Introduction
One of the most common types of cancer affecting the
digestive system is esophageal cancer (EC). EC is a malignant
cancer listed globally as the sixth highest in mortality rate and
seventh highest in frequency of occurrence (1–3).
Esophageal squamous cell cancer (ESCC) is the predominant type of
EC, as nine out of 10 people living with EC in China are diagnosed
with this subtype (4). The other
subtype is esophageal adenocarcinoma (EA) (5,6). The
majority of patients are diagnosed in an advanced stage of the
disease due to indiscernible early symptoms and a lack of
efficacious screening methods (7,8).
Extensive research focused on the elucidation of the
pathophysiological mechanisms of EC has been performed (9,10);
however, the 5-year survival rate of patients with EC is only 15 to
25% and the recurrence rate is ~38.14%, which results in the
continued severity of this disease (11,12).
Various treatment options are available for EC, including
chemotherapy, radiotherapy, surgery and comprehensive therapy
(13). The optimum treatment plan
for patients with advanced EC is a combination of surgery and
radiotherapy (14,15). However, reduced treatment efficacy is
a consequence of the development of radioresistant tumor cells
following radiotherapy, which is the major clinical obstacle in the
treatment of EC (16). Thus, the
establishment of efficacious means of improving tumor cell
radiosensitivity is crucial to successful EC therapy.
At present, an increasing number of studies have
reported the abnormal activation of the Sonic Hedgehog (Shh)
signaling pathway in several types of human cancer, including
hepatic, breast, bladder and pancreatic cancers (17–20). The
majority of these studies focused on the association between tumor
cell radioresistance and the Shh signaling pathway; however, there
are a few studies on radioresistance in EC and Shh signaling
pathway. A previous study established the inhibitory effect of
P162, which is a new peptide with anti-cancer mechanism, on
glioma-associated oncogene family zinc finger 1 (Gli1), which
resulted in noticeable improvements in the level of
radiosensitivity in EC cells (21).
The present study aimed to establish the association between Shh
signaling pathway activation and EC cell radiosensitivity.
The current study examined the contribution of Gli1,
a factor in the Shh signaling pathway, to the radiosensitivity of
EC cells. The results of the present study may form the foundation
for enhancing radiotherapy efficacy as part of the treatment
regimen for future patients with EC.
Materials and methods
Cell culture
The Eca109 cell line was supplied by Taihe Hospital,
an affiliate of Hubei Medical College (Shiyan, China). Cells were
maintained in liquid nitrogen and cultured in Dulbecco's modified
Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented
with fetal bovine serum (10%) and penicillin (100 µg/ml) in a
humidified atmosphere at 37°C and 5% CO2.
Eca109R cell line establishment
X-ray irradiation of the Eca109 parental cell line
was performed during the logarithmic growth phase at an absorbance
dose rate of 2 Gy/min using the Varian 2300 linear accelerator
(6MV). On reaching 90% confluence, the cells were digested with
trypsin in 0.25% EDTA, reinoculated and treated with repeated
irradiation (2 Gy) during the logarithmic growth phase. A
cumulative irradiation dose of 60 Gy was achieved by repeating the
process 30 times. Subculture of the cells was performed to
establish stable morphology, resulting in the formation of the
radiation-resistant Eca109R cell line. Following continuous
culturing over two weeks, the cells were separated into two
batches, one of which was stored in liquid nitrogen, whereas the
other continued to be subcultured. The Eca109R cell line was still
resistant to radiation through 50 generations of subculturing
without irradiation.
Clone formation assay
A single cell suspension of Eca109 and Eca109R cells
was prepared following trypsinization of each cell line in the
logarithmic growth phase. Cells were seeded at a range of
concentrations (400, 400, 800, 1,000 and 1,200 cells/well) in
six-well plates and irradiated with 0, 2, 4, 6 and 8 Gy, with an
average dose rate of 200 cGy/min. Following a two-week incubation
period, the cells were fixed with pure methanol at room temperature
for 15 min, stained with crystal violet at room temperature for 30
min and examined under a light microscope. Colony numbers were
counted; a cell number ≥50 cells was used as a clone. The cell
survival curve was established using a single hit multi-target
model, which allowed the calculation of various parameters,
including the average lethal dose (D0), the quasi-threshold dose
(Dq), the value of curve extension to the Y-axis intercept (N) and
the cell survival fraction following irradiation with 2 Gy
(SF2). These experiments were performed three times.
Immunofluorescence assay
Immunofluorescence was used to determine Gli1
protein expression. Following fixation of Eca109 and Eca109R cells
onto microscope slides with pure methanol at room temperature for
15 min, the cells were blocked for 10 min with 10% goat serum
(ZSBG-Bio), followed by incubation with the primary antibody
against Gli1 (1:150; cat. no. NBP2-45872; Novus Biologicals) for 1
h at 37°C. The cells were then washed with PBS three times prior to
incubation with goat anti-mouse IgG H&L secondary antibody
(1:500; cat. no. ab150113; Abcam) for 1.5 hat 37°C in the dark.
Following counterstaining with DAPI at room temperature for 10 min,
the cells were examined using fluorescence microscopy (eight fields
examined) and the experiment was repeated three times. Image J 15.0
software (National Institutes of Health) was used to measure the
expression level of Gli1.
Western blotting analysis
Eca109 and Eca109R cells were lysed using RIPA lysis
buffer (Beyotime Institute of Biotechnology) for 30 min at room
temperature. Cells were centrifuged at 12,000 × g for 15 min at 4°C
and the supernatant was collected. The BCA Protein Quantitative
Assay kit (Servicebio) was used to determine the protein
concentration. The samples were denatured by boiling prior to
separation (35 µg per lane) using 10% SDS-PAGE followed by transfer
to nitrocellulose membranes. Skimmed milk (5%) was used as a
blocking agent for 1.5 h at room temperature. Next, the membranes
were incubated on a shaker with the primary antibodies against Gli1
(1:2,000; cat. no. NBP2-45872; Novus Biologicals), Shh (1:5,000;
cat. no. ab53281; Abcam), Smo (1:200; cat. no. GTX60154; GeneTex,
Inc.), Ptch (1:500; cat. no. GTX83771; GeneTex, Inc.) and β-actin
(1:5,000; cat. no. GTX629630; GeneTex, Inc.), overnight at 4°C,
washed with Tris-buffered saline with 0.05% Tween-20, and incubated
with secondary antibodies (1:4,000; cat. nos. 1031-05 and 4050-05;
Southern Biotech) for 1 h at 37°C. Bands were detected using
enhanced chemiluminescence substrate (Bio-Rad Laboratories, Inc.),
and grayscale images were analyzed with ImageJ 15.0 software
(National Institutes of Health). This procedure was performed three
times.
Cell transfection
For Gli1 overexpression, the full-length Gli1
sequence was synthesized by cloning the Gli1 gene into the GV146
vector (Shanghai GeneChem Co., Ltd.) via XhoI and
EcoRI sites, and was referred to as pGli1-IRES-EGFP. Empty
GV146 vector acted as a negative control (NC) of pGli1-IRES-EGFP
and was referred to as pIRES-EGFP. For knockdown of Gli1, short
hairpin RNA (shRNA) directed against Gli1 was ligated into the
GV102 vector (Shanghai GeneChem Co., Ltd.) and was referred to as
sh-Gli1; a non-targeting sequence was ligated into the GV102 vector
as the NC of sh-Gli1, and was referred to as sh-NC. The target
sequence was 5′-CCTCTGTCTACTCACCACA-3′ and the NC sequence was
5′-TTCTCCGAACGTGTCACGT-3′. Eca109 and Eca109R cells were seeded in
6-well plates at a density of 6×105 cells/well one day
before transfection. Briefly, 5 µl Lipofectamine® 3000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and 2.5 µg
plasmid were diluted into 125 µl Opti-MEM (Gibco; Thermo Fisher
Scientific, Inc.) and mixed for 5 min at room temperature. Cells
were then incubated with this solution, and complete medium was
added to the final volume of 2 ml. Cell transfection was performed
for 24 h following the manufacturer's protocols prior to further
experiments.
Statistical analysis
SPSS software (version 17.0; SPSS, Inc.) was used
for statistical analysis. Data are expressed as the mean ± standard
deviation. Differences among multiple groups were analyzed by
one-way ANOVA with Duncan's multiple range test; the unpaired
Student' t-test was used for comparisons between two groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
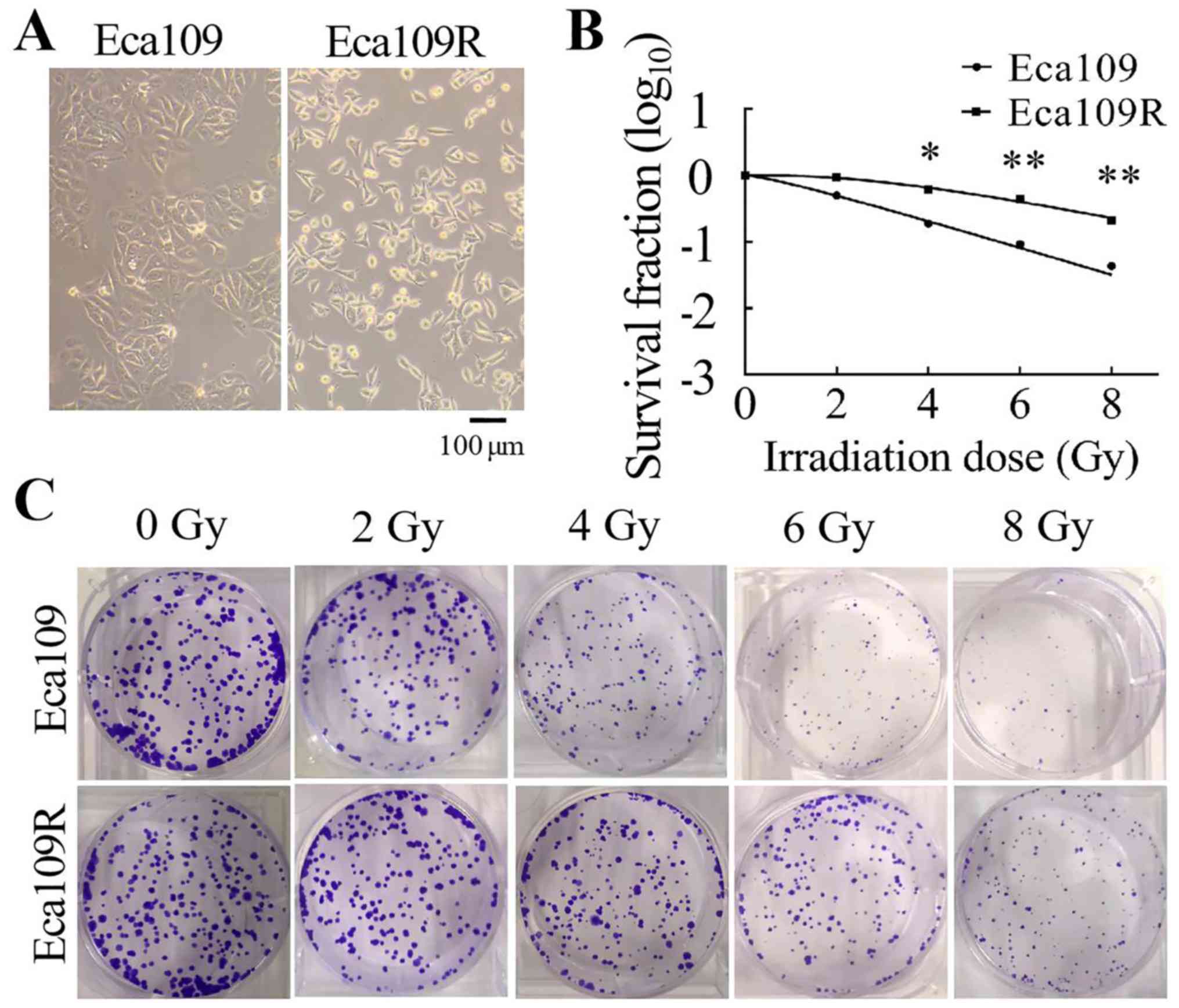
Differences between the Eca109
parental and Eca109R radiation-resistant cell lines
The Eca109R radiation-resistant cell line was
established by repeated irradiation of the Eca109 parental cell
line with 2 Gy/time (cumulative 60 Gy, 30 times in total). The
morphology of the cells was observed using a light microscope
(Olympus Corporation). At a higher magnification, Eca109 cells
appeared as spherical or irregular polygons with a high degree of
nuclear chromosome refractivity, whereas Eca109R cells appeared as
irregular shuttle shapes with dense granular cytoplasm and small
nuclei (Fig. 1A).
A colony formation assay was performed to
characterize the radiosensitivity of Eca109 and Eca109R cells. The
cells were seeded into six-well plates at different concentrations
(400, 400, 800, 1,000 and 1,200 cells/well) and received
irradiation at different doses (0, 2, 4, 6 and 8 Gy). Following
irradiation, the survival fraction of the two cell lines was
measured. Cell survival curves were generated using the single-hit
multi-target mode (Fig. 1B). The
results demonstrated that with the radiation dose increasing, cell
survival rate gradually decreased and Eca109 cells were more
sensitive to radiation than Eca109R cells (Fig. 1C). Radiological measurements of Dq,
D0 and N values were higher in Eca109R cells compared with those in
Eca109 cells (Table I). Thus, the
Eca109R cell line exhibited higher resistance to irradiation
compared with the Eca109 cell line.
 | Table I.Radiobiological parameters of Eca109
and Eca109R cells. |
Table I.
Radiobiological parameters of Eca109
and Eca109R cells.
| Group | Eca109 | Eca109R | P-value |
|---|
| N | 1.426±0.395 | 3.310±0.200 | 0.032 |
| D0 (Gy) | 2.142±0.332 | 3.092±0.153 | 0.024 |
| Dq (Gy) | 0.944±0.157 | 3.692±0.101 | 0.041 |
| SF2
(%) | 0.499±0.042 | 0.937±0.013 | 0.027 |
Different expression of key factors in
Shh signaling pathway between the Eca109 parental and Eca109R
radiation-resistant cell lines
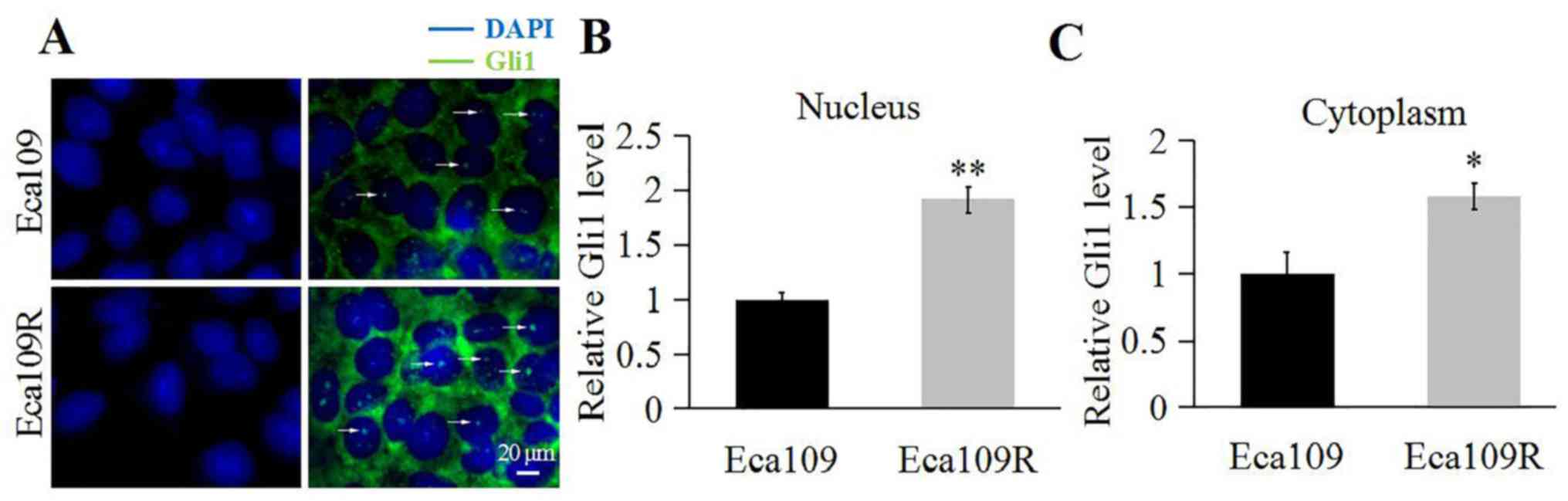
Gli1 expression and localization were determined
through immunofluorescence. The results demonstrated that although
Gli1 was present in the nucleus and cytoplasm of Eca109 and Eca109R
cells (Fig. 2), and that Gli1
appeared abundant in the nucleus and the cytoplasm of Eca109R cells
(Fig. 2A) compared with Eca109 cells
(Fig. 2B and C).
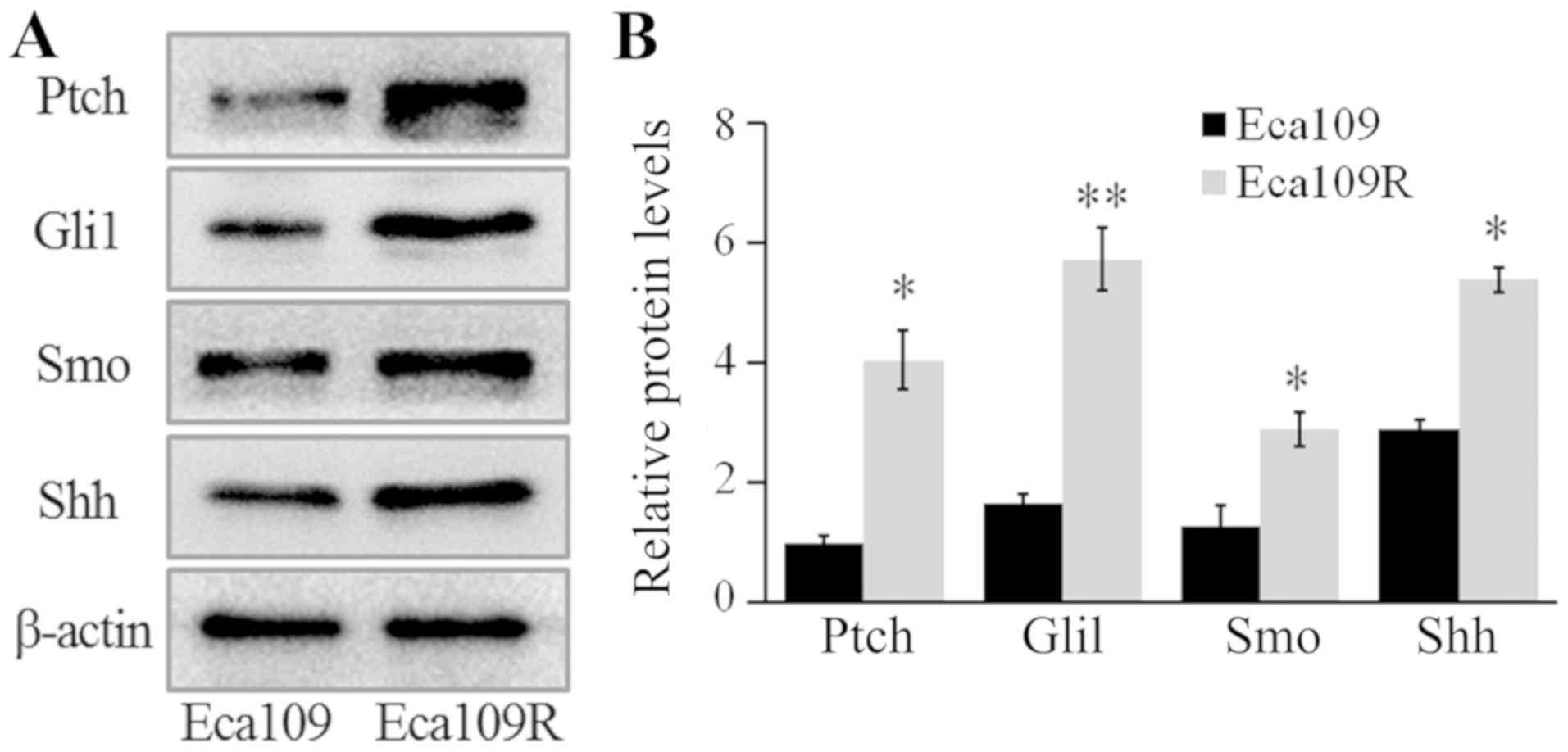
The expression levels of Gli1, patched 1 (Ptch),
smoothened frizzled class receptor (Smo) and Shh were detected by
western blotting, and the results revealed that the expression
levels of these proteins were significantly higher in Eca109R,
compared with Eca109 cells (P<0.05; Fig. 3). Thus, these results demonstrated a
significant upregulation of the proteins in the Shh signaling
pathway in radiation-resistant cells.
Decreased Eca109 cell radiosensitivity
following Gli1 overexpression
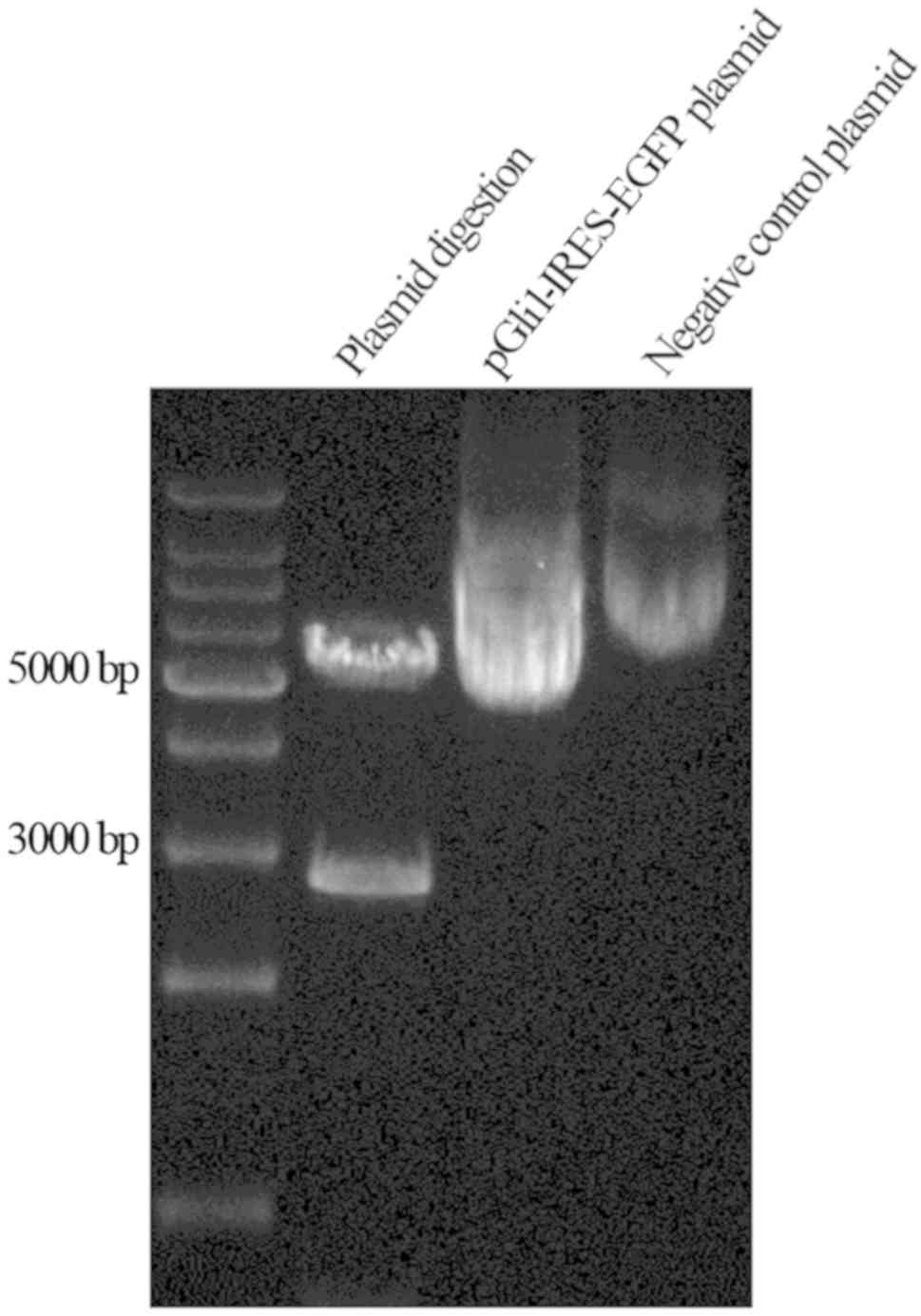
The results of plasmid construction were confirmed
by sequencing and through enzyme digestion using both EcoRI
and XhoI, generating fragments of 3 and 5 kb in length
(Fig. 4). This confirmed the correct
insertion of the target fragment and accurate construction of the
pGli1-IRES-EGFP plasmid. Gli1 overexpression was established by
transfecting the pGli1-IRES-EGFP plasmid into the Eca109 cell line,
while the NC (empty plasmid) was transfected as a negative control,
and the control group contained untransfected Eca109 cells.
Following 48 h of transfection, the cells were identified using
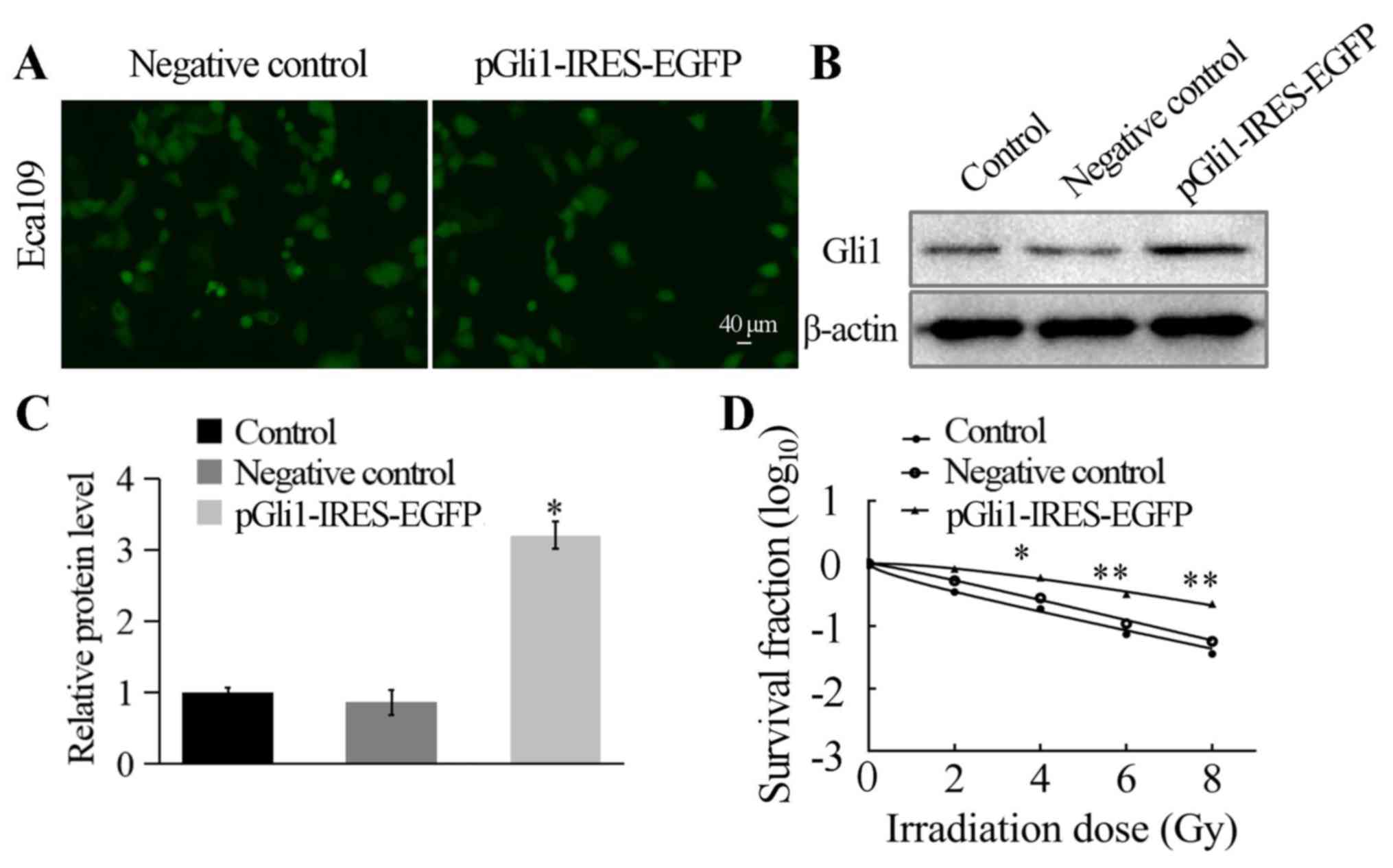
fluorescence microscopy. The transfection efficiency reached ~70%
(Fig. 5A). Next, the protein
expression level of Gli1 was detected by western blotting analysis;
the results indicated significantly higher levels of Gli1
expression in cells transfected with pGli1-IRES-EGFP compared with
those in the two control groups (both P<0.05; Fig. 5B and C).
Transfection of Eca109 cells with the
pGli1-IRES-EGFPplasmid established the association between
overexpression of Gli1 and the radiosensitivity of the cells. The
three cell types were seeded into 6-well plates and then irradiated
at 0, 2, 4, 6 and 8 Gy, with a mean dose rate of 200 cGy/min,
followed by 2 weeks of culture. A clone formation assay was
performed to measure the survival fraction of each experimental
group, and the single-hit multi-target mode was used to establish
cell survival curves (Fig. 5D). The
results demonstrated that with the radiation dose increasing, cell
survival rate gradually decreased and pGli1-IRES-EGFP-transfected
Eca109 cells were more resistant to radiation. The two control
groups exhibited significantly lower D0 and Dq values compared with
the pGli1-IRES-EGFP-transfected Eca109 cells (P<0.05; Table II). The SF2 in
pGli1-IRES-EGFP-transfected Eca109 cells was 0.760±0.033, whereas
those in the NC and control groups were 0.532±0.062 and
0.350±0.029, respectively (Table
II). These findings demonstrated a reduction in radiation
sensitivity in Eca109 cells following Gli1 overexpression (Table II). Therefore, the transfection of
Eca109 cells with the pGli1-IRES-EGFP established an association
between overexpression of Gli1 and the radiosensitivity of the
cells.
 | Table II.Comparison of radiobiological
parameters of transfected Eca109 cells. |
Table II.
Comparison of radiobiological
parameters of transfected Eca109 cells.
| Group | Control | Negative
control |
pGli1-IRES-EGFP | P-value |
|---|
| N | 0.736±0.157 | 1.732±0.810 | 1.909±0.662 | 0.113 |
| D0 (Gy) | 1.945±1.266 | 2.084±0.525 | 3.516±0.722 | 0.049 |
| Dq (Gy) | 0.432±1.186 | 0.826±0.752 | 1.984±0.604 | 0.007 |
| SF2
(%) | 0.350±0.029 | 0.532±0.062 | 0.760±0.033 | 0.001 |
Increased Eca109R cell
radiosensitivity may be attributed to Gli1 knockdown
Reduced Gli1 expression was established by
transfecting the Gli1-silencing plasmid into the Eca109R cell line.
The non-targeted Gli1 plasmid was transfected into the Eca109R cell
line to create the NC, and the control group contained
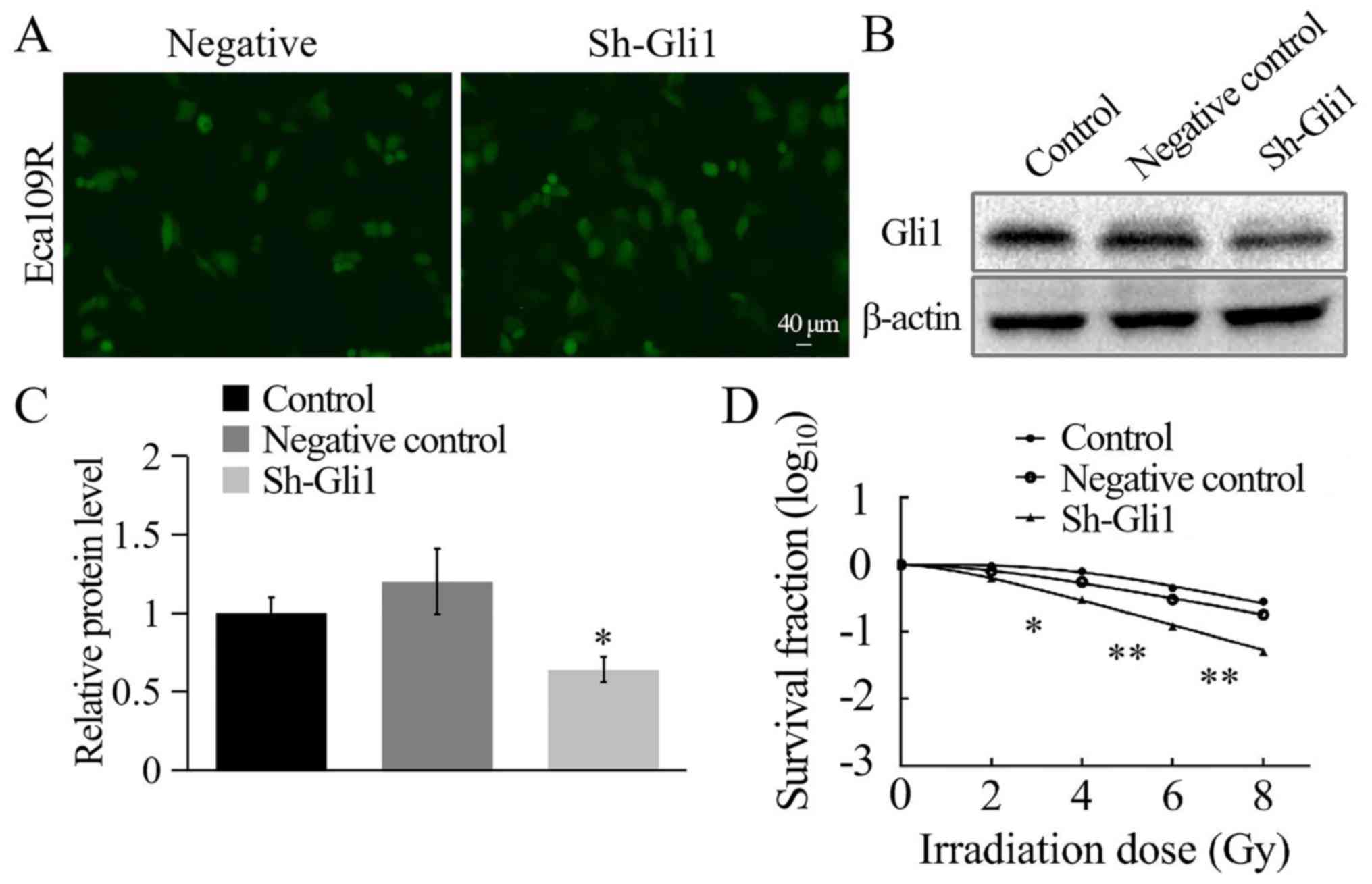
untransfected Eca109R cells. The transfection efficiency of the
Gli1-silencing plasmid in Eca109R cells was analyzed using
fluorescence microscopy after 48 h. Green fluorescence confirmed
successful transfection, and the transfection efficiency reached
~70% (Fig. 6A). Gli1 expression
levels were significantly lower in sh-Gli1-transfected Eca109R
cells compared with the NC and control groups, as determined by
western blotting (P<0.05; Fig. 6B and
C). These results indicated the effective generation of a
Gli1-silenced Eca109R cell line.
The cells were harvested, seeded into six-well
plates, irradiated and cultured over two weeks, followed by
staining and counting. SF2 was measured, and the
single-hit multi-target model was used to establish cell survival
curves (Fig. 6D). The results
indicated that with the radiation dose increasing, cell survival
rate gradually decreased and Gli1-silenced Eca109R cells were more
sensitive to radiation. The Gli1-silenced Eca109R cells exhibited
significantly lower levels of the radiation-associated biological
measurements (Dq, D0 and N values) compared with the two control
groups (P<0.05; Table III). The
SF2 in the sh-Gli1 plasmid-transfected Eca109R cell line
was 0.446±0.010, whereas in the NC and the control groups it was
0.867±0.070 and 0.916±0.128, respectively (Table III). These results indicated that
the radiosensitivity of Eca109R Gli1-silenced cell line was
significantly enhanced and that the radioresistance was
weakened.
 | Table III.Comparison of radiobiological
parameters of transfected Eca109R cells. |
Table III.
Comparison of radiobiological
parameters of transfected Eca109R cells.
| Group | Control | Negative
control | sh-Gli1 | P-value |
|---|
| N | 3.456±0.297 | 4.087±0.121 | 2.134±0.221 | 0.047 |
| D0 (Gy) | 3.698±0.331 | 1.916±0.193 | 0.902±0.038 | 0.039 |
| Dq (Gy) | 3.057±0.222 | 2.978±0.227 | 1.282±0.081 | 0.021 |
| SF2
(%) | 0.916±0.128 | 0.867±0.070 | 0.446±0.010 | 0.034 |
Discussion
EC frequently occurs as a primary tumor in the upper
gastrointestinal tract, and its malignant nature is harmful to
humans (22). A primary form of
treatment for EC is radiation therapy (15,23).
However, reduced radiosensitivity develops with gradual increases
in the irradiation dose and frequency, leading to the development
of radiotherapy-resistant tumors (24). The recurrence and metastasis of
tumors have been widely attributed to radiotherapy resistance
(25,26), and the options available to
individuals suffering from inoperable forms of EC are further
reduced by the development of radioresistance (27,28).
To enhance the efficacy radiotherapy, it is crucial
to research radioresistance mechanisms. Various studies have been
performed on the mechanisms of radioresistance, including DNA
damage and repair (29), cell
proliferation and apoptosis (30),
abnormal expression of microRNA and lncRNA (31–34) and
disruption of the cell cycle (30)
and altered expression of Shh signaling pathway (35). Xie et al (35) demonstrated that Raf kinase inhibitory
protein reduction enhances radioresistance by activating the Shh
signaling pathway. The present study also tried to explore whether
radioresistance was associated with Shh signaling pathway
activation.
The Sonic Hedgehog signaling pathway consists of Shh
ligands, the transmembrane proteins Ptch and Smo, and the
downstream Gli transcription factors (Gli1, Gli2 and Gli3)
(36). Abnormal activation of the
Shh signaling pathway is reliably detected through the expression
of Gli1 (37,38). An earlier study established the
activation of the Shh signaling pathway during tissue repair and an
absence of this signaling in normally-functioning adult tissues and
organs (39). Furthermore, a
previous study suggested an association between Shh signaling
pathway activation and the development of resistance in a range of
human cancer types, including EC (40).
In normal tissues, Smo protein activity is inhibited
by Ptch (41). However, when Shh
associates with Ptch, the Gli1 protein enters the nucleus to
activate the transcription of the downstream target genes (42). Increased Gli1 expression levels were
demonstrated in EC tissues and adjacent tissues compared with
normal tissues (43), and Gli1 has
been detected in the nuclei of a number of tumor-cell types. A
recent study by Huang et al (44) demonstrated that Hh signaling pathway
is activated in Hela-RR and Siha-RR, which was also demonstrated in
the present study. Furthermore, the expression of Shh, Ptch and Smo
has been detected in 34 ESCC cell lines, and Gli1 was highly
expressed in 31 EC cell lines (45).
In addition, silencing of Gli1 expression was achieved through
specific inhibitors of Smo, which led to the inhibition of fission,
recurrence and metastasis in ESCC (45). Gli1 transcription efficacy is
positively associated with its expression, which can be used to
efficiently detect abnormal activation of the Shh signaling pathway
(46). The results of the current
study revealed higher Gli1 protein expression levels in Eca109R
cells, compared with Eca109 cells. Furthermore, Gli1 in
radiation-resistant cells was aggregated around the nucleus, as
determined by immunofluorescence. These results suggested an
association between radioresistance in EC and the Shh signaling
pathway.
The radiation-resistant cell line Eca109R was
generated through low-level X-ray irradiation of the human EC cell
line Eca109. Colony formation assays demonstrated higher
measurements of the radiation-related biological parameters (D0, Dq
and N) in Eca109R, compared with Eca109 cells, which indicated
increased levels of resistance in the Eca109R cell line compared
with the parental cells. Furthermore, the expression of Gli1, Ptch,
Shh and Smo was confirmed by western blotting in Eca109 and Eca109R
cells; all of the tested proteins exhibited significantly higher
expression levels in Eca109R cells compared with Eca109 cells, and
immunofluorescence displayed Gli1 protein aggregation around the
nucleus. A previous study reported that Shh signaling pathway
activation is associated with the development of esophageal
squamous cell carcinoma (ESCC) (47). High expression of Shh signaling
pathway-related genes is present in ESCC, and patients with high
Gli1 expression in ESCC are not sensitive to radiation therapy
(47), which was the case in the
present study. A Gli1 overexpression plasmid was constructed and
subsequently transfected into Eca109 cells, and analyses confirmed
that Gli1 protein expression was increased. In addition, the clone
formation assay showed that radiosensitivity was decreased in
Gli1-overexpressing Eca109 cells compared with untransfected cells.
Furthermore, the Eca109R cell line was transfected with a Gli-1
silencing plasmid. These cells exhibited significantly lower Gli1
expression levels and higher levels of radiosensitivity compared
with the control groups. A previous study reported that Hh
signaling pathway can influence the radiation response in some
patient-derived murine xenograft (PDX) model of esophageal
adenocarcinoma, and that inhibition of this pathway could increase
the radiation efficacy (48). In
conclusion, these findings demonstrated an association between Gli1
and radioresistance in EC.
The mechanism underlying radioresistance, which
contributes to the pathogenicity of EC, is a complex process;
further research into how the Shh signaling pathway impacts
radioresistance is required. The present study described novel
research that may be used as a foundation for future studies into
the processes regulating Shh signaling pathway activation-induced
radioresistance.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Foundation of Hubei Province of China (grant no.
2014CFB818).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
QW, HLi, QL and HLo designed the experiments. FY,
JY, YH and JL performed the experiments and analyzed all data. FY
and JY drafted the manuscript and all authors approved the final
version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lagergren J, Smyth E, Cunningham D and
Lagergren P: Oesophageal cancer. Lancet. 390:2383–2396. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sohda M and Kuwano H: Current status and
future prospects for esophageal cancer treatment. Ann Thorac
Cardiovasc Surg. 23:1–11. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Abnet CC, Arnold M and Wei WQ:
Epidemiology of esophageal squamous cell carcinoma.
Gastroenterology. 154:360–373. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lin Y, Totsuka Y, He Y, Kikuchi S, Qiao Y,
Ueda J, Wei W, Inoue M and Tanaka H: Epidemiology of esophageal
cancer in Japan and China. J Epidemiol. 23:233–242. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Keditsu KK, Jiwnani S, Karimundackal G and
Pramesh CS: Multimodality management of esophageal cancer. Indian J
Surg Oncol. 4:96–104. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Januszewicz W and Fitzgerald RC: Early
detection and therapeutics. Mol Oncol. 13:599–613. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bird-Lieberman EL and Fitzgerald RC: Early
diagnosis of oesophageal cancer. Br J Cancer. 101:1–6. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guo X, Duan Y, Ye X, Hu L, Xu T, Tong L
and Yu M: Stable silencing of dll4 gene suppresses the growth and
metastasis of esophagus cancer cells by attenuating Akt
phosphorylation. Oncol Rep. 40:495–503. 2018.PubMed/NCBI
|
|
10
|
Alexandre L, Long E and Beales IL:
Pathophysiological mechanisms linking obesity and esophageal
adenocarcinoma. World J Gastrointest Pathophysiol. 5:534–549. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qian X, Tan C, Wang F, Yang B, Ge Y, Guan
Z and Cai J: Esophageal cancer stem cells and implications for
future therapeutics. Onco Targets Ther. 9:2247–2254.
2016.PubMed/NCBI
|
|
12
|
Lou F, Sima CS, Adusumilli PS, Bains MS,
Sarkaria IS, Rusch VW and Rizk NP: Esophageal cancer recurrence
patterns and implications for surveillance. J Thoracic Oncol.
8:1558–1562. 2013. View Article : Google Scholar
|
|
13
|
Domper Arnal MJ, Ferrandez Arenas A and
Lanas Arbeloa A: Esophageal cancer: Risk factors, screening and
endoscopic treatment in Western and Eastern countries. World J
Gastroenterol. 21:7933–7943. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu SG, Xie WH, Zhang ZQ, Sun JY, Li FY,
Lin HX, Yong Bao and He ZY: Surgery combined with radiotherapy
improved survival in metastatic esophageal cancer in a surveillance
epidemiology and end results population-based study. Sci Rep.
6:282802016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ordu AD, Nieder C, Geinitz H, Kup PG,
Deymann LF, Scherer V, Combs SE and Fakhrian K: Radio(chemo)therapy
for locally advanced squamous cell carcinoma of the esophagus:
Long-term outcome. Strahlenther Onkol. 191:153–160. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Malhotra A, Sharma U, Puhan S, Chandra
Bandari N, Kharb A, Arifa PP, Thakur L, Prakash H, Vasquez KM and
Jain A: Stabilization of miRNAs in esophageal cancer contributes to
radioresistance and limits efficacy of therapy. Biochimie.
156:148–157. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hu YT, Li BF, Zhang PJ, Wu D, Li YY, Li
ZW, Shen L, Dong B, Gao J and Zhu X: Dbx2 exhibits a
tumor-promoting function in hepatocellular carcinoma cell lines via
regulating Shh-Gli1 signaling. World J Gastroenterol. 25:923–940.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu X, Zhao T, Bai X, Li M, Ren J, Wang M,
Xu R, Zhang S, Li H, Hu Y, et al: LOC101930370/MiR-1471 axis
modulates the hedgehog signaling pathway in breast cancer. Cell
Physiol Biochem. 48:1139–1150. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Islam SS, Mokhtari RB, Noman AS, Uddin M,
Rahman MZ, Azadi MA, Zlotta A, van der Kwast T, Yeger H and Farhat
WA: Sonic hedgehog (Shh) signaling promotes tumorigenicity and
stemness via activation of epithelial-to-mesenchymal transition
(EMT) in bladder cancer. Mol Carcinog. 55:537–551. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Z, Jiang J, Qin T, Xiao Y and Han L:
EIF5A regulates proliferation and chemoresistance in pancreatic
cancer through the SHH signalling pathway. J Cell Mol Med.
23:2678–2688. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen J, Wu QM, Long H, Zhang H and Chen
JH: P162 enhances the radiosensitivity of esophageal cancer Eca109
cells by inhibiting Gli-1, the transcription factor of Hedgehog
signaling pathway. World Chin J Digestology. 22:615–623. 2014.
|
|
22
|
Lin Y, Totsuka Y, Shan B, Wang C, Wei W,
Qiao Y, Kikuchi S, Inoue M, Tanaka H and He Y: Esophageal cancer in
high-risk areas of China: Research progress and challenges. Ann
Epidemiol. 27:215–221. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xi M and Lin SH: Recent advances in
intensity modulated radiotherapy and proton therapy for esophageal
cancer. Expert Rev Anticancer Ther. 17:635–646. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang YY, Wang LB, Zhao Q, Li G, Gong SL
and Dong LH: Advanced research on relationship between tumor
microenvironment and radiosensitivity of tumor cells. J Jilin
University (Medical Edition). 42:1038–1044. 2016.
|
|
25
|
Shen C, Chen F, Wang H, Li G, Yu C, Wang X
and Wen Z: The Pinx1 gene downregulates telomerase and inhibits
proliferation of CD133+ cancer stem cells isolated from a
nasopharyngeal carcinoma cell line by regulating Trfs and
Mad1/C-Myc/p53 pathways. Cell Physiol Biochem. 49:282–294. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sharma BK, Manglik V, O'Connell M,
Weeraratna A, McCarron EC, Broussard JN, Divito KA,
Simbulan-Rosenthal CM, Rosenthal DS and Zapas JL: Clonal dominance
of CD133+ subset population as risk factor in tumor progression and
disease recurrence of human cutaneous melanoma. Int J Oncol.
41:1570–1576. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Siersema PD and Van Hillegersberg R:
Treatment of locally advanced esophageal cancer with surgery and
chemoradiation. Curr Opin Gastroenterol. 24:535–540. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Koike R, Nishimura Y, Nakamatsu K,
Kanamori S and Shibata T: Concurrent chemoradiotherapy for
esophageal cancer with malignant fistula. Int J Radiat Oncol Biol
Phys. 70:1418–1422. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou Y, Chu L, Wang Q, Dai W, Zhang X,
Chen J, Li L, Ding P, Zhang L, Gu H, et al: CD59 is a potential
biomarker of esophageal squamous cell carcinoma radioresistance by
affecting DNA repair. Cell Death Dis. 9:8872018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pan S, Sun Y, Sui D, Yang T, Fu S, Wang J,
Hui B, Xi R, He C and Zhang X: Lobaplatin promotes
radiosensitivity, induces apoptosis, attenuates cancer stemness and
inhibits proliferation through PI3K/AKT pathway in esophageal
squamous cell carcinoma. Biomed Pharmacother. 102:567–574. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Park M, Yoon HJ, Kang MC, Kwon J and Lee
HW: MiR-338-5p enhances the radiosensitivity of esophageal squamous
cell carcinoma by inducing apoptosis through targeting survivin.
Sci Rep. 7:109322017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zang C, Zhao F, Hua L and Pu Y: The
miR-199a-3p regulates the radioresistance of esophageal cancer
cells via targeting the AK4 gene. Cancer Cell Int. 18:1862018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang H, Hua Y, Jiang Z, Yue J, Shi M,
Zhen X, Zhang X, Yang L, Zhou R and Wu S: Cancer-associated
fibroblast-promoted LncRNA DNM3OS confers radioresistance by
regulating DNA damage response in esophageal squamous cell
carcinoma. Clin Cancer Res. 25:1989–2000. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li Z, Zhou Y, Tu B, Bu Y, Liu A and Kong
J: Long noncoding RNA MALAT1 affects the efficacy of radiotherapy
for esophageal squamous cell carcinoma by regulating Cks1
expression. J Oral Pathol Med. 46:583–590. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xie SY, Li G, Han C, Yu YY and Li N: RKIP
reduction enhances radioresistance by activating the Shh signaling
pathway in non-small-cell lung cancer. Onco Targets Ther.
10:5605–5619. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu G, Chen G, Zhou J, Zhu H, Chu J and
Zhang F: Liriodenine enhances radiosensitivity in esophageal cancer
ECA109 cells by inducing apoptosis and G2/M arrest. Oncol Lett.
16:5020–5026. 2018.PubMed/NCBI
|
|
37
|
Hu L, Lin X, Lu H, Chen B and Bai Y: An
overview of hedgehog signaling in fibrosis. Mol Pharmacol.
87:174–182. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhu SL, Luo MQ, Peng WX, Li QX, Feng ZY,
Li ZX, Wang MX, Feng XX, Liu F and Huang JL: Sonic hedgehog
signalling pathway regulates apoptosis through Smo protein in human
umbilical vein endothelial cells. Rheumatology (Oxford).
54:1093–1102. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tanaka T, Arai M, Minemura S, Oyamada A,
Saito K, Jiang X, Tsuboi M, Sazuka S, Maruoka D, Matsumura T, et
al: Expression level of sonic hedgehog correlated with the speed of
gastric mucosa regeneration in artificial gastric ulcers. J
Gastroenterol Hepatol. 29:736–741. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wadhwa R, Wang X, Baladandayuthapani V,
Liu B, Shiozaki H, Shimodaira Y, Lin Q, Elimova E, Hofstetter WL,
Swisher SG, et al: Nuclear expression of Gli-1 is predictive of
pathologic complete response to chemoradiation in trimodality
treated oesophageal cancer patients. Br J Cancer. 117:648–655.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Briscoe J and Thérond PP: The mechanisms
of hedgehog signalling and its roles in development and disease.
Nat Rev Mol Cell Biol. 14:416–429. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Miele E, Po A, Begalli F, Antonucci L,
Mastronuzzi A, Marras CE, Carai A, Cucchi D, Abballe L, Besharat
ZM, et al: b-arrestin1-mediated acetylation of Gli1 regulates
Hedgehog/Gli signaling and modulates self-renewal of SHH
medulloblastoma cancer stem cells. BMC Cancer. 17:4882017.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wei L and Xu Z: Cross-signaling among
phosphinositide-3 kinase, mitogen-activated protein kinase and
sonic hedgehog pathways exists in esophageal cancer. Int J Cancer.
129:275–284. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Huang C, Lu H, Li J, Xie X, Fan L, Wang D,
Tan W, Wang Y, Lin Z and Yao T: SOX2 regulates radioresistance in
cervical cancer via the hedgehog signaling pathway. Gynecol Oncol.
151:533–541. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mori Y, Okumura T, Tsunoda S, Sakai Y and
Shimada Y: Gli-1 expression is associated with lymph node
metastasis and tumor progression in esophageal squamous cell
carcinoma. Oncology. 70:378–389. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Cheng J, Gao J and Tao K: Prognostic role
of Gli1 expression in solid malignancies: A meta-analysis. Sci Rep.
6:221842016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhu W, You Z, Li T, Yu C, Tao G, Hu M and
Chen X: Correlation of hedgehog signal activation with
chemoradiotherapy sensitivity and survival in esophageal squamous
cell carcinomas. Jpn J Clin Oncol. 41:386–393. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Teichman J, Dodbiba L, Thai H, Fleet A,
Morey T, Liu L, McGregor M, Cheng D, Chen Z, Darling G, et al:
Hedgehog inhibition mediates radiation sensitivity in mouse
xenograft models of human esophageal adenocarcinoma. PLoS One.
13:e01948092018. View Article : Google Scholar : PubMed/NCBI
|