu C, Kim TK, Etheridge A, Lin M, et al: A systematic study on
dysregulated microRNAs in cervical cancer development. Int J
Cancer. 138:1312–1327. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pardini B, De Maria D, Francavilla A, Di
Gaetano C, Ronco G and Naccarati A: MicroRNAs as markers of
progression in cervical cancer: A systematic review. BMC Cancer.
18:6962018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Di Meo A, Bartlett J, Cheng Y, Pasic MD
and Yousef GM: Liquid biopsy: A step forward towards precision
medicine in urologic malignancies. Mol Cancer. 16:802017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Finotti A, Allegretti M, Gasparello J,
Giacomini P, Spandidos DA, Spoto G and Gambari R: Liquid biopsy and
PCR-free ultrasensitive detection systems in oncology (Review). Int
J Oncol. 53:1395–1434. 2018.PubMed/NCBI
|
|
9
|
Muinelo-Romay L, Casas-Arozamena C and
Abal M: Liquid biopsy in endometrial cancer: New opportunities for
personalized oncology. Int J Mol Sci. 19:E23112018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yao T, Rao Q, Liu L, Zheng C, Xie Q, Liang
J and Lin Z: Exploration of tumor-suppressive microRNAs silenced by
DNA hypermethylation in cervical cancer. Virol J. 10:1752013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang Y, Xie YJ, Xu Q, Chen JX, Shan NC and
Zhang Y: Down-regulation of miR-1246 in cervical cancer tissues and
its clinical significance. Gynecol Oncol. 138:683–688. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yu Q, Liu SL, Wang H, Shi G, Yang P and
Chen XL: miR-126 Suppresses the proliferation of cervical cancer
cells and alters cell sensitivity to the chemotherapeutic drug
bleomycin. Asian Pac J Cancer Prev. 14:6569–6572. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang L, Wang Q, Li HL and Han LY:
Expression of MiR200a, miR93, metastasis-related gene RECK and
MMP2/MMP9 in human cervical carcinoma-relationship with prognosis.
Asian Pac J Cancer Prev. 14:2113–2118. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yin ZL, Wang YL, Ge SF, Guo TT, Wang L,
Zheng XM and Liu J: Reduced expression of miR-503 is associated
with poor prognosis in cervical cancer. Eur Rev Med Pharmacol Sci.
19:4081–4085. 2015.PubMed/NCBI
|
|
15
|
Wang LQ, Zhang Y, Yan H, Liu KJ and Zhang
S: MicroRNA-373 functions as an oncogene and targets YOD1 gene in
cervical cancer. Biochem Biophys Res Commun. 459:515–520. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Song X, Shi B, Huang K and Zhang W:
miR-133a inhibits cervical cancer growth by targeting EGFR. Oncol
Rep. 34:1573–1580. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li QQ, Zhang L, Wan HY, Liu M, Li X and
Tang H: CREB1-driven expression of miR-320a promotes mitophagy by
down-regulating VDAC1 expression during serum starvation in
cervical cancer cells. Oncotarget. 6:34924–34940. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wei Q, Li YX, Liu M, Li X and Tang H:
MiR-17-5p targets TP53INP1 and regulates cell proliferation and
apoptosis of cervical cancer cells. IUBMB Life. 64:697–704. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen J, Yao D, Li Y, Chen H, He C, Ding N,
Lu Y, Ou T, Zhao S, Li L and Long F: Serum microRNA expression
levels can predict lymph node metastasis in patients with
early-stage cervical squamous cell carcinoma. Int J Mol Med.
32:557–567. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yue C, Wang M, Ding B, Wang W, Fu S, Zhou
D, Zhang Z and Han S: Polymorphism of the pre-miR-146a is
associated with risk of cervical cancer in a Chinese population.
Gynecol Oncol. 122:33–37. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou Q, Han LR, Zhou YX and Li Y: MiR-195
suppresses cervical cancer migration and invasion through targeting
Smad3. Int J Gynecol Cancer. 26:817–824. 2016. View Article : Google Scholar
|
|
22
|
Chen XF and Liu Y: MicroRNA-744 inhibited
cervical cancer growth and progression through apoptosis induction
by regulating Bcl-2. Biomed Pharmacother. 81:379–387. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun Y, Yang X, Liu M and Tang H: B4GALT3
up-regulation by miR-27a contributes to the oncogenic activity in
human cervical cancer cells. Cancer Lett. 375:284–292. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang J, Wang F, Xu J, Wang X, Ye F and
Xie X: Micro ribonucleic acid-93 promotes oncogenesis of cervical
cancer by targeting RAB11 family interacting protein 1. J Obstet
Gynaecol Res. 42:1168–1179. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xiao S, Liao S, Zhou Y, Jiang B, Li Y and
Xue M: High expression of octamer transcription factor 1 in
cervical cancer. Oncol Lett. 7:1889–1894. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yi Y, Li H, Lv Q, Wu K and Zhang W, Zhang
J, Zhu D, Liu Q and Zhang W: miR-202 inhibits the progression of
human cervical cancer through inhibition of cyclin D1. Oncotarget.
7:72067–72075. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cheng YX, Zhang QF, Hong L, Pan F, Huang
JL, Li BS and Hu M: MicroRNA-200b suppresses cell invasion and
metastasis by inhibiting the epithelial-mesenchymal transition in
cervical carcinoma. Mol Med Rep. 13:3155–3160. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun L, Jiang R, Li J, Wang B, Ma C, Lv Y
and Mu N: MicoRNA-425-5p is a potential prognostic biomarker for
cervical cancer. Ann Clin Biochem. 54:127–133. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu Y, Zhang Y and Zhang S: MicroRNA-92
regulates cervical tumorigenesis and its expression is upregulated
by human papillomavirus-16 E6 in cervical cancer cells. Oncol Lett.
6:468–474. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang Y, Liu H, Wang X and Chen L:
Up-regulation of microRNA-664 inhibits cell growth and increases
cisplatin sensitivity in cervical cancer. Int J Clin Exp Med.
8:18123–18129. 2015.PubMed/NCBI
|
|
31
|
Qin X, Wan Y, Wang S and Xue M:
MicroRNA-125a-5p modulates human cervical carcinoma proliferation
and migration by targeting ABL2. Drug Des Devel Ther. 10:71–79.
2015.PubMed/NCBI
|
|
32
|
Sun P, Shen Y, Gong JM, Zhou LL, Sheng JH
and Duan FJ: A new MicroRNA expression signature for cervical
cancer. Int J Gynecol Cancer. 27:339–343. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chandrasekaran KS, Sathyanarayanan A and
Karunagaran D: MicroRNA-214 suppresses growth, migration and
invasion through a novel target, high mobility group AT-hook 1, in
human cervical and colorectal cancer cells. Br J Cancer.
115:741–751. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lin W, Feng M, Chen G, Zhou Z, Li J and Ye
Y: Characterization of the microRNA profile in early-stage cervical
squamous cell carcinoma by next-generation sequencing. Oncol Rep.
37:1477–1486. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Azizmohammadi S, Safari A, Azizmohammadi
S, Kaghazian M, Sadrkhanlo M, Yahaghi E, Farshgar R and
Seifoleslami M: Molecular identification of miR-145 and miR-9
expression level as prognostic biomarkers for early-stage cervical
cancer detection. QJM. 110:11–15. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jin X, Chen X, Hu Y, Ying F, Zou R, Lin F,
Shi Z, Zhu X, Yan X, Li S and Zhu H: LncRNA-TCONS_00026907 is
involved in the progression and prognosis of cervical cancer
through inhibiting miR-143-5p. Cancer Med. 6:1409–1423. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu J and Ni S: Association between
genetic polymorphisms in the promoters of let-7 and risk of
cervical squamous cell carcinoma. Gene. 642:256–260. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou Y, Hao Y, Li Y, Li R, Wu R, Wang S
and Fang Z: Amplification and up-regulation of MIR30D was
associated with disease progression of cervical squamous cell
carcinomas. BMC Cancer. 17:2302017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Song R, Cong L, Ni G and Chen M, Sun H,
Sun Y and Chen M: MicroRNA-195 inhibits the behavior of cervical
cancer tumors by directly targeting HDGF. Oncol Lett. 14:767–775.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhao J, Li B, Shu C, Ma Y and Gong Y:
Downregulation of miR-30a is associated with proliferation and
invasion via targeting MEF2D in cervical cancer. Oncol Lett.
14:7437–7442. 2017.PubMed/NCBI
|
|
41
|
He S, Liao B, Deng Y, Su C, Tuo J, Liu J,
Yao S and Xu L: MiR-216b inhibits cell proliferation by targeting
FOXM1 in cervical cancer cells and is associated with better
prognosis. BMC Cancer. 17:6732017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li S, Yang F, Wang M, Cao W and Yang Z:
miR-378 functions as an onco-miRNA by targeting the
ST7L/Wnt/β-catenin pathway in cervical cancer. Int J Mol Med.
40:1047–1056. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hu Y, Xie H, Liu Y, Liu W, Liu M and Tang
H: miR-484 suppresses proliferation and epithelial-mesenchymal
transition by targeting ZEB1 and SMAD2 in cervical cancer cells.
Cancer Cell Int. 17:362017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Luo S, Li N, Yu S, Chen L, Liu C and Rong
J: MicroRNA-92a promotes cell viability and invasion in cervical
cancer via directly targeting Dickkopf-related protein 3. Exp Ther
Med. 14:1227–1234. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang Z, Wang J, Wang X, Song W, Shi Y and
Zhang L: MicroRNA-21 promotes proliferation, migration, and
invasion of cervical cancer through targeting TIMP3. Arch Gynecol
Obstet. 297:433–442. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li M, Li BY, Xia H and Jiang LL:
Expression of microRNA-142-3p in cervical cancer and its
correlation with prognosis. Eur Rev Med Pharmacol Sci.
21:2346–2350. 2017.PubMed/NCBI
|
|
47
|
Zhao Y, Liu X and Lu YX: MicroRNA-143
regulates the proliferation and apoptosis of cervical cancer cells
by targeting HIF-1α. Eur Rev Med Pharmacol Sci. 21:5580–5586.
2017.PubMed/NCBI
|
|
48
|
Tao L, Zhang CY, Guo L, Li X, Han NN, Zhou
Q and Liu ZL: MicroRNA-497 Accelerates Apoptosis While Inhibiting
Proliferation, Migration, and Invasion through negative regulation
of the MAPK/ERK signaling pathway via RAF-1. J Cell Physiol.
233:6578–6588. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gocze K, Gombos K, Kovacs K, Juhasz K,
Gocze P and Kiss I: MicroRNA expressions in HPV-induced cervical
dysplasia and cancer. Anticancer Res. 35:523–530. 2015.PubMed/NCBI
|
|
50
|
Zhang J, Zheng F, Yu G, Yin Y and Lu Q:
miR-196a targets netrin 4 and regulates cell proliferation and
migration of cervical cancer cells. Biochem Biophys Res Commun.
440:582–588. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chen Y, Ma C, Zhang W, Chen Z and Ma L:
Down regulation of miR-143 is related with tumor size, lymph node
metastasis and HPV16 infection in cervical squamous cancer. Diagn
Pathol. 9:882014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang C and Jiang T: MicroRNA-335
represents an independent prognostic marker in cervical cancer.
Tumour Biol. 36:5825–5830. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zeng K, Zheng W, Mo X, Liu F, Li M, Liu Z,
Zhang W and Hu X: Dysregulated microRNAs involved in the
progression of cervical neoplasm. Arch Gynecol Obstet. 292:905–913.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ma L, Hong Y, Lu C, Chen Y and Ma C: The
occurrence of cervical cancer in Uygur women in Xinjiang Uygur
Autonomous Region is correlated to microRNA-146a and ethnic factor.
Int J Clin Exp Pathol. 8:9368–9375. 2015.PubMed/NCBI
|
|
55
|
Zheng W, Liu Z, Zhang W and Hu X: miR-31
functions as an oncogene in cervical cancer. Arch Gynecol Obstet.
292:1083–1089. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Qian K, Pietilä T, Rönty M, Michon F,
Frilander MJ, Ritari J, Tarkkanen J, Paulín L, Auvinen P and
Auvinen E: Identification and Validation of Human Papillomavirus
Encoded microRNAs. PLoS One. 8:e702022013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang J, Jia J, Zhao L, Li X, Xie Q, Chen
X, Wang J and Lu F: Down-regulation of microRNA-9 leads to
activation of IL-6/Jak/STAT3 pathway through directly targeting
IL-6 in HeLa cell. Mol Carcinog. 55:732–742. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhao S, Yao D, Chen J and Ding N:
Circulating miRNA-20a and miRNA-203 for screening lymph node
metastasis in early stage cervical cancer. Genet Test Mol Biomark.
17:631–636. 2013. View Article : Google Scholar
|
|
59
|
Jia W, Wu Y, Zhang Q, Gao GE, Zhang C and
Xiang Y: Expression profile of circulating microRNAs as a promising
fingerprint for cervical cancer diagnosis and monitoring. Mol Clin
Oncol. 3:851–858. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Jiang W, Pan JJ, Deng YH, Liang MR and Yao
LH: Down-regulated serum microRNA-101 is associated with aggressive
progression and poor prognosis of cervical cancer. J Gynecol Oncol.
28:e752017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhou LL, Shen Y, Gong JM, Sun P and Sheng
JH: MicroRNA-466 with tumor markers for cervical cancer screening.
Oncotarget. 8:70821–70827. 2017.PubMed/NCBI
|
|
62
|
Yu J, Wang Y, Dong R, Huang X, Ding S and
Qiu H: Circulating microRNA-218 was reduced in cervical cancer and
correlated with tumor invasion. J Cancer Res Clin Oncol.
138:671–674. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
You W, Wang Y and Zheng J: Plasma miR-127
and miR-218 Might Serve as Potential Biomarkers for Cervical
Cancer. Reprod Sci. 22:1037–1041. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Luo M, Shen D, Wang W and Xian J: Aberrant
expression of microRNA-26b and its prognostic potential in human
cervical cancer. Int J Clin Exp Pathol. 8:5542–5548.
2015.PubMed/NCBI
|
|
65
|
Liu S, Pan X, Yang Q, Wen L, Jiang Y, Zhao
Y and Li G: MicroRNA-18a enhances the radiosensitivity of cervical
cancer cells by promoting radiation-induced apoptosis. Oncol Rep.
33:2853–2862. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Xie H, Lee L, Scicluna P, Kavak E, Larsson
C, Sandberg R and Lui WO: Novel functions and targets of miR-944 in
human cervical cancer cells. Int J Cancer. 136:E230–E241. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Shen S, Wang L, Jia Y, Hao Y, Zhang L and
Wang H: Upregulation of microRNA-224 is associated with aggressive
progression and poor prognosis in human cervical cancer. Diagn
Pathol. 8:692013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Huang L, Lin JX, Yu YH, Zhang MY, Wang HY
and Zheng M: Downregulation of Six MicroRNAs Is Associated with
Advanced Stage, Lymph Node Metastasis and Poor Prognosis in Small
Cell Carcinoma of the Cervix. PLoS One. 7:e337622012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Xing AY, Wang B, Shi DB, Zhang XF, Gao C,
He XQ, Liu WJ and Gao P: Deregulated expression of miR-145 in
manifold human cancer cells. Exp Mol Pathol. 95:91–97. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Lee H, Kim KR, Cho NH, Hong SR, Jeong H,
Kwon SY, Park KH, An HJ, Kim TH, Kim I, et al: MicroRNA expression
profiling and Notch1 and Notch2 expression in minimal deviation
adenocarcinoma of uterine cervix. World J Surg Oncol. 12:3342014.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ivanov MK, Titov SE, Glushkov SA,
Dzyubenko VV, Malek AV, Arkhangelskaya PA, Samsonov RB, Mikhetko
AA, Bakhidze EV, Berlev IV and Kolesnikov NN: Detection of
high-grade neoplasia in air-dried cervical PAP smears by a
microRNA-based classifier. Oncol Rep. 39:1099–1111. 2018.PubMed/NCBI
|
|
72
|
Yu X, Zhao W, Yang X, Wang Z and Hao M:
miR-375 affects the proliferation, invasion, and apoptosis of
HPV16-positive human cervical cancer cells by targeting IGF-1R. Int
J Gynecol Cancer. 26:851–858. 2016. View Article : Google Scholar
|
|
73
|
Hao M, Zhao W, Zhang L, Wang H and Yang X:
Low folate levels are associated with methylation-mediated
transcriptional repression of miR-203 and miR-375 during cervical
carcinogenesis. Oncol Lett. 11:3863–3869. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Nagamitsu Y, Nishi H, Sasaki T, Takaesu Y,
Terauchi F and Isaka K: Profiling analysis of circulating microRNA
expression in cervical cancer. Mol Clin Oncol. 5:189–194. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Kogo R, How C, Chaudary N, Bruce J, Shi W,
Hill RP, Zahedi P, Yip KW and Liu FF: The microRNA-218~Survivin
axis regulates migration, invasion, and lymph node metastasis in
cervical cancer. Oncotarget. 6:1090–1100. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Lajer CB, Garnæs E, Friis-Hansen L,
Norrild B, Therkildsen MH, Glud M, Rossing M, Lajer H, Svane D,
Skotte L, et al: The role of miRNAs in human papilloma virus
(HPV)-associated cancers: Bridging between HPV-related head and
neck cancer and cervical cancer. Br J Cancer. 106:1526–1534. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Huang YW, Kuo CT, Chen JH, Goodfellow PJ,
Huang TH, Rader JS and Uyar DS: Hypermethylation of miR-203 in
endometrial carcinomas. Gynecol Oncol. 133:340–345. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Myklebust MP, Bruland O, Fluge Ø,
Skarstein A, Balteskard L and Dahl O: MicroRNA-15b is induced with
E2F-controlled genes in HPV-related cancer. Br J Cancer.
105:1719–1725. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Sharma Saha S, Roy Chowdhury R, Mondal NR,
Chakravarty B, Chatterjee T, Roy S and Sengupta S: Identification
of genetic variation in the lncRNA HOTAIR associated with
HPV16-related cervical cancer pathogenesis. Cell Oncol (Dordr).
39:559–572. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Andersen CL, Jensen JL and Ørntoft TF:
Normalization of real-time quantitative reverse transcription-PCR
data: A model-based variance estimation approach to identify genes
suited for normalization, applied to bladder and colon cancer data
sets. Cancer Res. 64:5245–5250. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Rossi ED, Bizzarro T, Martini M,
Capodimonti S, Sarti D, Cenci T, Bilotta M, Fadda G and Larocca LM:
The evaluation of miRNAs on thyroid FNAC: The promising role of
miR-375 in follicular neoplasms. Endocrine. 54:723–732. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Kottaridi C, Spathis A, Margari N, Koureas
N, Terzakis E, Chrelias C, Pappas A, Bilirakis E, Pouliakis A,
Panayiotides IJ and Karakitsos P: Evaluation analysis of miRNAs
overexpression in liquid-based cytology endometrial samples. J
Cancer. 8:2699–2703. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Barodawala SM, Chadha K, Kavishwar V,
Murthy A and Shetye S: Cervical cancer screening by molecular
Pap-transformation of gynecologic cytology. Diagn Cytopathol.
47:374–381. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Bonde J, Ejegod DM, Cuschieri K, Dillner
J, Heideman DAM, Quint W, Pavon Ribas MA, Padalko E, Christiansen
IK, Xu L and Arbyn M: The Valgent4 protocol: Robust analytical and
clinical validation of 11 HPV assays with genotyping on cervical
samples collected in SurePath medium. J Clin Virol. 108:64–71.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Tota JE, Bentley J, Blake J, Coutlée F,
Duggan MA, Ferenczy A, Franco EL, Fung-Kee-Fung M, Gotlieb W,
Mayrand MH, et al: Approaches for triaging women who test positive
for human papillomavirus in cervical cancer screening. Prev Med.
98:15–20. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Derveaux S, Vandesompele J and Hellemans
J: How to do successful gene expression analysis using real-time
PCR. Methods. 50:227–230. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Kiss T: Small nucleolar RNAs: An abundant
group of noncoding RNAs with diverse cellular functions. Cell.
109:145–148. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Dieci G, Preti M and Montanini B:
Eukaryotic snoRNAs: A paradigm for gene expression flexibility.
Genomics. 94:83–88. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zubillaga-Guerrero MI, Alarcón-Romero Ldel
C, Illades-Aguiar B, Flores-Alfaro E, Bermúdez-Morales VH, Deas J
and Peralta-Zaragoza O: MicroRNA miR-16-1 regulates CCNE1 (cyclin
E1) gene expression in human cervical cancer cells. Int J Clin Exp
Med. 8:15999–16006. 2015.PubMed/NCBI
|