Cervical cancer (CC) is the fourth most common
cancer among women, with an estimated 527,600 new cases and 265,700
deaths worldwide (1). In developed
countries, the incidence and mortality rates have decreased
significantly in the past decades. In contrast, the burden from CC
remains high in developing and underdeveloped countries due to
continuing challenges implementing effective prevention and control
programs. Difficulties in less developed countries include barriers
to accessing health care services, which are compounded by
inadequate cytological examination, usually involving screening
with the low coverage of Papanicolaou (Pap) test (2). To implement additional strategies for
improving CC screening, some programs have focused on introducing
innovative molecular diagnostic tests such as the molecular HPV
testing that could provide more sensitive and specific detection of
precursor lesions once validated.
MicroRNAs (miRNAs) are small non-coding RNAs (19 to
24 nucleotides) involved in the post-transcriptional regulation of
gene expression (3), where they play
a critical role in several cellular processes, such as
proliferation, cell growth and apoptosis (4). Many studies have reported aberrant
expression of miRNAs in cancer (5),
suggesting that these molecules could be used as potential tumor
biomarkers. Indeed, a recent systematic review identified
differentially expressed miRNAs in precursors cervical lesions and
CC that could be associated with tumor progression (6). However, most analyses of miRNA
expression in CC have used tumor tissue samples obtained from
invasive procedures that cause patient discomfort, such as cervical
tissue biopsies or surgery (7). The
development of minimally invasive liquid biopsy cytology (LBC)
based on miRNA expression (7) is a
new approach to identify non-invasive biomarkers for early
diagnosis, monitoring response to therapies and for tumor
progression (7–9). LBC are considered as an accurate and
promising low-cost method for clinical practice but the
standardization of miRNA expression analysis remains a major
challenge. Thus, standardization of this detection technique is
fundamental for the reproducible use of miRNA biomarkers in
clinical practice.
Reverse transcription-quantititative PCR (RT-qPCR)
is a robust technique frequently used in the diagnosis of many
neoplasms and infectious diseases due to its high sensitivity and
specificity (9,10). Analysis of miRNAs is considered an
important new biomarker because miRNAs are specific and stable in
diverse types of clinical samples. However, identification of a
constitutively expressed housekeeping genes for adequate
normalization of miRNAs expression analysis is a crucial step for
better accuracy with this technique. The use of housekeeping genes
as endogenous control is the most common method for normalizing
RT-qPCR data for miRNA expression (11). Housekeeping genes are internal
reaction controls used to gene expression normalization of distinct
miRNAs, and they can have different isoforms. For a gene to be
considered a reliable housekeeping transcript, it needs to meet
some stringent performance criteria (12), such as minimal expression variability
between tissues and physiological states of the organism. Moreover,
the normalization control being used must faithfully measure any
technical variability resulting from differences in the quantity or
quality of genetic material being tested (13). Above all else, the most important of
these criteria is that the pattern of expression of the normalizer
does not interfere or produce artifactual changes in the test
samples. Satisfying these basic conditions are the essential
properties of a good housekeeping gene for transcript normalization
from specific biological samples of interest. The identification of
suitable housekeeping genes is a crucial step for deriving
reproducible results when investigating the differential expression
of miRNAs. The use of unreliable normalization control genes can
lead to an incorrect estimate of the expression levels of miRNAs of
interest (14,15). For this reason, the choice of
appropriate housekeeping genes for normalizing the expression of
miRNAs analysis using LBC is an important issue to be solved. No
housekeeping gene is unique and constitutively expressed in all
sample types, as well as different types of diseases in all
experimental designs, which indicates that the stability of
housekeeping gene expression should be checked rigorously (16,17).
This is the first study to evaluate control
housekeeping genes for miRNA RT-qPCR data normalization in LBC
cervical samples. Since there is little consensus on the best
choice of normalizers, we performed a literature review to identify
housekeeping genes most commonly used in miRNA RT-qPCR data
normalization. In addition, we evaluated their relative expression
levels in LBC samples from patients who underwent routine cervical
cancer screening.
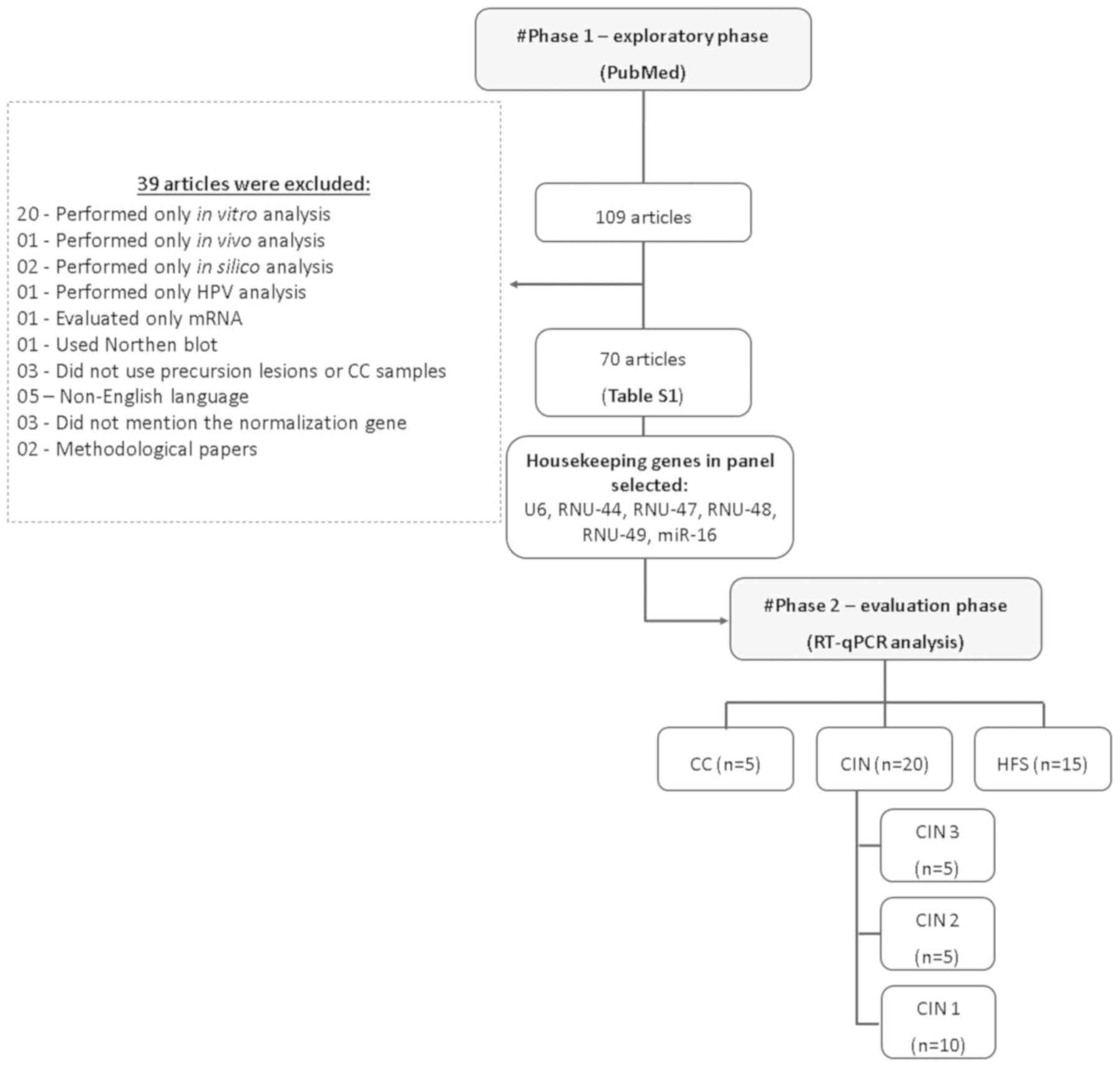
In order to select suitable housekeeping genes, we
conducted a systematic two-phase analysis including an initial
exploratory review of the literature, followed by a laboratory
evaluation phase of selected genes (Fig.
1).
All candidate housekeeping genes identified by our
literature review were selected for expression analysis by RT-qPCR
and tested in the LBC samples from 5 CC (5), 20 CIN (5 CIN3; 5 CIN2 and 10 CIN1) and
15 healthy women (HSF-without CIN). We considered a housekeeping
gene to be suitable for normalization purposes when it was stably
expressed across all samples independently of the histological
condition, and when the cycle quantification (Cq) values did not
exceed 35. Finally, the best housekeeping gene was evaluated using
the NormFinder algorithm, which is software designed to identify
the optimal normalization gene among a set of candidates (80).
We analyzed a total of 40 LBC samples randomly
obtained from women who had undergone routine colposcopy in the
Department of Prevention of the Barretos Cancer Hospital in 2014.
All samples were collected immediately before colposcopy and
preserved in ThinPrep™ Pap test (Hologic) for subsequent molecular
analyses. ThinPrep™ samples were classified into five groups: HFS;
low-grade CIN (CIN1); high-grade CIN (CIN2 group or CIN3 group);
and CC. All CIN- and CC-histological diagnoses from women who
presented with suspicious/abnormal areas during colposcopy were
subsequently confirmed by analyses of tissue samples collected for
the Department of Pathology of the Barretos Cancer Hospital using a
colposcopy-guided cervical biopsy.
ThinPrep™ samples were manually washed to remove the
buffered preservative solution and to lyse blood cells, which could
inhibit downstream molecular analyses. Total RNA was performed
using the RecoverAll Total Nucleic Acid Isolation Kit (Thermo
Fisher Scientific), according to the manufacturer's protocol. The
purity of total RNA was evaluated by NanoDrop®
Spectrophotometer v3.7 (Thermo Fisher Scientific).
Considering that the focus of this study is LBC
cervical samples we do not use housekeeping genes to analyze the
expression of these miRNAs in tissue samples. To perform RT-qPCR
reactions we used TaqMan microRNA assays (Thermo Fisher Scientific)
using LBC cervical samples. Initially, a target-specific stem-loop
reverse transcription RT-PCR was performed using a High Capacity
cDNA Reverse Transcription kit (Thermo Fisher Scientific),
following the protocol provided by the manufacturer. Briefly, for
each sample 10 ng of total RNA was reverse transcribed using
miRNA-specific primers and TaqMan Assays (Table SI) in a 15 µl reaction volume for 30
min at 16°C, 20 min at 42°C and 5 min at 85°C. All RT-PCR reactions
were performed using the Proflex™ 3×32-well PCR system (Thermo
Fisher Scientific). Then 2 µl of the reverse transcription products
(cDNA) was amplified in the QuantStudio 6 Flex Real-Time PCR system
(Thermo Fisher Scientific) using the TaqMan Universal PCR Master
Mix II (Thermo Fisher Scientific), according to the manufacturer's
protocol. All RT-qPCR reactions were performed in triplicate using
Taqman probes. The PCR protocol comprised 40 cycles of 2 min at
50°C, 10 min at 95°C, 15 sec at 95°C and 1 min at 60°C. The
Threshold Cycle (Cq) values were determined using the same
threshold setting and analyzed according to a previously reported
method (81).
All variables were presented using mean values and
standard deviation (SD). ANOVA with a Bonferroni post hoc test and
the Kruskall-Wallis tests were used to compare the mean values of
continuous variables across the histologic groups. P-values of
<0.05 were considered statistically significant. All statistical
analyses were performed with SPSS for Windows, v.21.0 (IBM
Corporation). All graphs was expressed just descriptive analysis
data.
We selected six candidate housekeeping genes based
on their expression profile across the reviewed studies (Table I): U6 (U6 small nuclear RNA); miR-16
(hsa-microRNA-16); RNU-44 (SNORD44 small nucleolar RNA); RNU-48
(SNORD48 small nucleolar RNA); RNU-47 (SNORD47 small nucleolar
RNA); and RNU-49 (SNORD49A small nucleolar RNA). Most of the
candidate genes have previously been described as housekeeping
transcripts for miRNA normalization for expression quantification
using different types of biological samples, such as fresh tissue
biopsies, formalin-fixed paraffin-embedded (FFPE) tissues, serum or
plasma, and air-dried cervical smears. Of the six selected
housekeeping genes, only miR-16 has previously been used as an
endogenous control for LBC expression analysis (28,74).
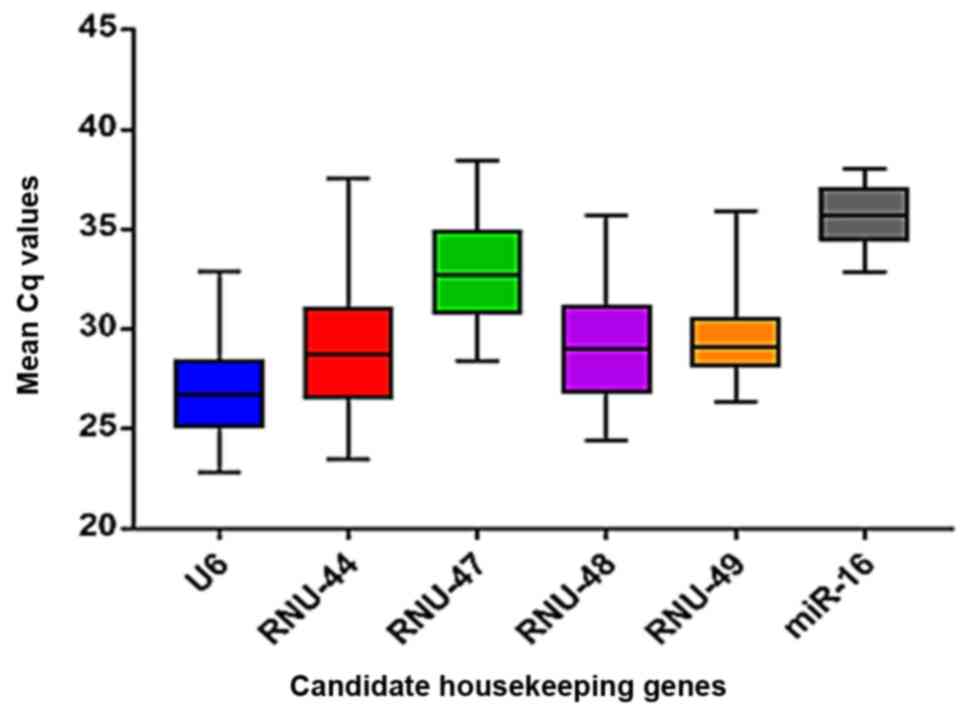
The technical performance of six selected candidate
housekeeping genes as normalization controls was investigated using
40 LBC samples. We found that U6 and RNU-49 had the lowest Cq value
variation among the six tested candidate housekeeping genes
(Fig. 2). Furthermore, both genes
amplified more efficiently than the other candidates and required
fewer amplification cycles to achieve Cq values above background
fluorescence levels. The lower Cq value for U6 (21,81)
indicated that their expression levels and PCR efficiencies
required fewer cycles of amplification to reach the detection
threshold. In addition, we found that the U6 gene was more
uniformly expressed in LBC samples than the other candidate
housekeeping genes. In contrast, more cycles of amplification were
required (Cq values >35) for RNU-44, RNU-47, RNU-48 e miR-16,
indicating that these genes might not be suitable housekeeping
genes for normalization using LBC samples.
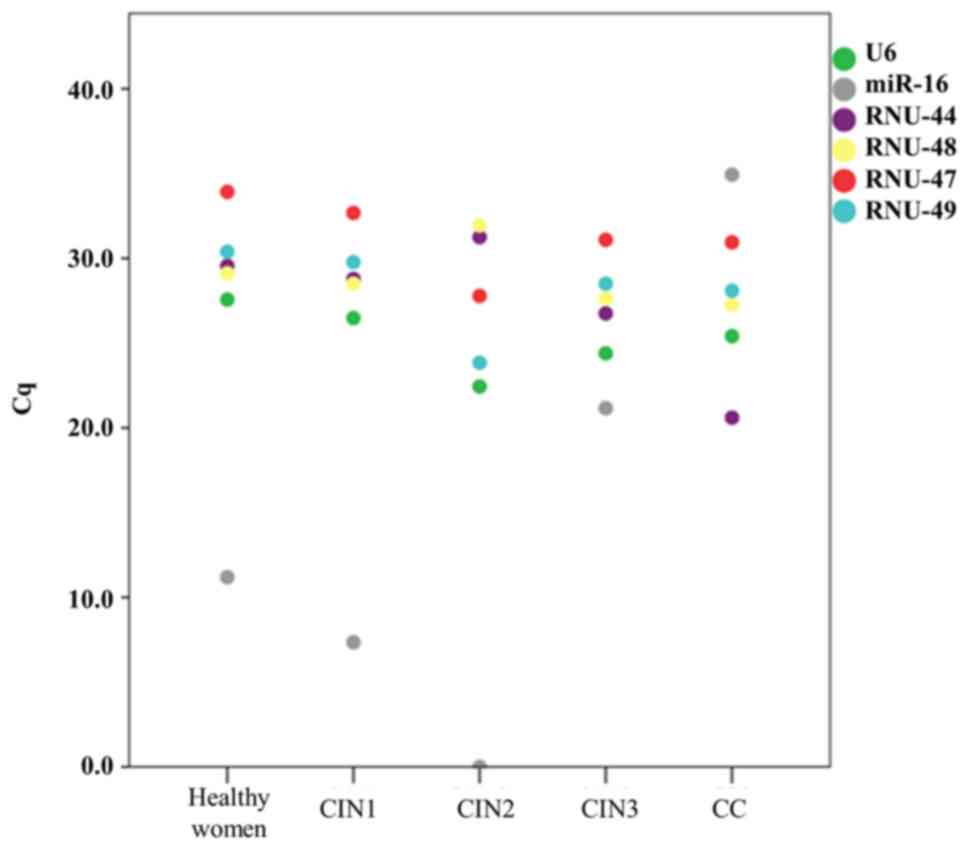
To assess whether the candidate housekeeping genes
were differentially expressed in varying histologic conditions, we
compared the mean Cq values of the candidate housekeeping genes
obtained from each of five histologic groups (Table II). We found no significant
differences in the expression of U6 (P-value, 0.06) and RNU-49
(P-value: 0.128) across all groups, supporting their stable
performance and potential as robust endogenous controls in RT-qPCR
normalization using LBC samples. We did not find a significant
association for RNU-47 (P-value, 0.064) and this gene required a
greater number of amplification cycles across the experimental
groups, especially in LBC samples from patients HFS (34.16±2.85).
We also observed significant differences for miR-16 (P-value,
0.045); RNU-44 (P-value, 0.004); and RNU-48 (P-value, 0.022),
indicating that these genes can be differentially expressed in LBC
samples from patients with different cervical histology and that
such variation could lead to inconsistent normalization. Indeed,
miR-16 exhibited the most variable expression across the groups
(Fig. 3), ranging from no
amplification at all in LBC samples from CIN2 patients to low
levels in CIN1 patients (7.35±15.51). Furthermore, RNU-44 and
RNU-48 were more abundant in LBC samples from CC patients (mean Cq
values: 20.60±11.60 and 27.27±2.30, respectively) in comparison to
other histologic groups indicating that they would likely bias
expression values.
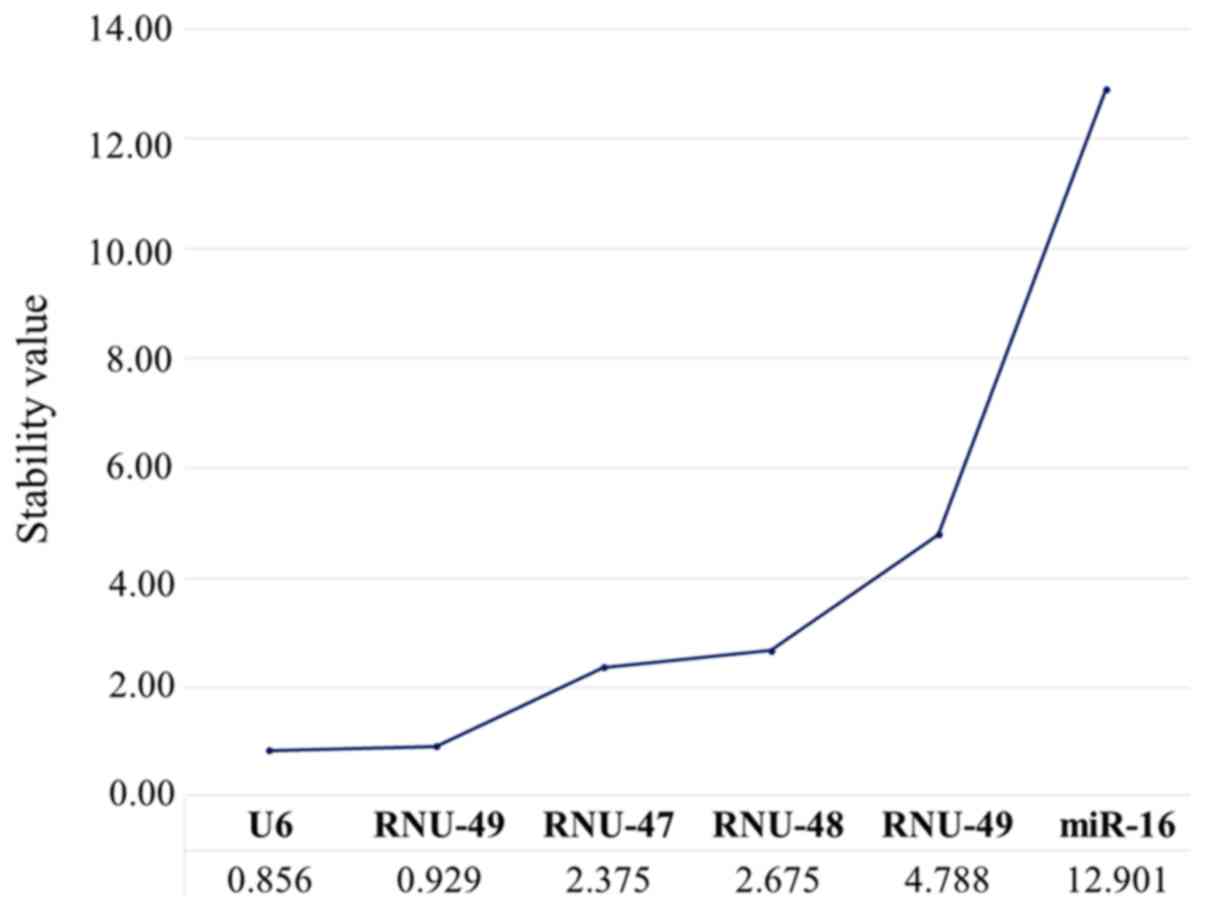
We further analyzed the stability values of each
candidate housekeeping gene using the NormFinder algorithm. We
found that among the six candidates, U6 was the most stable gene
(stability value, 0.856), followed by RNU-49 (0.929) (Fig. 4). In contrast, the other candidate
housekeeping genes presented inadequate stability values, ranging
from 2.375 to 12.901. These findings suggested that U6 and RNU-49
were the best housekeeping genes in LBC samples, whereas miR-16,
RNU-44, RNU-47, and RNU-48 should not be considered suitable for
use as endogenous controls for RT-qPCR normalization.
There have been several studies investigating the
utility of miRNA in translational research, considering deregulated
expression in diverse diseases, variation in tissue-specific
distribution and the overall stability of miRNA in different
clinical samples (19–22). Indeed, there is emerging evidence
demonstrating the feasibility of using miRNAs as non- or minimally
invasive diagnostic biomarkers in cancer. For instance, Rossi et
al (82) evaluated a five-miRNA
expression signature developed for thyroid lesions using fine
needle aspiration cytology (FNAC). Their analysis suggested miR-375
as a promising preoperative biomarker for distinguishing benign
from malignant follicular neoplasms. In another study, Kottaridi
et al (83) designed a panel
of seven overexpressed miRNAs for use in histologically confirmed
LBC malignant endometrial samples to discriminate between
non-malignant and malignant specimens and to identify any samples
with inadequate RNA. There are many studies that have reported
promising molecular approaches to LBC samples for clinical
laboratories (84–86). None of the studies to date have
focused on evaluating miRNAs in LBC cervical samples, which could
be considered an important minimally invasive approach for cancer
detection by miRNA expression data. For this reason, RT-qPCR is now
one of the most commonly used new methods for the evaluation of
miRNA expression due to its high sensitivity and reproducibility
(87,88).
Since reliable normalization is fundamental to
RT-qPCR, there is a need to choose a suitable gene for use as an
endogenous control in order to obtain an accurate miRNA expression
and to ensure consistency. The selection of housekeeping genes as
normalizers for miRNA has relied on choosing from distinct miRNAs
and other small RNAs, such as U6, RNU6B, miR-16, and RNU-44
(37,68,76,82).
However, the choice of housekeeping gene remains quite empirical
because, to the best of our knowledge, there are no previous
studies that have validated endogenous housekeeping control genes
for miRNA normalization in LBC cervical samples.
In this study, we evaluated six candidate
housekeeping genes for miRNA RT-qPCR data using LBC samples from
patients who underwent cervical cancer screening. We analyzed the
expression of five small nucleolar (sno) RNAs: RNU-44, RNU-47,
RNU-48, RNU-49 and U6. The snoRNAs are a group of non-coding RNAs
with variable length (80 to 1000 nt in yeast), mainly required for
ribosomal RNA (rRNA) maturation (89). Many types of snoRNAs have been
described in eukaryotes and each of them corresponds to a specific
mode of transcription (90) and have
been used as housekeeping genes for miRNA normalization (30,31).
Some studies that have used miRNA profiling to discriminate
cervical cancer from benign lesions selected RNU-44 and RNU-48 as
endogenous controls for normalization of miRNA RT-qPCR data, mostly
using tissue (26,32–34) and
serum samples (35). However, our
findings suggest that these snoRNAs are unsuitable for miRNA
normalization in LBC samples, due to the higher number of
amplification cycles required and the differential expression
across distinct histologic groups. In addition, RNU-47 also
required more amplification, confirming that it may not be an
appropriate housekeeping gene. In contrast, RNU-49 and U6 could be
amplified with fewer cycles and both have smaller variation
according to NormFinder algorithm. Several studies have used U6 as
housekeeping gene for RT-qPCR data normalization in cervical
tissues (10–13), whole blood (62), serum (19,58,59) and
air-dried Pap smears (71), but not
previously in LBC samples. In agreement with other studies, our
analyses indicate that U6 is the best housekeeping gene for LBC
samples.
We also evaluated the miR-16 expression in LBC
samples because it has been suggested as a housekeeping gene for
cervical samples in other studies (25,45).
There are doubts about the reliability of this gene for
normalization because some studies have reported miR-16 as
differentially expressed in CC. Zubillaga-Guerrero et al
(91) demonstrated altered
expression of miR-16 in CC, with miR-16 downregulating cyclin E1
(CCNE1) gene expression in cervical cancer cell lines. These data
suggest a potential role of miR-16 in modulating cell cycle in CC
and make it less likely to be a suitable control housekeeping gene.
A recent systematic review also shown that miR-16 was deregulated
and associated with cervical cancer progression (6). In our study, we confirmed that miR-16
was not a good endogenous control for RT-qPCR normalization in LBC
samples because it presented higher variability expression across
all samples-including amplification under background fluorescence
in some cases-and altered expression in different histologic
conditions.
In summary, our data demonstrates that U6 and RNU-49
are suitable housekeeping genes that can be used for miRNA RT-qPCR
analyzes in LBC samples from patients who underwent cervical cancer
screening. This is the first study that provide comprehensive
information on the analytical performance of these genes for future
normalizations of miRNA expression studies in LBC cervical samples
that can be very useful for application of miRNAs in screening with
LBC and had a clinical significance. However, future studies using
RT-qPCR may to demonstrate in practice housekeepings U6 and RNU-49
may be excellent normalizers for liquid-based cytology (LBC)
cervical samples. Thus, continued research efforts should be made
about miRNA expression analysis wisely differentially expressed in
these histological subtypes and LBC cervical samples using other
tools for the stability analysis of housekeepings, such as
geNorm.
The authors would like to thank Dr Jeremy Squire
(Genetics Department, University of Sao Paulo, Ribeirão Preto, São
Paulo, Brazil) for carefully proofreading the manuscript and for
providing constructive criticism.
The present study was supported by internal funding
from FAPESP (grant no. 2016/15831-3) and the Public Ministry of
Work.
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
RLC developed and led the overall study, conducted
the data reviews and the analysis, and prepared the manuscript. DPP
participated in setting up the PCR assay and prepared the
manuscript. KCBS participated in setting up the PCR assay and
critically revised the manuscript. AFE, RMVR and JHTGF designed and
developed the study, and critically revised the manuscript. MMCMS
conceived the study, provided advice during the study development
and prepared the manuscript.
The present study was approved by the Research
Ethics Committee of the Barretos Cancer Hospital (approval no.
784/2014). Each research participant provided written informed
consent for the publication of any data associated with the present
study. All information that could be used to identify the study
participants was kept confidential and encrypted in a secure
database to ensure full confidentiality of clinical information,
laboratory findings and the anonymity of each participant.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parkin DM and Bray F: The burden of
HPV-related cancers. Vaccine. 24 (Suppl 3):S11–S25. 2006.
View Article : Google Scholar
|
|
3
|
Iorio MV and Croce CM: Causes and
consequences of microRNA dysregulation. Cancer J. 18:215–222. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
He Y, Lin J, Ding Y, Liu G, Luo Y, Huang
M, Xu C, Kim TK, Etheridge A, Lin M, et al: A systematic study on
dysregulated microRNAs in cervical cancer development. Int J
Cancer. 138:1312–1327. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pardini B, De Maria D, Francavilla A, Di
Gaetano C, Ronco G and Naccarati A: MicroRNAs as markers of
progression in cervical cancer: A systematic review. BMC Cancer.
18:6962018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Di Meo A, Bartlett J, Cheng Y, Pasic MD
and Yousef GM: Liquid biopsy: A step forward towards precision
medicine in urologic malignancies. Mol Cancer. 16:802017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Finotti A, Allegretti M, Gasparello J,
Giacomini P, Spandidos DA, Spoto G and Gambari R: Liquid biopsy and
PCR-free ultrasensitive detection systems in oncology (Review). Int
J Oncol. 53:1395–1434. 2018.PubMed/NCBI
|
|
9
|
Muinelo-Romay L, Casas-Arozamena C and
Abal M: Liquid biopsy in endometrial cancer: New opportunities for
personalized oncology. Int J Mol Sci. 19:E23112018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yao T, Rao Q, Liu L, Zheng C, Xie Q, Liang
J and Lin Z: Exploration of tumor-suppressive microRNAs silenced by
DNA hypermethylation in cervical cancer. Virol J. 10:1752013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang Y, Xie YJ, Xu Q, Chen JX, Shan NC and
Zhang Y: Down-regulation of miR-1246 in cervical cancer tissues and
its clinical significance. Gynecol Oncol. 138:683–688. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yu Q, Liu SL, Wang H, Shi G, Yang P and
Chen XL: miR-126 Suppresses the proliferation of cervical cancer
cells and alters cell sensitivity to the chemotherapeutic drug
bleomycin. Asian Pac J Cancer Prev. 14:6569–6572. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang L, Wang Q, Li HL and Han LY:
Expression of MiR200a, miR93, metastasis-related gene RECK and
MMP2/MMP9 in human cervical carcinoma-relationship with prognosis.
Asian Pac J Cancer Prev. 14:2113–2118. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yin ZL, Wang YL, Ge SF, Guo TT, Wang L,
Zheng XM and Liu J: Reduced expression of miR-503 is associated
with poor prognosis in cervical cancer. Eur Rev Med Pharmacol Sci.
19:4081–4085. 2015.PubMed/NCBI
|
|
15
|
Wang LQ, Zhang Y, Yan H, Liu KJ and Zhang
S: MicroRNA-373 functions as an oncogene and targets YOD1 gene in
cervical cancer. Biochem Biophys Res Commun. 459:515–520. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Song X, Shi B, Huang K and Zhang W:
miR-133a inhibits cervical cancer growth by targeting EGFR. Oncol
Rep. 34:1573–1580. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li QQ, Zhang L, Wan HY, Liu M, Li X and
Tang H: CREB1-driven expression of miR-320a promotes mitophagy by
down-regulating VDAC1 expression during serum starvation in
cervical cancer cells. Oncotarget. 6:34924–34940. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wei Q, Li YX, Liu M, Li X and Tang H:
MiR-17-5p targets TP53INP1 and regulates cell proliferation and
apoptosis of cervical cancer cells. IUBMB Life. 64:697–704. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen J, Yao D, Li Y, Chen H, He C, Ding N,
Lu Y, Ou T, Zhao S, Li L and Long F: Serum microRNA expression
levels can predict lymph node metastasis in patients with
early-stage cervical squamous cell carcinoma. Int J Mol Med.
32:557–567. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yue C, Wang M, Ding B, Wang W, Fu S, Zhou
D, Zhang Z and Han S: Polymorphism of the pre-miR-146a is
associated with risk of cervical cancer in a Chinese population.
Gynecol Oncol. 122:33–37. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou Q, Han LR, Zhou YX and Li Y: MiR-195
suppresses cervical cancer migration and invasion through targeting
Smad3. Int J Gynecol Cancer. 26:817–824. 2016. View Article : Google Scholar
|
|
22
|
Chen XF and Liu Y: MicroRNA-744 inhibited
cervical cancer growth and progression through apoptosis induction
by regulating Bcl-2. Biomed Pharmacother. 81:379–387. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun Y, Yang X, Liu M and Tang H: B4GALT3
up-regulation by miR-27a contributes to the oncogenic activity in
human cervical cancer cells. Cancer Lett. 375:284–292. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang J, Wang F, Xu J, Wang X, Ye F and
Xie X: Micro ribonucleic acid-93 promotes oncogenesis of cervical
cancer by targeting RAB11 family interacting protein 1. J Obstet
Gynaecol Res. 42:1168–1179. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xiao S, Liao S, Zhou Y, Jiang B, Li Y and
Xue M: High expression of octamer transcription factor 1 in
cervical cancer. Oncol Lett. 7:1889–1894. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yi Y, Li H, Lv Q, Wu K and Zhang W, Zhang
J, Zhu D, Liu Q and Zhang W: miR-202 inhibits the progression of
human cervical cancer through inhibition of cyclin D1. Oncotarget.
7:72067–72075. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cheng YX, Zhang QF, Hong L, Pan F, Huang
JL, Li BS and Hu M: MicroRNA-200b suppresses cell invasion and
metastasis by inhibiting the epithelial-mesenchymal transition in
cervical carcinoma. Mol Med Rep. 13:3155–3160. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun L, Jiang R, Li J, Wang B, Ma C, Lv Y
and Mu N: MicoRNA-425-5p is a potential prognostic biomarker for
cervical cancer. Ann Clin Biochem. 54:127–133. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu Y, Zhang Y and Zhang S: MicroRNA-92
regulates cervical tumorigenesis and its expression is upregulated
by human papillomavirus-16 E6 in cervical cancer cells. Oncol Lett.
6:468–474. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang Y, Liu H, Wang X and Chen L:
Up-regulation of microRNA-664 inhibits cell growth and increases
cisplatin sensitivity in cervical cancer. Int J Clin Exp Med.
8:18123–18129. 2015.PubMed/NCBI
|
|
31
|
Qin X, Wan Y, Wang S and Xue M:
MicroRNA-125a-5p modulates human cervical carcinoma proliferation
and migration by targeting ABL2. Drug Des Devel Ther. 10:71–79.
2015.PubMed/NCBI
|
|
32
|
Sun P, Shen Y, Gong JM, Zhou LL, Sheng JH
and Duan FJ: A new MicroRNA expression signature for cervical
cancer. Int J Gynecol Cancer. 27:339–343. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chandrasekaran KS, Sathyanarayanan A and
Karunagaran D: MicroRNA-214 suppresses growth, migration and
invasion through a novel target, high mobility group AT-hook 1, in
human cervical and colorectal cancer cells. Br J Cancer.
115:741–751. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lin W, Feng M, Chen G, Zhou Z, Li J and Ye
Y: Characterization of the microRNA profile in early-stage cervical
squamous cell carcinoma by next-generation sequencing. Oncol Rep.
37:1477–1486. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Azizmohammadi S, Safari A, Azizmohammadi
S, Kaghazian M, Sadrkhanlo M, Yahaghi E, Farshgar R and
Seifoleslami M: Molecular identification of miR-145 and miR-9
expression level as prognostic biomarkers for early-stage cervical
cancer detection. QJM. 110:11–15. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jin X, Chen X, Hu Y, Ying F, Zou R, Lin F,
Shi Z, Zhu X, Yan X, Li S and Zhu H: LncRNA-TCONS_00026907 is
involved in the progression and prognosis of cervical cancer
through inhibiting miR-143-5p. Cancer Med. 6:1409–1423. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu J and Ni S: Association between
genetic polymorphisms in the promoters of let-7 and risk of
cervical squamous cell carcinoma. Gene. 642:256–260. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou Y, Hao Y, Li Y, Li R, Wu R, Wang S
and Fang Z: Amplification and up-regulation of MIR30D was
associated with disease progression of cervical squamous cell
carcinomas. BMC Cancer. 17:2302017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Song R, Cong L, Ni G and Chen M, Sun H,
Sun Y and Chen M: MicroRNA-195 inhibits the behavior of cervical
cancer tumors by directly targeting HDGF. Oncol Lett. 14:767–775.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhao J, Li B, Shu C, Ma Y and Gong Y:
Downregulation of miR-30a is associated with proliferation and
invasion via targeting MEF2D in cervical cancer. Oncol Lett.
14:7437–7442. 2017.PubMed/NCBI
|
|
41
|
He S, Liao B, Deng Y, Su C, Tuo J, Liu J,
Yao S and Xu L: MiR-216b inhibits cell proliferation by targeting
FOXM1 in cervical cancer cells and is associated with better
prognosis. BMC Cancer. 17:6732017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li S, Yang F, Wang M, Cao W and Yang Z:
miR-378 functions as an onco-miRNA by targeting the
ST7L/Wnt/β-catenin pathway in cervical cancer. Int J Mol Med.
40:1047–1056. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hu Y, Xie H, Liu Y, Liu W, Liu M and Tang
H: miR-484 suppresses proliferation and epithelial-mesenchymal
transition by targeting ZEB1 and SMAD2 in cervical cancer cells.
Cancer Cell Int. 17:362017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Luo S, Li N, Yu S, Chen L, Liu C and Rong
J: MicroRNA-92a promotes cell viability and invasion in cervical
cancer via directly targeting Dickkopf-related protein 3. Exp Ther
Med. 14:1227–1234. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang Z, Wang J, Wang X, Song W, Shi Y and
Zhang L: MicroRNA-21 promotes proliferation, migration, and
invasion of cervical cancer through targeting TIMP3. Arch Gynecol
Obstet. 297:433–442. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li M, Li BY, Xia H and Jiang LL:
Expression of microRNA-142-3p in cervical cancer and its
correlation with prognosis. Eur Rev Med Pharmacol Sci.
21:2346–2350. 2017.PubMed/NCBI
|
|
47
|
Zhao Y, Liu X and Lu YX: MicroRNA-143
regulates the proliferation and apoptosis of cervical cancer cells
by targeting HIF-1α. Eur Rev Med Pharmacol Sci. 21:5580–5586.
2017.PubMed/NCBI
|
|
48
|
Tao L, Zhang CY, Guo L, Li X, Han NN, Zhou
Q and Liu ZL: MicroRNA-497 Accelerates Apoptosis While Inhibiting
Proliferation, Migration, and Invasion through negative regulation
of the MAPK/ERK signaling pathway via RAF-1. J Cell Physiol.
233:6578–6588. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gocze K, Gombos K, Kovacs K, Juhasz K,
Gocze P and Kiss I: MicroRNA expressions in HPV-induced cervical
dysplasia and cancer. Anticancer Res. 35:523–530. 2015.PubMed/NCBI
|
|
50
|
Zhang J, Zheng F, Yu G, Yin Y and Lu Q:
miR-196a targets netrin 4 and regulates cell proliferation and
migration of cervical cancer cells. Biochem Biophys Res Commun.
440:582–588. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chen Y, Ma C, Zhang W, Chen Z and Ma L:
Down regulation of miR-143 is related with tumor size, lymph node
metastasis and HPV16 infection in cervical squamous cancer. Diagn
Pathol. 9:882014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang C and Jiang T: MicroRNA-335
represents an independent prognostic marker in cervical cancer.
Tumour Biol. 36:5825–5830. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zeng K, Zheng W, Mo X, Liu F, Li M, Liu Z,
Zhang W and Hu X: Dysregulated microRNAs involved in the
progression of cervical neoplasm. Arch Gynecol Obstet. 292:905–913.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ma L, Hong Y, Lu C, Chen Y and Ma C: The
occurrence of cervical cancer in Uygur women in Xinjiang Uygur
Autonomous Region is correlated to microRNA-146a and ethnic factor.
Int J Clin Exp Pathol. 8:9368–9375. 2015.PubMed/NCBI
|
|
55
|
Zheng W, Liu Z, Zhang W and Hu X: miR-31
functions as an oncogene in cervical cancer. Arch Gynecol Obstet.
292:1083–1089. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Qian K, Pietilä T, Rönty M, Michon F,
Frilander MJ, Ritari J, Tarkkanen J, Paulín L, Auvinen P and
Auvinen E: Identification and Validation of Human Papillomavirus
Encoded microRNAs. PLoS One. 8:e702022013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang J, Jia J, Zhao L, Li X, Xie Q, Chen
X, Wang J and Lu F: Down-regulation of microRNA-9 leads to
activation of IL-6/Jak/STAT3 pathway through directly targeting
IL-6 in HeLa cell. Mol Carcinog. 55:732–742. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhao S, Yao D, Chen J and Ding N:
Circulating miRNA-20a and miRNA-203 for screening lymph node
metastasis in early stage cervical cancer. Genet Test Mol Biomark.
17:631–636. 2013. View Article : Google Scholar
|
|
59
|
Jia W, Wu Y, Zhang Q, Gao GE, Zhang C and
Xiang Y: Expression profile of circulating microRNAs as a promising
fingerprint for cervical cancer diagnosis and monitoring. Mol Clin
Oncol. 3:851–858. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Jiang W, Pan JJ, Deng YH, Liang MR and Yao
LH: Down-regulated serum microRNA-101 is associated with aggressive
progression and poor prognosis of cervical cancer. J Gynecol Oncol.
28:e752017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhou LL, Shen Y, Gong JM, Sun P and Sheng
JH: MicroRNA-466 with tumor markers for cervical cancer screening.
Oncotarget. 8:70821–70827. 2017.PubMed/NCBI
|
|
62
|
Yu J, Wang Y, Dong R, Huang X, Ding S and
Qiu H: Circulating microRNA-218 was reduced in cervical cancer and
correlated with tumor invasion. J Cancer Res Clin Oncol.
138:671–674. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
You W, Wang Y and Zheng J: Plasma miR-127
and miR-218 Might Serve as Potential Biomarkers for Cervical
Cancer. Reprod Sci. 22:1037–1041. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Luo M, Shen D, Wang W and Xian J: Aberrant
expression of microRNA-26b and its prognostic potential in human
cervical cancer. Int J Clin Exp Pathol. 8:5542–5548.
2015.PubMed/NCBI
|
|
65
|
Liu S, Pan X, Yang Q, Wen L, Jiang Y, Zhao
Y and Li G: MicroRNA-18a enhances the radiosensitivity of cervical
cancer cells by promoting radiation-induced apoptosis. Oncol Rep.
33:2853–2862. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Xie H, Lee L, Scicluna P, Kavak E, Larsson
C, Sandberg R and Lui WO: Novel functions and targets of miR-944 in
human cervical cancer cells. Int J Cancer. 136:E230–E241. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Shen S, Wang L, Jia Y, Hao Y, Zhang L and
Wang H: Upregulation of microRNA-224 is associated with aggressive
progression and poor prognosis in human cervical cancer. Diagn
Pathol. 8:692013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Huang L, Lin JX, Yu YH, Zhang MY, Wang HY
and Zheng M: Downregulation of Six MicroRNAs Is Associated with
Advanced Stage, Lymph Node Metastasis and Poor Prognosis in Small
Cell Carcinoma of the Cervix. PLoS One. 7:e337622012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Xing AY, Wang B, Shi DB, Zhang XF, Gao C,
He XQ, Liu WJ and Gao P: Deregulated expression of miR-145 in
manifold human cancer cells. Exp Mol Pathol. 95:91–97. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Lee H, Kim KR, Cho NH, Hong SR, Jeong H,
Kwon SY, Park KH, An HJ, Kim TH, Kim I, et al: MicroRNA expression
profiling and Notch1 and Notch2 expression in minimal deviation
adenocarcinoma of uterine cervix. World J Surg Oncol. 12:3342014.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ivanov MK, Titov SE, Glushkov SA,
Dzyubenko VV, Malek AV, Arkhangelskaya PA, Samsonov RB, Mikhetko
AA, Bakhidze EV, Berlev IV and Kolesnikov NN: Detection of
high-grade neoplasia in air-dried cervical PAP smears by a
microRNA-based classifier. Oncol Rep. 39:1099–1111. 2018.PubMed/NCBI
|
|
72
|
Yu X, Zhao W, Yang X, Wang Z and Hao M:
miR-375 affects the proliferation, invasion, and apoptosis of
HPV16-positive human cervical cancer cells by targeting IGF-1R. Int
J Gynecol Cancer. 26:851–858. 2016. View Article : Google Scholar
|
|
73
|
Hao M, Zhao W, Zhang L, Wang H and Yang X:
Low folate levels are associated with methylation-mediated
transcriptional repression of miR-203 and miR-375 during cervical
carcinogenesis. Oncol Lett. 11:3863–3869. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Nagamitsu Y, Nishi H, Sasaki T, Takaesu Y,
Terauchi F and Isaka K: Profiling analysis of circulating microRNA
expression in cervical cancer. Mol Clin Oncol. 5:189–194. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Kogo R, How C, Chaudary N, Bruce J, Shi W,
Hill RP, Zahedi P, Yip KW and Liu FF: The microRNA-218~Survivin
axis regulates migration, invasion, and lymph node metastasis in
cervical cancer. Oncotarget. 6:1090–1100. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Lajer CB, Garnæs E, Friis-Hansen L,
Norrild B, Therkildsen MH, Glud M, Rossing M, Lajer H, Svane D,
Skotte L, et al: The role of miRNAs in human papilloma virus
(HPV)-associated cancers: Bridging between HPV-related head and
neck cancer and cervical cancer. Br J Cancer. 106:1526–1534. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Huang YW, Kuo CT, Chen JH, Goodfellow PJ,
Huang TH, Rader JS and Uyar DS: Hypermethylation of miR-203 in
endometrial carcinomas. Gynecol Oncol. 133:340–345. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Myklebust MP, Bruland O, Fluge Ø,
Skarstein A, Balteskard L and Dahl O: MicroRNA-15b is induced with
E2F-controlled genes in HPV-related cancer. Br J Cancer.
105:1719–1725. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Sharma Saha S, Roy Chowdhury R, Mondal NR,
Chakravarty B, Chatterjee T, Roy S and Sengupta S: Identification
of genetic variation in the lncRNA HOTAIR associated with
HPV16-related cervical cancer pathogenesis. Cell Oncol (Dordr).
39:559–572. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Andersen CL, Jensen JL and Ørntoft TF:
Normalization of real-time quantitative reverse transcription-PCR
data: A model-based variance estimation approach to identify genes
suited for normalization, applied to bladder and colon cancer data
sets. Cancer Res. 64:5245–5250. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Rossi ED, Bizzarro T, Martini M,
Capodimonti S, Sarti D, Cenci T, Bilotta M, Fadda G and Larocca LM:
The evaluation of miRNAs on thyroid FNAC: The promising role of
miR-375 in follicular neoplasms. Endocrine. 54:723–732. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Kottaridi C, Spathis A, Margari N, Koureas
N, Terzakis E, Chrelias C, Pappas A, Bilirakis E, Pouliakis A,
Panayiotides IJ and Karakitsos P: Evaluation analysis of miRNAs
overexpression in liquid-based cytology endometrial samples. J
Cancer. 8:2699–2703. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Barodawala SM, Chadha K, Kavishwar V,
Murthy A and Shetye S: Cervical cancer screening by molecular
Pap-transformation of gynecologic cytology. Diagn Cytopathol.
47:374–381. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Bonde J, Ejegod DM, Cuschieri K, Dillner
J, Heideman DAM, Quint W, Pavon Ribas MA, Padalko E, Christiansen
IK, Xu L and Arbyn M: The Valgent4 protocol: Robust analytical and
clinical validation of 11 HPV assays with genotyping on cervical
samples collected in SurePath medium. J Clin Virol. 108:64–71.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Tota JE, Bentley J, Blake J, Coutlée F,
Duggan MA, Ferenczy A, Franco EL, Fung-Kee-Fung M, Gotlieb W,
Mayrand MH, et al: Approaches for triaging women who test positive
for human papillomavirus in cervical cancer screening. Prev Med.
98:15–20. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Derveaux S, Vandesompele J and Hellemans
J: How to do successful gene expression analysis using real-time
PCR. Methods. 50:227–230. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Kiss T: Small nucleolar RNAs: An abundant
group of noncoding RNAs with diverse cellular functions. Cell.
109:145–148. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Dieci G, Preti M and Montanini B:
Eukaryotic snoRNAs: A paradigm for gene expression flexibility.
Genomics. 94:83–88. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zubillaga-Guerrero MI, Alarcón-Romero Ldel
C, Illades-Aguiar B, Flores-Alfaro E, Bermúdez-Morales VH, Deas J
and Peralta-Zaragoza O: MicroRNA miR-16-1 regulates CCNE1 (cyclin
E1) gene expression in human cervical cancer cells. Int J Clin Exp
Med. 8:15999–16006. 2015.PubMed/NCBI
|