Introduction
Cholangiocarcinoma (CCA) is a malignant tumor that
originates in the mucosal epithelial cells of the biliary system.
With a high degree of malignancy, rapid progression, low surgical
resection rates and high recurrence rates, CCA prognoses are
unsatisfactory (1–4). Recent studies of the molecular
pathological mechanisms of CCA have demonstrated that tumor
formation, growth, invasion, metastasis and other processes in CCA
are regulated by a variety of molecules and signal transduction
pathways, such as the aPKC-ι/P-Sp1/Snail signaling pathway and the
Merlin/YAP/c-Myc/mTOR signaling pathway (5,6). This
information has assisted early diagnosis, prognosis predictions and
drug development. However, due to the complexity of CCA
development, there are currently no specific biomarkers that meet
clinical standards (7–10). Therefore, there is an urgent need to
identify suitable biomolecular markers that can screen for CCA and
assist in prognostic evaluations.
Eukaryotic mRNA precursors are known to remove
certain unused fragments during the maturation process of mRNAs,
allowing the remaining fragments to be rejoined. The fragments that
are removed or left in place vary in their effects, depending on
the cell type and status. Thus, a gene can encode multiple proteins
in a process called alternative splicing (AS) (11,12).
Previous studies have demonstrated that AS events occur
ubiquitously in eukaryotes, with >95% of genes undergoing AS
events (13–15). However, AS events are more common in
cancer cells and affect cancer development and treatment resistance
(16–24). In addition, AS events in tumor cells
may be used as molecular markers that can differentially diagnose
tumor types and predict prognosis (25–28), as
well as serve as potential targets for cancer treatment (29–33).
Application of AS events as a means diagnosing and predicting
prognosis has been demonstrated in in prostate adenocarcinoma,
uteri corpus endometrial carcinoma and colorectal cancer (34–36).
CCA involves a high frequency of gene mutations and
abnormal epigenetic changes, including DNA methylation, and histone
and RNA modifications (37), all of
which may lead to the occurrence of AS events (19,38–47). AS
events modify gene expression and influence protein structure by
changing coding regions, thus regulating certain biological
processes. For example, the widely reported tumor suppressor genes
tumor protein p53, AT-rich interaction domain 1A, PTEN and PI3K, as
well as the proto-oncogenes NOTCH1 and MET proto-oncogene, receptor
tyrosine kinase, cause variations in gene function through AS
events, thus affecting cancer development (30). Similarly, the apoptosis-related genes
BCL-X and modulator of VRAC current 1 have been demonstrated to
serve opposite functions in promoting and resisting apoptosis due
to the occurrence of AS events (48–51). AS
can also alter the amino acid sequence of a protein, which may
destroy the target for certain antitumor drugs and result in drug
resistance (20,30,52). AS
events therefore serve a unique role in the diagnosis and treatment
of tumors.
Previous studies on the association between AS
events and tumors have focused on the single-gene level, and few
large-scale data mining studies based on high-throughput sequencing
exist (53–55). The Cancer Genome Atlas (TCGA)
database (portal.gdc.cancer.gov/) contains high-throughput
sequencing data and comprehensive clinical information from a large
number of cancer samples. Previous studies using TCGA data have
reported that AS events may be used as prognostic indicators for
lung (56), ovarian (57), bladder (58) cancer and gastrointestinal
pan-adenocarcinomas (59,60). However, the prognostic value of AS
events has not yet been reported for CCA. In the present study, the
overall survival (OS)-related AS events in CCA were systemically
evaluated. These findings may facilitate development of novel
genomic models for clinical cancer management, and construction of
novel models based on the prognostic index (PI) to predict CCA
survival.
Materials and methods
Data collection
The percent spliced index (PSI) values for CCA AS
events were downloaded from the TCGASpliceSeq database (http://bioinformatics.mdanderson.org/TCGASpliceSeq/)
(61). PSI is the ratio of
normalized read counts indicating inclusion of a transcript element
to the total normalized read counts for that event (i.e., both
inclusion and exclusion read counts). PSI values range from 0 to 1,
indicating the likelihood of the existence of an exon. Information
related to seven types of AS events: Exon skip (ES), mutually
exclusive exons (ME), retained intron (RI), alternate promoter
(AP), alternate terminator (AT), alternate donor (AD) site and
alternate acceptor (AA) site events, was included in the analysis.
Corresponding clinical information and gene expression levels of
the samples were obtained from TCGA database. The database included
data from 32 patients with CCA with complete clinical information
and total survival time >90 days. Of the 32 patients with CCA,
13 were male and 19 were female. The age of the patients was 29–82,
and the median age was 66.5, with 27 patients older than 50 and 5
patients younger than 50.
Association between AS events and
survival
R software (R version 3.4.2) (62) was used to perform univariate Cox
regression analysis to investigate the PSI prognostic values from
the seven types of AS events and the differentially expressed genes
in the 32 CCA cases. AS events with P<0.05 were selected for
further analysis. The event-dependent survival curve for the top
three AS events was plotted (collating P-values from low to
high).
Systematic review
To confirm that the identified AS events were
CCA-specific, all prognostic AS events from different types of
cancer were extracted from published articles on PubMed (https://www.ncbi.nlm.nih.gov/), Wiley Online Library
(https://onlinelibrary.wiley.com/), EBSCO
(https://www.ebsco.com/), Web of Science
(https://www.webofknowledge.com/), and
Google Scholar (https://scholar.google.com/) on or prior to 1st May
2019 for comparison. The key words were as follows: ‘cancer OR
carcinoma OR adenocarcinoma OR tumour OR tumor OR malignanc* OR
neoplas*’ AND ‘prognostic OR prognosis OR predict*’ AND ‘
‘alternative splicing’ OR AS’. The studies which were included
needed to meet the following criteria: i) Studies describing cancer
prognosis with a predicted model constructed using prognostic AS
events; and ii) the prognostic AS events could be extracted from
the studies. All the included studies were assessed using the bias
assessment for studies of diagnostic accuracy (QUADAS) guidelines
(63) and had a QUADAS score of ≤7.
The following data were extracted from the included studies: First
author's name, year of publication and prognostic AS events.
AS event interaction analysis and gene
network construction
The interactions of the seven types of AS events
were analyzed using the UpSetR package for R (64), and an UpSet plot was drawn. The
network was constructed using the Reactome FI plug-in (65) for Cytoscape 3.6.0 (66), with AS events identified using
univariate Cox regression analysis (P<0.01). Genes with AS
events were processed using the Kyoto Encyclopedia of Genes and
Genomes (KEGG) and Gene Ontology (GO) gene enrichment analyses. Due
to the occurrence of a large number of significant GO pathways,
only the top five pathways per group were presented using the
ggplot2 package for R (67).
PI construction and prognostic value
assessment
To assess the association between AS events and OS,
univariate Cox regression was carried out with a significance
threshold of P<0.05. The top ten most promising AS events, based
on their prognostic value, AS events selected for each splicing
type. Multivariate Cox regression analysis was performed, splicing
events with P<0.05 were selected as PIs. The model was
established by grouping together all splicing events of the seven
splicing event types. The samples were divided into two groups
according to the median PSI value of the splicing events. The
formula used to calculate the PI was as follows:
Risk score =
∑inPSIi*βi
The clinical prognostic value of the PI was assessed
using Kaplan-Meier (K-M) curve analysis and time-dependent receiver
operator characteristic (ROC) curve analysis. The time-dependent
ROC curve was drawn using the survival ROC package for R (68).
Experimental validation of AS events
in clinical CCA tissue samples
In addition to the aforementioned analyses, the
existence of the AS events predicted by the TCGASpliceSeq database
was verified using PCR analyses with six samples of CCA tumor
tissues separately collected from a 37-year-old male patient and a
38-year-old female patient during surgery at the Pathology
Department of the First Affiliated Hospital of Guangxi Medical
University (Nanning, China) between February 2017 and October 2017.
The study was approved by the Ethics Committee of the First
Affiliated Hospital of Guangxi Medical University, and written
informed consent was obtained from all patients. Total RNA was
extracted using the AxyPrep Multisource Total RNA Miniprep kit
(Axygen; Corning, Inc.), quantified using a NanoDrop 2000 (Thermo
Fisher Scientific, Inc.) and 500 ng total RNA was
reverse-transcribed into cDNA using MiScript® II RT
SuperMix kit (Vazyme). AS events in phytanoyl-CoA 2-hydroxylase
(PHYH)_100582_ES and transferrin receptor 2 (TFR2)_80979_ES were
selected for verification. The mean PSI of PHYH_100582_ES in
TCGASpliceSeq database was 0.78, and ES was present in exons 7 and
8. The forward primer (primer 1) was designed based on exon 6, and
the sequence was 5′-GATACTGCACTCTCCCCGAG-3′. The reverse primer
(primer 2) was designed based on exon 9, and the primer sequence
was 5′-GACCAGATCCGTGGATGAGC-3′. The PCR system contained 10 µl 2X
PCR Master mix (Thermo Fisher Scientific, Inc), 1 µl each of the
forward and reverse primers (10 µM), 1 µl cDNA and 7 µl
nuclease-free water (total volume, 20 µl). The thermocycling
conditions were as follows: 95°C for 3 min, followed by 35 cycles
of 95°C for 30 sec, 60°C for 30 sec and 72°C for 1 min. PCR
products were subjected to electrophoresis on 2% agarose gels. The
mean PSI of TFR2_80979_ES in the TCGASpliceSeq database was 1, and
ES was present in exon 10. The forward primer (primer 3) was
designed based on exon 9, and the primer sequence was
5′-CAGCCCATCAGTGCAGACAT-3′. The reverse primer (primer 4) was
designed based on exon 11, and the primer sequence was
5′-TTGTTGACCACTAGCCGCAG-3′. The PCR system and thermocycling
conditions were as mentioned above.
Correlation analysis between splicing
factors and prognosis-related AS events
A splicing factor that serves as a splicing
activator when bound to an intronic enhancer element may function
as a repressor when bound to its splicing element in the context of
an exon (69). Therefore, the
correlations between splicing factors and the prognosis-related AS
events were investigated. Splicing factor information was obtained
from the SpliceAid 2 database (http://193.206.120.249/splicing_tissue.html) (70). Univariate Cox regression analysis was
performed on the splicing events and splicing factors, followed by
Pearson's correlation analysis between the significant splicing
events, splicing factors and the OS-related AS events identified by
multivariate Cox analysis. The results were visualized using
Cytoscape version 3.4.0 (66).
Results
CCA AS events
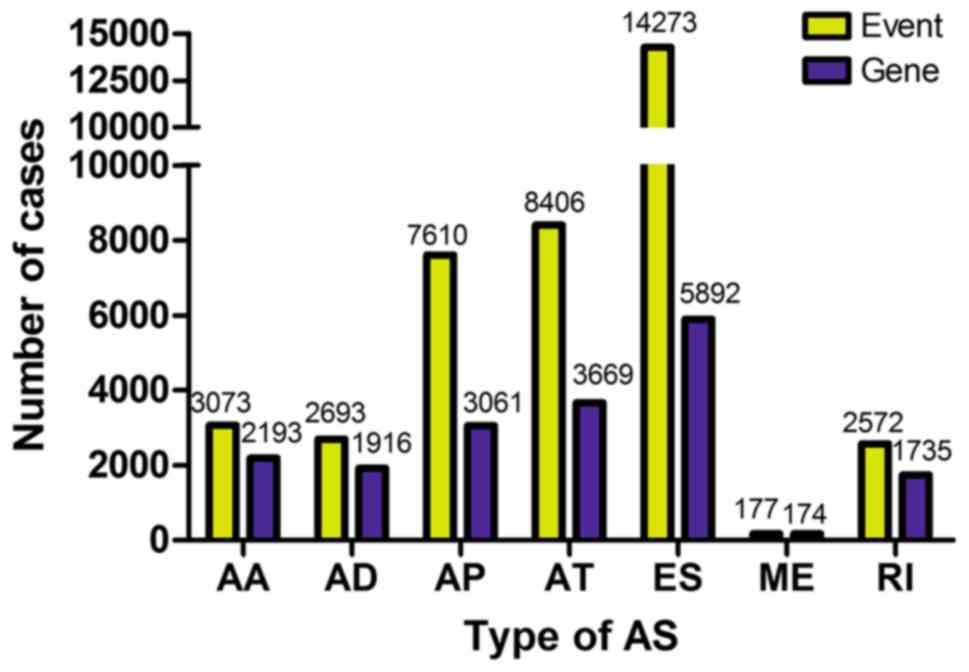
A total of 38,804 AS events were obtained from 9,673
genes in 32 CCA cases from TCGA SpliceSeq dataset. The numbers of
each type of AS event and the corresponding genes are presented in
Fig. 1. The results demonstrated
that a single gene may undergo multiple AS events simultaneously.
ES was the most frequent AS event in this dataset.
AS events associated with CCA OS
rates
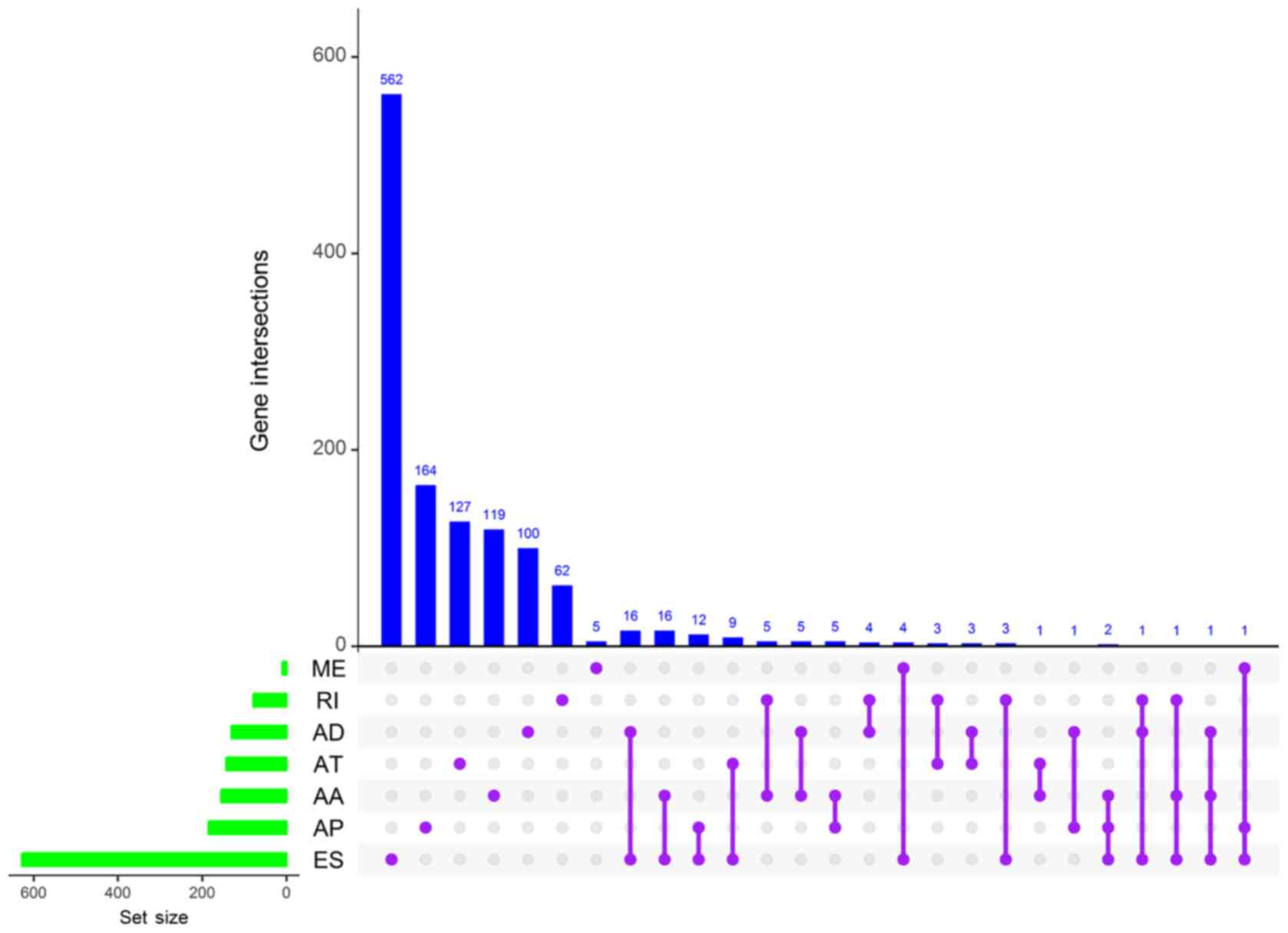
Univariate Cox analysis revealed that 1,639 AS
events were associated with CCA OS rates (P<0.05) and that
certain genes had undergone multiple OS-related AS events.
Visualization of the intersecting sets was performed using UpSet
plotting software (Fig. 2), and ≤3
AS events were identified to be associated with OS rates within the
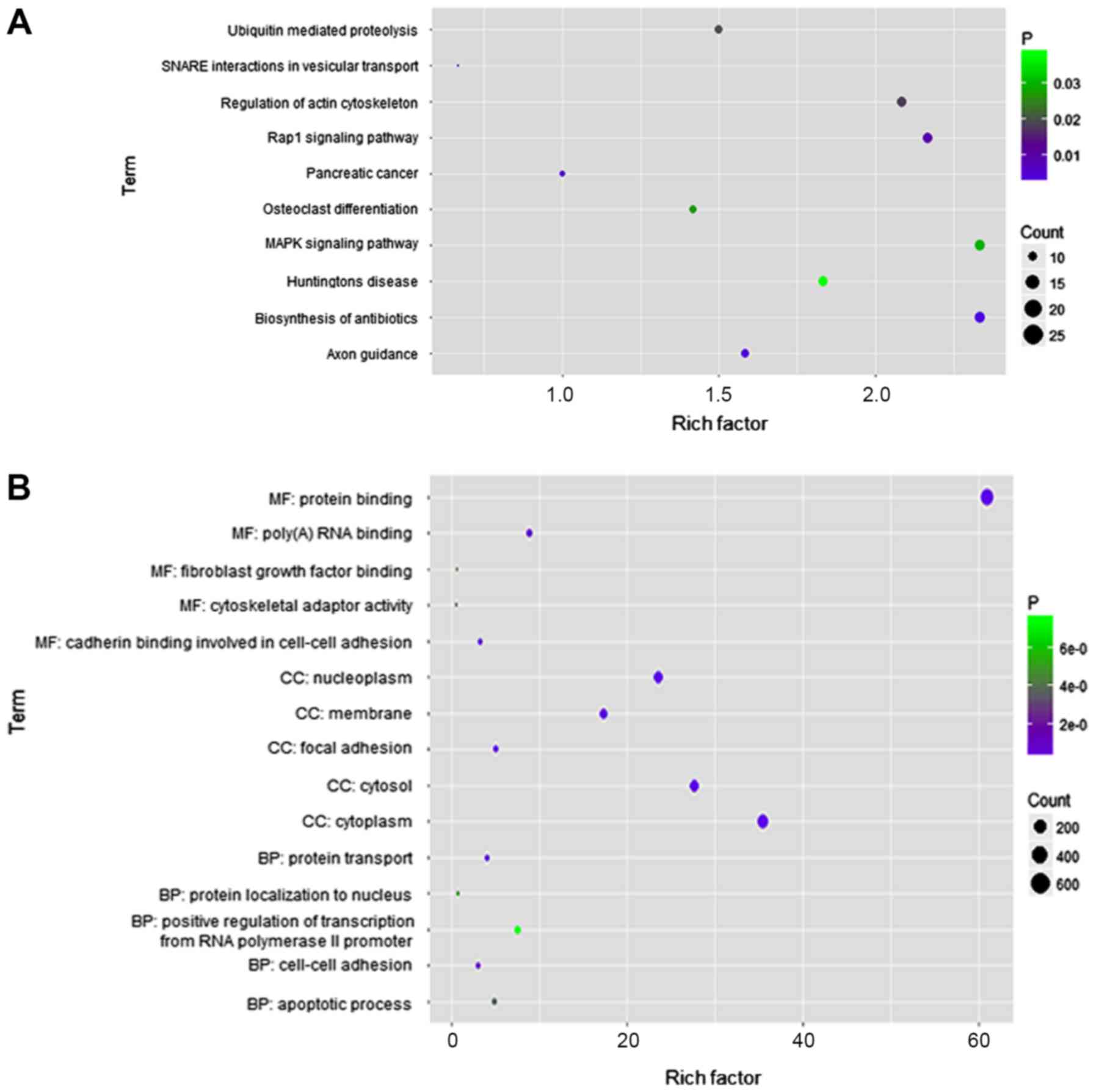
same gene. KEGG pathway enrichment analysis revealed that the genes
corresponding to these AS events were primarily enriched in
‘biosynthesis of antibiotics’, ‘axon guidance’, ‘pancreatic
cancer’, ‘RAP1 signaling pathway’ and ‘SNARE interactions in
vesicular transport’ (Fig. 3A). GO
gene enrichment analysis demonstrated that the genes with
OS-related AS events corresponded to ‘protein transport’,
‘cell-cell adhesion’, ‘apoptotic process’, ‘protein localization to
the nucleus’ and ‘positive regulation of transcription from RNA
polymerase II promoters’ (Fig. 3B).
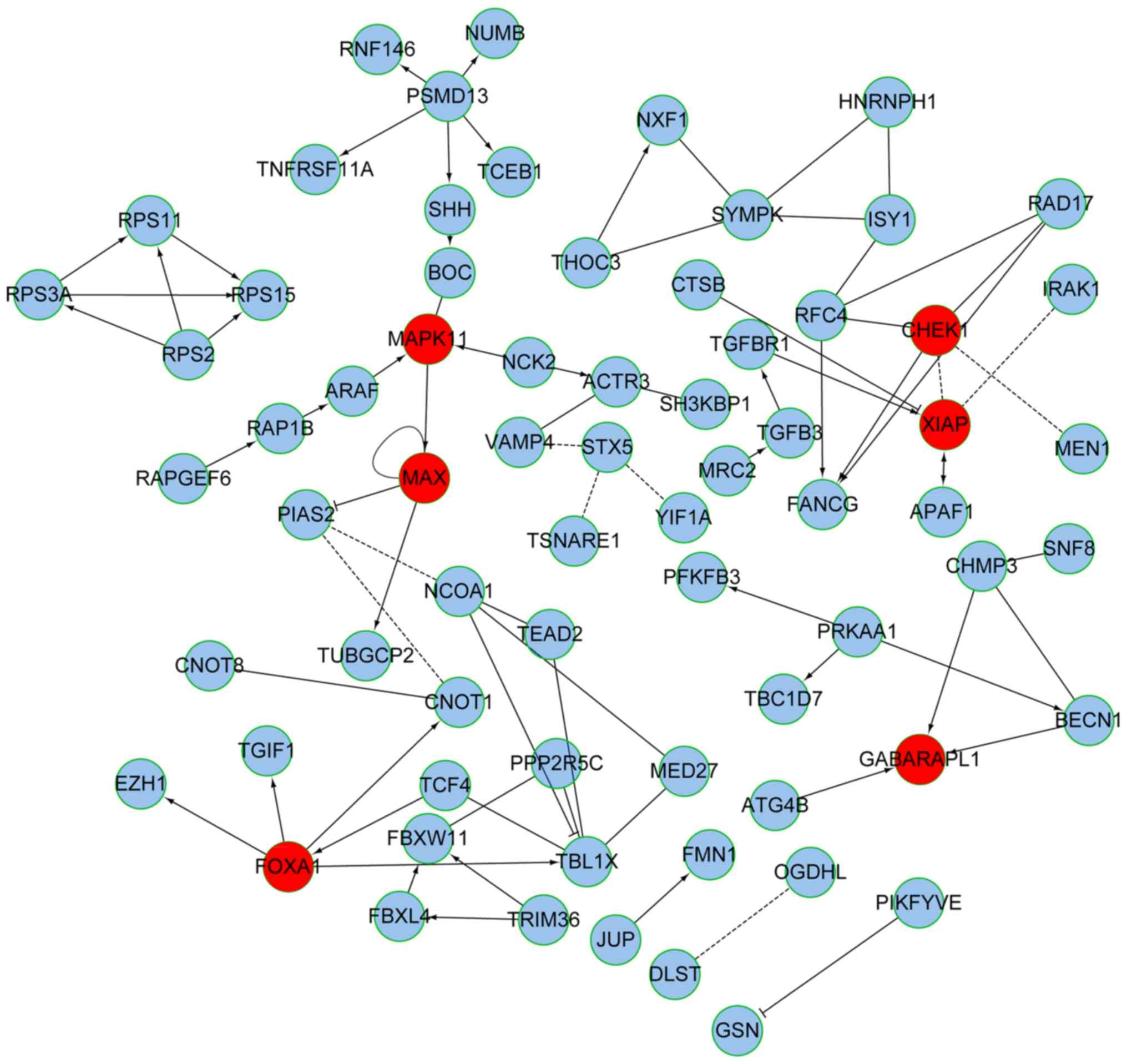
Using a network map of these genes, MYC associated factor X (MAX),
mitogen-activated protein kinase 11 (MAPK11), γ-aminobutyric acid
type A receptor-associated protein-like 1 (GABARAPL1), checkpoint
kinase 1 (CHEK1), X-linked inhibitor of apoptosis (XIAP) and
forkhead box A1 (FOXA1) exhibited the highest degrees of
connectivity and were thus considered critical positions of the
network (Fig. 4).
OS-related AS events are
CCA-specific
Comparisons with other types of cancer demonstrated
that ME-GTF2H3-306194 occurred in papillary thyroid cancer and CCA,
whereas the other identified AS events were CCA-specific (Table I).
 | Table I.Prognostic alternative splicing
events in different types of cancer. |
Table I.
Prognostic alternative splicing
events in different types of cancer.
| Author, year | Type | PMID | Alternative
splicing events | (Refs.) |
|---|
| Lin et al,
2019 | Papillary thyroid
cancer | 30986203 | AA-SHPRH-78032,
AA-CASK-88861, AD-FBXL19-36205, AD-SAT2-39030, AD-TRO-89255,
AD-CSTF2-89611, AP-ZC3H11A-9456, AP-STK32C-13483, AP-GRB2-43439,
AP-CRTC1-48500, AP-ERCC1-50440, AP-ESR1-78161, AT-MAGI3-4271,
AT-TPM1-30982, AT-ATP8B3-46544, AT-MAST1-47878, AT-SPAG16-57327,
AT-CBWD5-86498, AT-OLFM1-88103, ME-NSMF-193275, ME-GTF2H3-306194,
RI-C11orf49-15609, RI-ZNF276-38138, RI-USP36-43917,
RI-NUDT18-82937, RI-NAPRT1-85430 | (95) |
| Gao et al,
2019 | Uteri corpus
endometrial carcinoma | 30640723 | AP-BDNF-14763,
AP-DDX58-86057, AP-FYTTD1-68310, AP-GNAL-44643, AP-GPATCH2L-28538,
AP-HUS1-79610, AP-MAP4-64545, AT-IPO11-72190, AT-ZFAND4-11368,
ES-CKMT2-72660, ES-CMC2-37735, ES-FBXL12-47421, ES-NDUFB1-28987,
ES-PSMD12-43112, ES-ZNF528-51457, RI-AP3M2-83565,
RI-DNASE1L3-65424, RI-GABARAP-38871 | (34) |
| Huang et al,
2018 | Prostate
adenocarcinoma | 30221674 | ES-TCEB2-33303,
AD-ABHD17A-46558, AP-FKBP2-16603, ES-TXN-87183, AP-FKBP2-16602,
AD-YPEL3-36074, ES-STXBP2-47124, AT-PTGDS-88235, AT-HMGA2-22879,
ES-NHLRC3-25701 | (36) |
| He et al, 2018 | Bladder urothelial
carcinoma | 30048970 | AA-B4GALT2-1228,
ES-TMTC2-9217, ES-TIMM9-11224, ES-APOBEC3D-26508, AP-TPD52-35921,
ES-MICU1-4164, ES-DDX11-8115, ES-SMC6-22132 | (94) |
| Zhang et al,
2019 | Breast
carcinoma | 30984247 | AA-CARM1-47598,
AA-ZBTB25-27884, AA-GPBP1-72126, AA-ZNRF1-37578, AA-DDX41-74796,
AA-CTDSP1-57478, AD-OS9-22701, AD-HN1-43371, AD-THTPA-26757,
AD-NTMT1-87866, AD-MGME1-58753, AD-SEC31A-69735, AP-SEC22A-66462,
AP-ALG3-67851, AP-PACS2-29630, AP-ECE2-67857, AP-HSP90AB1-76378,
AT-MAGT1-89535, AT-RCBTB1-25898, AT-SIN3B-48214, AT-SARNP-22252,
AT-ZNF675-48822, AT-STOX2-71289, AT-NIPAL3-1110, ES-NDUFA12-23737,
ES-UBR4-880, ES-COPS3-39468, ES-ABCE1-70753, ES-CCNI-69628,
ES-RPAP1-30096, ME-HLCS-96019, RI-RBM48-80441, RI-RBM6-64936,
RI-RPAP1-30095, RI-METTL17-26476, RI-POMGNT1-2787, RI-TRABD-62792,
RI-WDR6-64794, RI-FASTK-82335, RI-NAA38-81579 | (96) |
| Lin et al,
2018 | Esophageal
adenocarcinoma | 30131306 | AA-U2AF1L4-49280,
AA-TICRR-32428, AA-RSRC2-24968, AA-PREPL-53439, AA-PPIL2-61247,
AA-FAM135A-76637, AA-CDV3-66839, AA-ABCB7-89517, AD-ZNF384-19927,
AD-RPP14-65434, AD-PQBP1-89028, AD-MFSD11-43690, AD-COX6C-84682,
AP-ZNF623-85469, AP-KIAA0513-37876, AP-FAM19A5-62732,
AP-ALDH6A1-28367, AT-TRIM4-80864, AT-RNASEH2B-25927,
AT-RNASEH2B-25926, AT-MCPH1-82574, AT-ARL6-65732, AT-AHI1-77886,
ES-TNC-87345, ES-PML-31651, ES-NBPF15-91080, ES-MYL6-22384,
ES-MRPL43-12857, ES-IRF9-117161, ME-SDR39U1-27012, ME-KLHL2-71038,
ME-CMC2-37707, RI-ZNF131-71926, RI-SLC52A3-58464,
RI-PPARGC1B-74051, RI-PCGF3-68404, RI-MDK-15570, RI-MAF-37687,
RI-FAM9C-88504 | (60) |
| Lin et al,
2018 | Stomach
adenocarcinoma | 30131306 | AA-RPLP0-24727,
AA-NAT6-64990, AA-MRVI1-14373, AA-LMO7-26065, AA-BDKRB2-29192,
AD-YIPF2-47605, AD-SPHK2-50793, AD-SENP1-21411, AD-PGAP2-14004,
AD-NFATC1-46241, AD-CCDC51-64653, AP-RCAN1-60494, AP-PLCD1-64009,
AP-LTBP1-53179, AP-FAM65B-75537, AP-ABL2-9101, AT-ZNF846-47399,
AT-ZFYVE28-68559, AT-STEAP4-80362, AT-STEAP4-80361, AT-KIF1B-602,
AT-KIF1B-601, AT-CXCL12-11344, AT-CLDN11-67617, AT-ABCB5-78909,
ES-UBXN11-1263, ES-TMEM230-58637, ES-SRSF3-75985, ES-SORBS1-12641,
ES-P4HA2-73263, ES-CREM-11245, ME-N4BP2L1-25590, ME-KDM6A-98323,
ME-FYN-77273, ME-CCDC53-106010, RI-TREX1-64682, RI-SRSF7-53276,
RI-RPS15-46490, RI-LDHA-14642, RI-BICD2-86883, RI-ALS2CL-64462 | (60) |
| Lin et al,
2018 | Colon
adenocarcinoma | 30131306 | AA-RASSF7-13691,
AA-PTGR1-87219, AA-FAM173A-32964, AA-DPP3-17040, AA-CDV3-66842,
AD-RNF14-73855, AD-IP6K2-64759, AD-HPS4-61506, AD-HDGF-8323,
AD-ANKRD46-84712, AD-ADPGK-31594, AP-TUBB3-38167, AP-RAB3IP-23345,
AP-MAZ-35938, AP-FADS2-16289, AP-ENO2-20011, AT-ZNF765-51718,
AT-UPK3B-80182, AT-RASEF-86677, AT-RASEF-86676, AT-NRG4-31911,
AT-AIG1-77972, ES-VTI1B-28083, ES-STRN3-27098, ES-RHOC-4236,
ES-PRMT1-51042, ES-PLEKHM2-767, ES-DMWD-50528, ES-D2HGDH-58423,
ME-CNOT10-63822, RI-ZNF226-50290, RI-NPIPA5-34148, RI-ELP5-38889,
RI-ALS2CL-64463 | (60) |
| Lin et al,
2018 | Rectal
adenocarcinoma | 30131306 | AA-ZNF467-82205,
AA-RNPC3-3907, AA-GGT1-61440, AA-BTN3A1-75660, AD-OSBPL9-2975,
AD-METTL23-43637, AD-BCS1L-57522, AP-TADA2B-68732, AP-PTCH1-86955,
AP-DAB2IP-87442, AT-PUS10-53676, AT-NOTCH2NL-4437, ES-SPAG9-42496,
ES-SERPINA1-29134, ES-PHB2-20048, ES-FGFR1OP2-20856,
ME-RBMS2-22465, RI-ZNF692-10557, RI-WDR33-55246, RI-TMEM91-50046,
RI-SIDT2-18886, RI-EXOSC9-70501, RI-ADARB1-60863 | (60) |
AS events that may be used as CCA
prognostic indicators
The three most statistically significant AS events
were selected, and their gene expression levels were used to
calculate their prognostic values in CCA (Fig. 5). K-M analysis revealed that these
prognostic signatures significantly separated patients with
distinct prognoses. To assess the ability of these splicing events
to predict CCA prognosis, a time-dependent ROC curve was
established. The results of the ROC curve suggested that a single
indicator was not enough to predict patient prognosis, as the area
under the curve (AUC) values were low for the AS events and gene
expression values (Fig. 5).
Therefore, ten AS events with the most significant prognostic
values in each splicing type were selected to construct the PIs
(Table II). The results suggested
that these PIs had higher predictive values compared with single
indicators (Fig. 6). The AUCs for
each of the seven subtypes of AS were as follows: AA, 0.901; AD,
0.858; AP, 0.986; AT, 0.839; ES, 0.939; ME, 0.779; and RI, 0.859.
In addition, the ten most significant AS events from all types
combined were selected to construct a final prognostic model; the
AUC was 0.984 (Fig. 6). It is worth
noting that the predictive efficacy of the prognostic model with
seven combined splicing events (0.984) was slightly lower than that
obtained with AP alone (0.986).
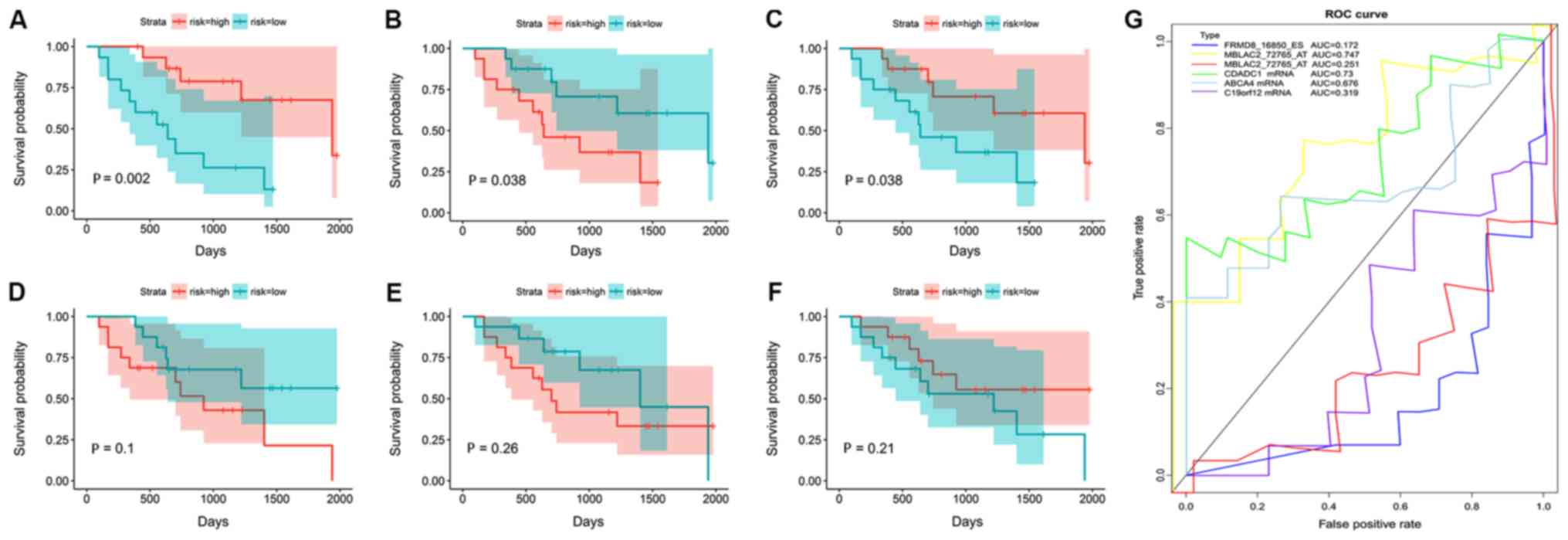 | Figure 5.Kaplan-Meier survival and ROC curves
of alternative splicing events and gene expression of the top three
factors identified using univariate Cox analysis. Blue, low-risk
group; red, high-risk group. (A-C) Kaplan-Meier curves of
alternative splicing events. (A) FRMD8_16850_ES (HR, 0.736; 95% CI,
0.627–0.824; P<0.001). (B) MBLAC2_72765_AT (HR, 1.186; 95% CI,
1.084–1.298; P<0.001). (C) MBLAC2_72766_AT (HR, 0.843; 95% CI,
0.771–0.923; P<0.001). (D-F) Kaplan-Meier curves of gene
expression levels. (D) CDADC1 (HR, 1060.208; 95% CI,
15.259–73662.548; P=0.001). (E) ABCA4 (HR, 13.302; 95% CI,
2.596–68.156; P=0.002). (F) C19orf12 (HR, 0.0004; 95%CI,
0.000001–0.081; P=0.004). (G) ROC curves of alternative splicing
events and gene expression levels: FRMD8_16850_ES, MBLAC2_72765_AT,
MBLAC2_72766_AT, CDADC1, ABCA4 and C19orf12. AA, alternate
acceptor; AD, alternate donor; AP, alternate promoter; AT,
alternate terminator; ROC, receiver operating characteristic; AUC,
area under the curve; ES, exon skip; ME, mutually exclusive exon;
RI, retained intron; HR, Hazard ratio; CI, confidence interval;
FRMD, FERM domain-containing 8; MBLAC2, metallo-β-lactamase
domain-containing 2; CDADC1, cytidine and DCMP deaminase
domain-containing 1; ABCA4, ATP-binding cassette subfamily A member
4; C19orf12, chromosome 19 open reading frame 12. |
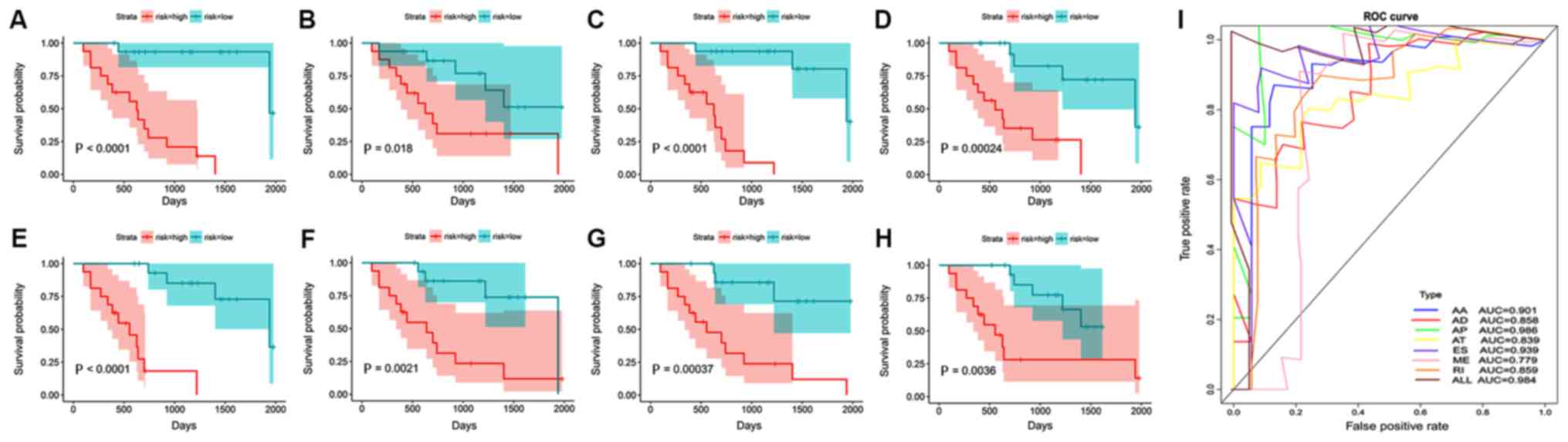 | Figure 6.Kaplan-Meier curves based on the
prognostic index and its ROC curves. Blue, low-risk group; red,
high-risk group. (A-H) Kaplan-Meier curves of alternative splicing
events. (A) AA site. (B) AD site. (C) AP. (D) AT. (E) ES. (F) ME
exons. (G) RI. (H) ALL. (I) ROC curves of AA, AD, AP, AT, ES, ME,
RI and ALL. AA, alternate acceptor; AD, alternate donor; AP,
alternate promoter; AT, alternate terminator; ROC, receiver
operating characteristic; AUC, area under the curve; ES, exon skip;
ME, mutually exclusive exon; RI, retained intron; ALL, all
alternative splicing types combined. |
 | Table II.Prediction models for
cholangiocarcinoma based on each type of splicing event. |
Table II.
Prediction models for
cholangiocarcinoma based on each type of splicing event.
| Risk score | Algorithm | HR (95% CI) | P-value | AUC |
|---|
| Risk score
(AA) | PSIST3GAL4_19399_AA
× 0.849 + | 10.990
(3.818–31.61) | <0.001 | 0.901 |
|
| PSITECR_47998_AA ×
(−6.242) + |
|
|
|
|
| PSITGIF1_44506_AA ×
(−0.713) |
|
|
|
| Risk score
(AD) | PSISYNGR1_62301_AD
× (−0.645) + | 3.375
(1.227–9.283) | 0.018 | 0.858 |
|
| PSIZHX3_59398_AD ×
(−0.277) |
|
|
|
| Risk score
(AP) |
PSIC12orf65_25058_AP × (−0.147) + | 17.910
(5.658–56.67) | <0.001 | 0.986 |
|
| PSICHMP3_54439_AP ×
1.274 + |
|
|
|
|
| PSISH3KBP1_88640_AP
× 0.432 |
|
|
|
| Risk score
(AT) | PSIMBLAC2_72765_AT
× 0.166 + | 7.622
(2.576–22.55) | <0.001 | 0.839 |
|
| PSITGFB3_28531_AT ×
(−1.244) |
|
|
|
| Risk score
(ES) | PSIACAD9_66674_ES ×
(−1.952) + | 19.090
(5.705–63.86) | <0.001 | 0.939 |
|
| PSIFRMD8_16850_ES ×
(−0.701) + |
|
|
|
|
| PSIPLEKHG2_49826_ES
× (−1.49) + |
|
|
|
|
| PSITP53I11_15489_ES
× 2.014 + |
|
|
|
|
| PSIUBE2F_58170_ES ×
8.384 |
|
|
|
| Risk score
(ME) | PSIFGFR3_68513_ME ×
(−0.15) + | 4.977
(1.790–13.84) | 0.002 | 0.779 |
|
| PSIGRB10_79717_ME ×
(−0.068) + |
|
|
|
|
| PSIGTF2H3_306194_ME
× 1.421 + |
|
|
|
|
| PSIRNF146_114496_ME
× (−0.085) + |
|
|
|
|
| PSISORBS2_71377_ME
× (−0.282) |
|
|
|
| Risk score
(RI) |
PSIC11orf88_18667_RI × (−0.073) + | 6.358
(2.295–17.61) | <0.001 | 0.859 |
|
| PSIDET1_32385_RI ×
(−0.214) + |
|
|
|
|
| PSIGAREML_52884_RI
× (−0.332) |
|
|
|
| Risk score
(merged) | PSITFDP1_26387_AD ×
(−0.57) + | 19.670
(5.842–66.22) | <0.001 | 0.984 |
|
| PSIMBLAC2_72766_AT
× (−0.838) + |
|
|
|
|
| PSIHACL1_63592_ES ×
(−0.62) + |
|
|
|
|
| PSIPLEKHG2_49826_ES
× (−4.314) + |
|
|
|
|
| PSIUBE2F_58170_ES ×
7.06 |
|
|
|
Confirmation of PHYH_100582_ES and
TFR2_80979_ES by clinical samples
Towards PHYH_100582_ES, the PCR products were
predicted to comprise two bands; the band without ES was predicted
to be 414 bp in length, whereas the band size following ES was
predicted to be 144 bp in length (Fig.
7A). If the TFR2_80979_ES event did not occur, the PCR product
was predicted to be 238 bp; following the ES event, the PCR product
size was predicted to be 140 bp (Fig.
7B). Since the PSI value of this ES event was 1, the ES event
occurred in all TFR2 mRNAs; if the TCGASpliceSeq database was
accurate, the PCR product was predicted to be a single band. The
results of PHYH_100582_ES and TFR2_80979_ES were consistent with
those predicted by the TCGASpliceSeq database. The PCR products of
PHYH_100582_ES were 414 and 144 bp (Fig.
7C), suggesting that, in CCA tissues, a proportion of PHYH mRNA
had undergone an ES event. The PSI of TFR2_80979_ES in the
TCGASpliceSeq database was 1, indicating that ES events occurred in
all TFR2 mRNAs. The electrophoresis result demonstrated that the
product of TFR2_80979_ES was a single band of 140 bp; no band of
238 bp was observed (Fig. 7D),
suggesting that ES events occurred in all TFR2 mRNA. These results
were consistent with those suggested by the RNA sequencing data
from TCGASpliceSeq.
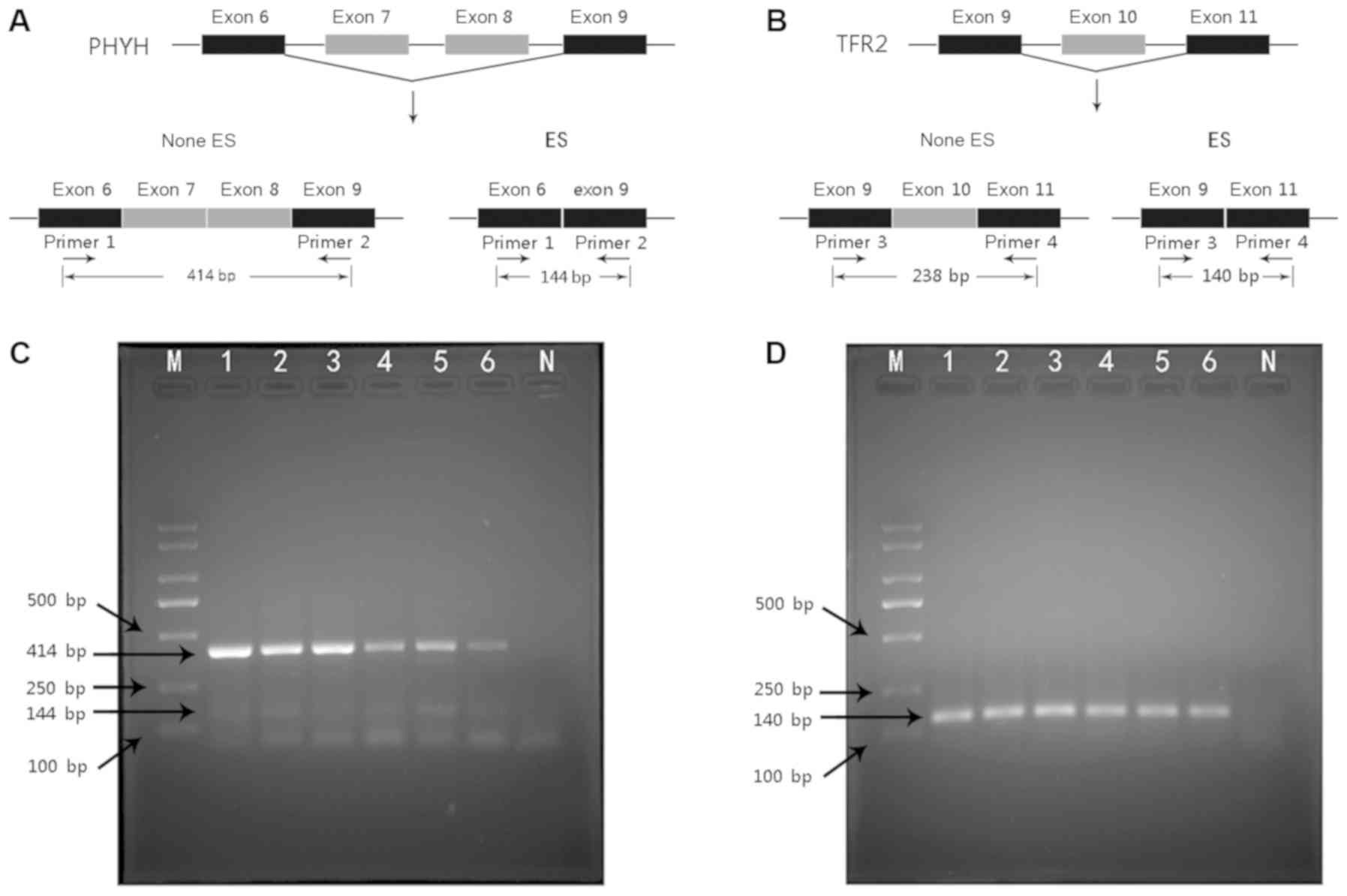 | Figure 7.Validation of predicted AS events in
clinical samples. (A) Illustration of PHYH_100582_ES AS event in
the present study. Following the ES event, exons 7 and 8 of PHYH
mRNA were cut out, and exons 6 and 9 were directly spliced. (B)
Illustration of TFR2_80979_ES AS event in the present study.
Following the ES event, exon 10 of TFR2 mRNA was cut out, and exons
9 and 11 exons were directly spliced. (C) PHYH_100582_ES event
electropherogram. Lanes 1–6 demonstrate the PCR amplification
results of cDNA from six CCA tissues with bands of 414 bp and 144
bp. (D) TFR2_80979_ES event electropherogram. Lanes 1–6 demonstrate
the PCR amplification results of cDNA from six CCA tissues with a
single band of 140 bp. M, DNA marker; N, negative control without a
template; AS, alternative splicing; ES, exon skip; PHYH,
phytanoyl-CoA 2-hydroxylase; TRF2, transferrin receptor 2; CCA,
cholangiocarcinoma. |
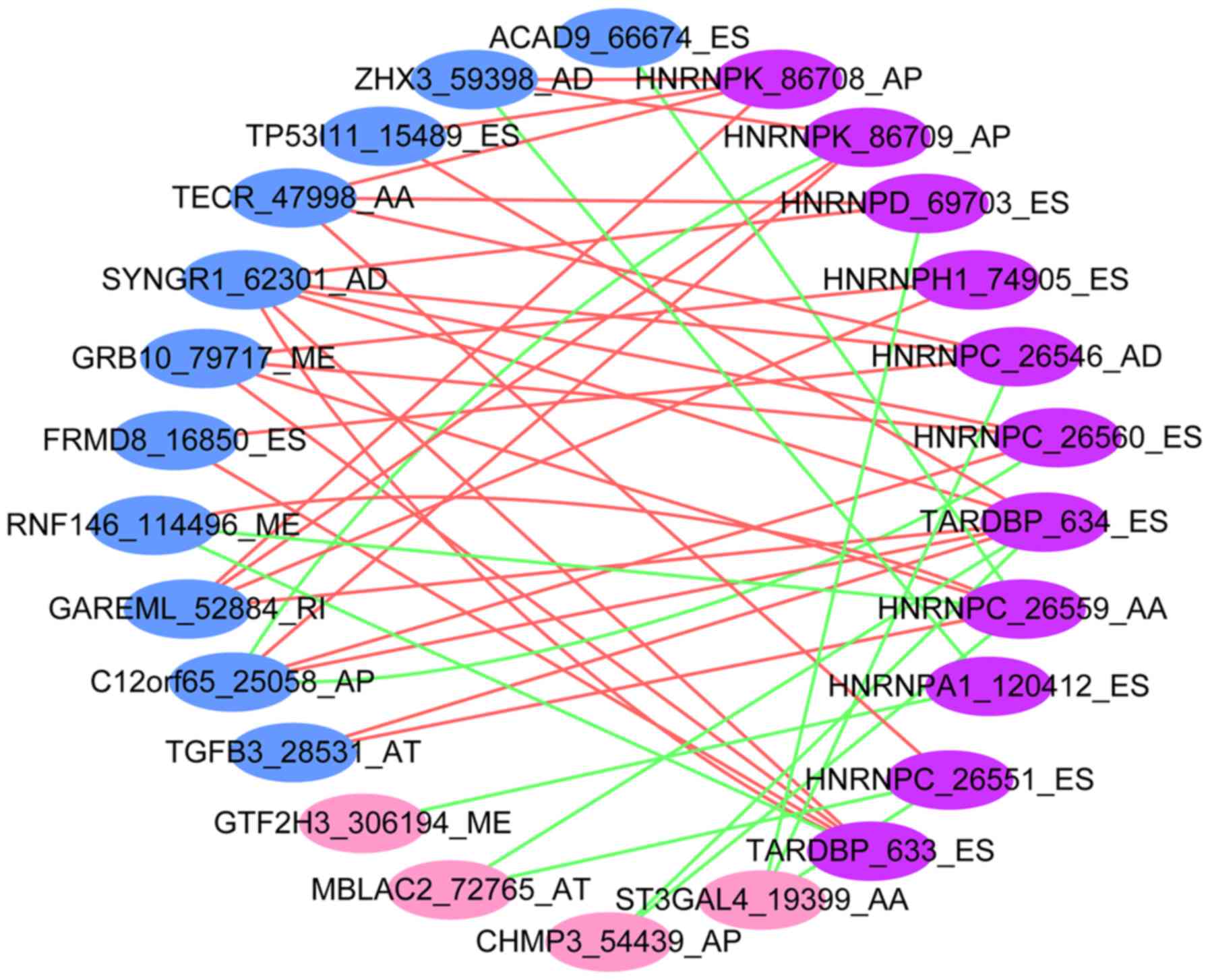
Correlation between splicing factors
and prognosis-related AS events
A total of 11 AS events and corresponding splicing
factors significantly related to CCA OS rates (P<0.05) were
identified. Multivariate Cox analysis identified 23
prognosis-related AS events (Table
III; P<0.001); 17 of these exhibited positive correlations
with OS, whereas 6 exhibited negative correlations with OS. Of
these 23 AS events, 15 were correlated with a group of splicing
factors, the expression of which was directly associated with AS
events (|r|>0.3; P<0.05). AS events are primarily regulated
by splicing factors, which often bind to pre-mRNAs and regulate RNA
splicing by influencing exon selection and splicing site.
Therefore, the associations between survival-related AS events and
splicing factors were determined (Fig.
8).
 | Table III.Predictive factors identified for
cholangiocarcinoma using multivariate logistic regression. |
Table III.
Predictive factors identified for
cholangiocarcinoma using multivariate logistic regression.
| Splicing type | Gene symbol | AS ID |
|---|
| AA | ST3GAL4 | 19399 |
| AA | TECR | 47998 |
| AA | TGIF1 | 44506 |
| AD | SYNGR1 | 62301 |
| AD | ZHX3 | 59398 |
| AP | C12orf65 | 25058 |
| AP | CHMP3 | 54439 |
| AP | SH3KBP1 | 88640 |
| AT | MBLAC2 | 72765 |
| AT | TGFB3 | 28531 |
| ES | PLEKHG2 | 49826 |
| ES | ACAD9 | 66674 |
| ES | TP53I11 | 15489 |
| ES | FRMD8 | 16850 |
| ES | UBE2F | 58170 |
| ME | FGFR3 | 68513 |
| ME | RNF146 | 114496 |
| ME | GRB10 | 79717 |
| ME | SORBS2 | 71377 |
| ME | GTF2H3 | 306194 |
| RI | C11orf88 | 18667 |
| RI | DET1 | 32385 |
| RI | GAREML | 52884 |
Discussion
Using data mined from TCGASpliceSeq and in silico
approaches, the present study identified that a number of AS events
are closely associated with survival in CCA. The present study is
the first to report this type of result. The results of the present
study also demonstrated that AS events may be used to construct a
PI model that effectively determines CCA prognosis.
The incidence of CCA has increased in recent years.
The prognosis of advanced CCA is poor, with an extremely low 5-year
survival rate (7). The prognosis
depends on the synergistic effects of various factors; however, no
clear and effective molecular markers have been identified for CCA
diagnosis and treatment. Carcinoembryonic antigens (CEAs) and
carbohydrate antigen (CA) 199, CA 125, CA 50 and CA 242 are
currently used as CCA tumor markers, but these biomarkers have
disadvantages in clinical application. For example, the majority of
studies that examined CA 199 as a biomarker for the detection of
CCA have reported its suboptimal accuracy, with wide variation in
reported sensitivity (38–93%) and specificity (67–98%) (71). CA 125 is upregulated in 65% of
patients with CCA and is of value in predicting survival (72); however, there are no further studies
on CA 125 in CCA. CA 50 exhibits cross-antigenicity with CA 199,
and elevated serum CA 50 levels are commonly used to diagnose or
prognose pancreatic and colorectal cancers (73). However, Shan et al (74) demonstrated that elevated serum CA 50
levels are not always associated with the expression of CA 50 in
cancer tissues. Despite the clinical utility of CA 242, it is not
sufficiently effective in the early detection of cancer, since
elevated classical tumor biomarker levels indicate the presence of
a significant number of cancer cells (75).
At the genetic level, a number of genes are
abnormally expressed in CCA. For example, the abnormal expression
of genes such as transforming growth factor β1, SMAD4, c-MET,
matrix metallopeptidase 7, vascular endothelial growth factor
(VEGF)-A, VEGF-B, VEGF-C and VEGF-D are associated with poor
prognosis (76–79). Mutations in genes such as human
epidermal growth factor receptor 2, TP53, KRAS, cytosolic
NADP-dependent isocitrate dehydrogenase and mitochondrial
NADP-dependent isocitrate dehydrogenase can also affect the
prognosis of patients with CCA, but the prognostic value of these
genes remains controversial and has not been applied clinically
(72). Due to the complexity of
cancer, diagnosis or prognosis based on a single molecule is
limited. The advent of high-throughput sequencing technologies
allows us to address this issue by identifying more genomic
abnormalities related to CCA.
Using the TCGASpliceSeq CCA data, 38,804 AS events
were identified in 9,673 CCA genes, some of which exhibited
multiple AS events occurring simultaneously. AS events are
widespread in CCA and may be related to its occurrence and
development. For instance, the first AS event reported to be
associated with CCA prognosis was exon 2 skipping of the trefoil
factor 2 (TFF2) gene (80). TFF2 is
highly expressed in a variety of tumors, including CCA. Exon 2
skipping AS events lead to the loss of exon 2, resulting in a
decrease in wild-type TFF2 proteins. A high proportion of this AS
event in TFF2 is associated with improved prognosis. Another
example is P53, as alterations in the N-terminus of this protein by
AS has been demonstrated to worsen the prognosis in patients with
CCA (81).
The present study identified a large number of CCA
AS events, 1,639 of which were correlated with patient prognosis
(P<0.05). Bioinformatics analyses of all genes with significant
prognostic values identified the following genes to be located at
the center of the gene network: MAX, MAPK11, GABARAPL1, CHEK1, XIAP
and FOXA1. These genes have been broadly reported to be involved in
cell cycle regulation, autophagy, proliferation, apoptosis and have
been previously demonstrated to be associated with cancer (82–87).
Tumor-specific AS events have been intensively studied by
investigating the differentially alternatively spliced genes
between tumor and adjacent normal tissues (88) in certain cancer types, such as breast
(89), lung (56) and ovarian (57) cancer. Yosudjai et al (90) also demonstrated that aberrant AS of
anterior gradient protein 2 homolog promoted cell proliferation,
migration, invasion and adhesive potential in CCA; however, there
has been a lack of research on its association with patient
prognoses. Among the genes identified above, MAX forms a
heterodimer with the proto-oncogene MYC, which binds to DNA to
regulate the transcription of multiple genes, thus regulating cell
proliferation, differentiation and apoptosis (82). The MAPK11 gene encodes a
serine/threonine kinase that is widely involved in various cellular
signal transduction pathways, such as p38 the MAPK signaling
pathway by phosphorylating multiple target proteins (87). CHEK1 is a cell cycle
monitoring-related protein, abnormalities of which result in DNA
damage and the bypassing of the cell cycle checkpoints (91). FOXA1 is a transcription factor, the
aberrant expression of which is closely associated with
hepatocellular carcinoma (84);
however, its association with CCA has not been previously
reported.
The current study is the first to identify the
characteristics of alternative CCA splicing events and establish a
model that can predict CCA prognosis using PIs from AS events. Li
et al (56), who studied
non-small cell lung cancer, were the first to demonstrate a
combined survival and correlation network between the expression of
splicing factors and AS events; the present study provides a more
accurate potential network by screening the significant prognostic
AS events identified by univariate Cox regression.
The present study attempted to predict the prognosis
of patients with CCA using the three most significant AS events and
genes. The results were not satisfactory (the maximum AUC was
0.747; data not shown), likely due to individual differences
between patients. Similarly, no single factor was able to
accurately predict patient prognosis. Therefore, ten AS events with
the most significant prognostic values, as calculated by
multivariate Cox regression analysis for each type of AS event,
were selected to construct a PI. The prognostic models based on the
combined seven types of splicing events demonstrated comparable
predictive efficacy to the model that used only the AP splicing
events. However, the combined model was more successful at
predicting patient prognosis compared with models based on any
other single type of splicing event.
The results of the present study were compared with
those reported in previous studies. All AS events identified in the
present study were uniquely associated with CCA, with the exception
of ME-GTF2H3-306194, which has been previously identified to have
prognostic value in papillary thyroid cancer and CCA, indicating
that ME-GTF2H3-306194 may serve similar functions in CCA and
papillary thyroid cancer and suggesting a new treatment possibility
for CCA and papillary thyroid cancer by targeting
ME-GTF2H3-306194.
In addition, the existence of two AS events
predicted by the TCGASpliceSeq database was verified using PCR
analysis of six CCA tumor tissue samples. Metastasis was common in
cholangiocarcinoma (7,92). The direct infiltration of
cholangiocarcinoma cells along the bile duct wall is one of the
main features of cholangiocarcinoma metastasis (7). Cancer cells diffuse invasively in the
bile duct wall and coexist with the bile duct and surrounding
connective tissue, making the infiltration range of
cholangiocarcinoma difficult to identify, and thus non-cancerous
bile duct tissues cannon be easily obtained during surgery
(93). Therefore, only two AS events
were verified, and the difference of the splicing events between
tumor and non-tumor tissues was not determined.
To investigate the causes of AS events in CCA, the
relationship between AS events and splicing factors was analyzed.
Splicing factors are a class of RNA-binding proteins that affect
the selection of cleavage sites by recognizing the cis-acting
elements of mRNA precursors (27).
Previous studies on lung (56),
ovarian (57) and bladder (94) cancer have demonstrated that the
expression of splicing factors was correlated with
prognosis-related AS events; however, this phenomenon was not
observed in the current study. Pearson's correlation analysis
between a group of splicing factors, the expression of which was
directly related to AS events, and general OS-related splicing
events identified 15 AS events that were mutually associated.
Therefore, in CCA, splicing factors may affect the structure of
their own proteins by creating their own AS event and then
subsequently affecting the AS events of other genes.
In summary, TCGASpliceSeq data were mined to
identify the characteristics of AS events associated with CCA to
establish a model that can predict CCA prognosis using PIs from AS
events. The prognostic effect was estimated using ROC curves and
was favorable, with AUCs >0.9. However, due to the small sample
size, the validity of this model should be verified with additional
clinical samples and in different populations. Several additional
limitations of the present study also need to be addressed. The
data in the current study were obtained from online databases and
thus remain at the bioinformatics level. Additionally, among the AS
events selected to construct the PI model, one event was not
CCA-specific, which reduced the specificity of the PI model. In
addition, although splicing factors have been demonstrated to
contribute to changes in the splicing patterns of target genes and
tumorigenesis, and certain cancer-specific splicing factors are
associated with OS rates, there has been a lack of research on
CCA-related splicing factors, and whether the splicing factors
identified in the present study are truly CCA-specific requires
additional research. Finally, although an accurate PI model was
constructed by screening the significant prognostic AS events
identified by univariate and multivariate Cox regression, the
results are based on a single cohort and should be validated in
additional cohorts; this is intended to be performed in our
subsequent study.
Acknowledgements
Not applicable.
Funding
The study was supported by the Promoting Project of
Basic Capacity for Young and Middle-aged University Teachers in
Guangxi (grant no. 2017KY0111), the Innovation Project of Guangxi
Graduate Education (grant no. YCBZ2017045), the National Natural
Science Foundation of China (grant nos. 31760319, 81360066 and
81660241) and the Guangxi First-class Discipline Project for
Pharmaceutical Sciences (grant no. GXFCDP-PS-2018).
Availability of data and materials
The datasets generated and analyzed during the
current study are available from the TCGA data portal (portal.gdc.cancer.gov/).
Authors' contributions
HYW analyzed and interpreted data, and drafted the
manuscript. YW, LML, ZBC, QPH and SLP obtained information from the
database, supervised the data mining, and revised the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the First Affiliated Hospital of Guangxi Medical University (ethics
approval no. 20170303-2). All the patients provided written
informed consent to participate in this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CCA
|
cholangiocarcinoma
|
|
TCGA
|
The Cancer Genome Atlas
|
|
K-M
|
Kaplan-Meier
|
|
AUC
|
area under the curve
|
|
ROC
|
receiver operator characteristic
|
|
PSI
|
percent spliced index
|
|
ES
|
exon skip
|
|
ME
|
mutually exclusive exons
|
|
RI
|
retained intron
|
|
AP
|
alternate promoter
|
|
AT
|
alternate terminator
|
|
AD
|
alternate donor site
|
|
AA
|
alternate acceptor site
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
References
|
1
|
Popat K, McQueen K and Feeley TW: The
global burden of cancer. Best Pract Res Clin Anaesthesiol.
27:399–408. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Verathamjamras C, Weeraphan C,
Chokchaichamnankit D, Watcharatanyatip K, Subhasitanont P,
Diskul-Na-Ayudthaya P, Mingkwan K, Luevisadpaibul V,
Chutipongtanate S, Champattanachai V, et al: Secretomic profiling
of cells from hollow fiber bioreactor reveals PSMA3 as a potential
cholangiocarcinoma biomarker. Int J Oncol. 51:269–280. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lu X, Zhou C, Li R, Deng Y, Zhao L and
Zhai W: Long noncoding RNA AFAP1-AS1 promoted tumor growth and
invasion in cholangiocarcinoma. Cell Physiol Biochem. 42:222–230.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cai X, Li J, Yuan X, Xiao J, Dooley S, Wan
X, Weng H and Lu L: CD133 expression in cancer cells predicts poor
prognosis of non-mucin producing intrahepatic cholangiocarcinoma. J
Transl Med. 16:502018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Luo G, Li B, Duan C, Cheng Y, Xiao B, Yao
F, Wei M, Tao Q, Feng C, Xia X, et al: cMyc promotes
cholangiocarcinoma cells to overcome contact inhibition via the
mTOR pathway. Oncol Rep. 38:2498–2506. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Qian Y, Yao W, Yang T, Yang Y, Liu Y, Shen
Q, Zhang J, Qi W and Wang J: aPKC-iota/P-Sp1/Snail signaling
induces epithelial-mesenchymal transition and immunosuppression in
cholangiocarcinoma. Hepatology. 66:1165–1182. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Razumilava N and Gores GJ:
Cholangiocarcinoma. Lancet. 383:2168–2179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou G, Yang Z, Wang X, Tao R and Zhou Y:
TRAIL enhances shikonin induced apoptosis through ROS/JNK signaling
in cholangiocarcinoma cells. Cell Physiol Biochem. 42:1073–1086.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tang J, Liao Y, He S, Shi J, Peng L, Xu X,
Xie F, Diao N, Huang J, Xie Q, et al: Autocrine parathyroid
hormone-like hormone promotes intrahepatic cholangiocarcinoma cell
proliferation via increased ERK/JNK-ATF2-cyclin D1 signaling. J
Transl Med. 15:2382017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chng KR, Chan SH, Ng AHQ, Li C, Jusakul A,
Bertrand D, Wilm A, Choo SP, Tan DMY, Lim KH, et al: Tissue
microbiome profiling identifies an enrichment of specific enteric
bacteria in opisthorchis viverrini associated cholangiocarcinoma.
Ebiomedicine. 8:195–202. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Breitbart RE, Andreadis A and Nadalginard
B: Alternative splicing: A ubiquitous mechanism for the generation
of multiple protein isoforms from single genes. Annu Rev Biochem.
56:467–495. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Blencowe BJ: Alternative splicing: New
insights from global analyses. Cell. 126:37–47. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Oltean S and Bates DO: Hallmarks of
alternative splicing in cancer. Oncogene. 33:5311–5318. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang ET, Rickard S, Luo S, Khrebtukova I,
Zhang L, Mayr C, Kingsmore SF, Schroth GP and Burge CB: Alternative
isoform regulation in human tissue transcriptomes. Nature.
456:470–476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pan Q, Shai O, Lee LJ, Frey BJ and
Blencowe BJ: Deep surveying of alternative splicing complexity in
the human transcriptome by high-throughput sequencing. Nat Genet.
40:1413–1415. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dvinge H and Bradley RK: Widespread intron
retention diversifies most cancer transcriptomes. Genome Medi.
7:452015. View Article : Google Scholar
|
|
17
|
Bechara EG, Sebestyén E, Bernardis I,
Eyras E and Valcárcel J: RBM5, 6 and 10 differentially regulate
NUMB alternative splicing to control cancer cell proliferation. Mol
Cell. 52:720–733. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ghigna C, Giordano S, Shen H, Benvenuto F,
Castiglioni F, Comoglio PM, Green MR, Riva S and Biamonti G: Cell
motility is controlled by SF2/ASF through alternative splicing of
the Ron Protooncogene. Mol Cell. 20:881–890. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jung H, Lee D, Lee J, Park D, Kim YJ, Park
WY, Hong D, Park PJ and Lee E: Intron retention is a widespread
mechanism of tumor-suppressor inactivation. Nat Genet.
47:1242–1248. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Goldstein L, Lee J, Gnad F, Klijn C,
Schaub A, Reeder J, Daemen A, Bakalarski CE, Holcomb T, Shames DS,
et al: Recurrent loss of NFE2L2 exon 2 is a mechanism for Nrf2
pathway activation in human cancers. Cell Rep. 16:2605–2617. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kornblihtt AR: Epigenetics at the base of
alternative splicing changes that promote colorectal cancer. J Clin
Invest. 127:3281–3283. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Poulikakos PI, Persaud Y, Janakiraman M,
Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, et al:
RAF inhibitor resistance is mediated by dimerization of aberrantly
spliced BRAF (V600E). Nature. 480:387–390. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sotillo E, Barrett DM, Black KL, Bagashev
A, Oldridge D, Wu G, Sussman R, Lanauze C, Ruella M, Gazzara MR, et
al: Convergence of acquired mutations and alternative splicing of
CD19 enables resistance to CART-19 immunotherapy. Cancer Discov.
5:1282–1295. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang BD, Ceniccola K, Hwang S, Andrawis R,
Horvath A, Freedman JA, Olender J, Knapp S, Ching T, Garmire L, et
al: Alternative splicing promotes tumour aggressiveness and drug
resistance in African American prostate cancer. Nat Commun.
8:159212017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sebestyén E, Zawisza M and Eyras E:
Detection of recurrent alternative splicing switches in tumor
samples reveals novel signatures of cancer. Nucleic Acids Res.
43:1345–1356. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Trincado JL, Sebestyén E, Pagés A and
Eyras E: The prognostic potential of alternative transcript
isoforms across human tumors. Genome Med. 8:852016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shen S, Wang Y, Wang C, Ying NW and Yi X:
SURVIV for survival analysis of mRNA isoform variation. Nat Commun.
7:115482016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Parsons DW, Li M, Zhang X, Jones S, Leary
RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, et al: The
genetic landscape of the childhood cancer medulloblastoma. Science.
331:435–439. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ghigna C, De Toledo M, Bonomi S, Valacca
C, Gallo S, Apicella M, Eperon I, Tazi J and Biamonti G:
Pro-metastatic splicing of Ron proto-oncogene mRNA can be reversed:
Therapeutic potential of bifunctional oligonucleotides and indole
derivatives. RNA Biol. 7:495–503. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee CW and Abdelwahab O: Therapeutic
targeting of splicing in cancer. Nat Med. 22:976–986. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Koh CM, Bezzi M, Low DH, Ang WX, Teo SX,
Gay FP, Al-Haddawi M, Tan SY, Osato M, Sabò A, et al: MYC regulates
the core pre-mRNA splicing machinery as an essential step in
lymphomagenesis. Nature. 523:96–100. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Havens MA and Hastings ML:
Splice-switching antisense oligonucleotides as therapeutic drugs.
Nucleic Acids Res. 44:6549–6563. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Salton M and Misteli T: Small molecule
modulators of Pre-mRNA splicing in cancer therapy. Trends Mol Med.
22:28–37. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gao L, Xie ZC, Pang JS, Li TT and Chen G:
A novel alternative splicing-based prediction model for uteri
corpus endometrial carcinoma. Aging (Albany NY). 11:263–283. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xiong Y, Deng Y, Wang K, Zhou H, Zheng X,
Si L and Fu Z: Profiles of alternative splicing in colorectal
cancer and their clinical significance: A study based on
large-scale sequencing data. EBioMedicine. 36:183–195. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang ZG, He RQ and Mo ZN: Prognostic
value and potential function of splicing events in prostate
adenocarcinoma. Int J Oncol. 53:2473–2487. 2018.PubMed/NCBI
|
|
37
|
Jusakul A, Cutcutache I, Yong CH, Lim JQ,
Huang MN, Padmanabhan N, Nellore V, Kongpetch S, Ng AWT, Ng LM, et
al: Whole-genome and epigenomic landscapes of etiologically
distinct subtypes of cholangiocarcinoma. Cancer Discov.
7:1116–1135. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Supek F, Miñana B, Valcárcel J, Gabaldón T
and Lehner B: Synonymous mutations frequently act as driver
mutations in human cancers. Cell. 156:1324–1335. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sterneweiler T and Sanford JR: Exon
identity crisis: Disease-causing mutations that disrupt the
splicing code. Genome Biol. 15:2012014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Diederichs S, Bartsch L, Berkmann JC,
Fröse K, Heitmann J, Hoppe C, Iggena D, Jazmati D, Karschnia P,
Linsenmeier M, et al: The dark matter of the cancer genome:
Aberrations in regulatory elements, untranslated regions, splice
sites, non-coding RNA and synonymous mutations. EMBO Mol Med.
8:442–457. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Singh S, Narayanan SP, Biswas K, Gupta A,
Ahuja N, Yadav S, Panday RK, Samaiya A, Sharan SK and Shukla S:
Intragenic DNA methylation and BORIS-mediated cancer-specific
splicing contribute to the Warburg effect. Proc Natl Acad Sci USA.
114:11440–11445. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gelfman S, Cohen N, Yearim A and Ast G:
DNA-methylation effect on cotranscriptional splicing is dependent
on GC architecture of the exon-intron structure. Genome Res.
23:789–799. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shukla S, Kavak E, Gregory M, Imashimizu
M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R and
Oberdoerffer S: CTCF-promoted RNA polymerase II pausing links DNA
methylation to splicing. Nature. 479:74–79. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yuan H, Li N, Fu D, Ren J, Hui J, Peng J,
Liu Y, Qiu T, Jiang M, Pan Q, et al: Histone methyltransferase
SETD2 modulates alternative splicing to inhibit intestinal
tumorigenesis. J Clin Invest. 127:3375–3391. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ding X, Liu S, Tian M, Zhang W, Zhu T, Li
D, Wu J, Deng H, Jia Y, Xie W, et al: Activity-induced histone
modifications govern Neurexin-1 mRNA splicing and memory
preservation. Nat Neurosci. 20:690–699. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sharma A, Nguyen H, Geng C, Hinman MN, Luo
G and Lou H: Calcium-mediated histone modifications regulate
alternative splicing in cardiomyocytes. Proc Natl Acad Sci USA.
111:E4920–E4928. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kim S, Kim H, Fong N, Erickson B and
Bentley DL: Pre-mRNA splicing is a determinant of histone H3K36
methylation. Proc Natl Acad Sci USA. 108:13564–13569. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Takehara T, Liu X, Fujimoto J, Friedman SL
and Takahashi H: Expression and role of Bcl-xL in human
hepatocellular carcinomas. Hepatology. 34:55–61. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Boise LH, Gonzálezgarcía M, Postema CE,
Ding L, Lindsten T, Turka LA, Mao X, Nuñez G and Thompson CB:
bcl-x, a bcl-2-related gene that functions as a dominant regulator
of apoptotic cell death. Cell. 74:597–608. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Bingle CD, Craig RW, Swales BM, Singleton
V, Zhou P and Whyte MK: Exon skipping in Mcl-1 results in a Bcl-2
homology domain 3 only gene product that promotes cell death. J
Biol Chem. 275:22136–22146. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bae J, Leo CP, Hsu SY and Hsueh AJ:
MCL-1S, a splicing variant of the antiapoptotic BCL-2 family member
MCL-1, encodes a proapoptotic protein possessing only the BH3
domain. J Biol Chem. 275:25255–25261. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Salton M, Kasprzak WK, Voss T, Shapiro BA,
Poulikakos PI and Misteli T: Inhibition of vemurafenib-resistant
melanoma by interference with pre-mRNA splicing. Nat Commun.
6:71032015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen K, Xiao H, Zeng J, Yu G, Zhou H,
Huang C, Yao W, Xiao W, Hu J, Guan W, et al: Alternative splicing
of EZH2 pre-mRNA by SF3B3 contributes to the tumorigenic potential
of renal cancer. Clin Cancer Res. 23:3428–3441. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Luo C, Cheng Y, Liu Y, Chen L, Liu L, Wei
N, Xie Z, Wu W and Feng Y: SRSF2 regulates alternative splicing to
drive hepatocellular carcinoma development. Cancer Res.
77:1168–1178. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Calabretta S, Bielli P, Passacantilli I,
Pilozzi E, Fendrich V, Capurso G, Fave GD and Sette C: Modulation
of PKM alternative splicing by PTBP1 promotes gemcitabine
resistance in pancreatic cancer cells. Oncogene. 35:2031–2039.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Li Y, Sun N, Lu Z, Sun S, Huang J, Chen Z
and He J: Prognostic alternative mRNA splicing signature in
non-small cell lung cancer. Cancer Lett. 393:40–51. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhu J, Chen Z and Yong L: Systematic
profiling of alternative splicing signature reveals prognostic
predictor for ovarian cancer. Gynecol Oncol. 148:368–374. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Miles S, Swift L and Leinster SJ: The
dundee ready education environment measure (DREEM): A review of its
adoption and use. Med Teach. 34:e620–e634. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Shi Y, Chen Z, Gao J, Wu S, Gao H and Feng
G: Transcriptome-wide analysis of alternative mRNA splicing
signature in the diagnosis and prognosis of stomach adenocarcinoma.
Oncol Rep. 40:2014–2022. 2018.PubMed/NCBI
|
|
60
|
Lin P, He RQ, Ma FC, Liang L, He Y, Yang
H, Dang YW and Chen G: Systematic analysis of survival-associated
alternative splicing signatures in gastrointestinal
pan-adenocarcinomas. EBioMedicine. 34:46–60. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ryan M, Wong WC, Brown R, Akbani R, Su X,
Broom B, Melott J and Weinstein J: TCGASpliceSeq a compendium of
alternative mRNA splicing in cancer. Nucleic Acids Res.
44:D1018–D1022. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
R Core Team. R, . A language and
environment for statistical computingR Foundation for Statistical
Computing; Vienna, Austria: 2012, ISBN 3-900051-07-0. http://www.R-project.org/
|
|
63
|
Smidt N DJaMT: Guide to the contents of a
Cochrane review and protocol. Cochrane handbook for systematic
reviews of diagnostic test accuracy. 2011.
|
|
64
|
Conway JR, Lex A and Gehlenborg N: UpSetR:
An R package for the visualization of intersecting sets and their
properties. Bioinformatics. 33:2938–2940. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wu G, Dawson E, Duong A, Haw R and Stein
L: ReactomeFIViz: A Cytoscape app for pathway and network-based
data analysis. F1000Res. 3:1462014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wickham H: ggplot2: Elegant Graphics for
Data AnalysisSpringer-Verlag; New York, NY: pp. 77–186. 2016
|
|
68
|
Heagerty PJ and Zheng Y: Survival model
predictive accuracy and ROC curves. Biometrics. 61:92–105. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Lee Y and Rio DC: Mechanisms and
regulation of alternative Pre-mRNA splicing. Annu Rev Biochem.
84:291–323. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Piva F, Giulietti M, Burini AB and
Principato G: SpliceAid 2: A database of human splicing factors
expression data and RNA target motifs. Hum Mutat. 33:81–85. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Rahnemai-Azar AA, Weisbrod A, Dillhoff M,
Schmidt C and Pawlik TM: Intrahepatic cholangiocarcinoma: Molecular
markers for diagnosis and prognosis. Surg Oncol. 26:125–137. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Khan SA, Davidson BR, Goldin RD, Heaton N,
Karani J, Pereira SP, Rosenberg WM, Tait P, Taylor-Robinson SD,
Thillainayagam AV, et al: Guidelines for the diagnosis and
treatment of cholangiocarcinoma: An update. Gut. 61:1657–1669.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Pasquali C, Sperti C, D'Andrea AA,
Costantino V, Filipponi C and Pedrazzoli S: CA50 as a serum marker
for pancreatic carcinoma: Comparison with CA19-9. Eur J Cancer 30A.
1042–1043. 1994. View Article : Google Scholar
|
|
74
|
Shan M, Tian Q and Zhang L: Serum CA50
levels in patients with cancers and other diseases. Prog Mol Biol
Transl Sci. 162:187–198. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Dou H, Sun G and Zhang L: CA242 as a
biomarker for pancreatic cancer and other diseases. Prog Mol Biol
Transl Sci. 162:229–239. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Huang CK, Aihara A, Iwagami Y, Yu T,
Carlson R, Koga H, Kim M, Zou J, Casulli S and Wands JR: Expression
of transforming growth factor β1 promotes cholangiocarcinoma
development and progression. Cancer Lett. 380:153–162. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Churi CR, Shroff R, Wang Y, Rashid A, Kang
HC, Weatherly J, Zuo M, Zinner R, Hong D, Meric-Bernstam F, et al:
Mutation profiling in cholangiocarcinoma: Prognostic and
therapeutic implications. PLoS One. 9:e1153832014. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Dana P, Kariya R, Vaeteewoottacharn K,
Sawanyawisuth K, Seubwai W, Matsuda K, Okada S and Wongkham S:
Upregulation of CD147 promotes metastasis of cholangiocarcinoma by
modulating the Epithelial-to-Mesenchymal transitional process.
Oncol Res. 25:1047–1059. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Schweitzer N and Vogel A: Systemic therapy
of cholangiocarcinoma: From chemotherapy to targeted therapies.
Best Pract Res Clin Gastroenterol. 29:345–353. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Kamlua S, Patrakitkomjorn S, Jearanaikoon
P, Menheniott TR, Giraud AS and Limpaiboon T: A novel TFF2 splice
variant (ΔEX2TFF2) correlates with longer overall survival time in
cholangiocarcinoma. Oncol Rep. 27:1207–1212. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Nutthasirikul N, Limpaiboon T, Leelayuwat
C, Patrakitkomjorn S and Jearanaikoon P: Ratio disruption of the
∆133p53 and TAp53 isoform equilibrium correlates with poor clinical
outcome in intrahepatic cholangiocarcinoma. Int J Oncol.
42:1181–1188. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Comino-Mendez I, Leandro-Garcia LJ,
Montoya G, Inglada-Pérez L, de Cubas AA, Currás-Freixes M, Tysoe C,
Izatt L, Letón R, Gómez-Graña Á, et al: Functional and in silico
assessment of MAX variants of unknown significance. J Mol Med
(Berl). 93:1247–1255. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Huang X, Wu Z, Mei Y and Wu M: XIAP
inhibits autophagy via XIAP-Mdm2-p53 signalling. EMBO J.
32:2204–2216. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Li Z, Tuteja G, Schug J and Kaestner KH:
Foxa1 and Foxa2 are essential for sexual dimorphism in liver
cancer. Cell. 148:72–83. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
McNeely S, Beckmann R and Bence Lin AK:
CHEK again: Revisiting the development of CHK1 inhibitors for
cancer therapy. Pharmacol Ther. 142:1–10. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Mo J, Zhang D and Yang R: MicroRNA-195
regulates proliferation, migration, angiogenesis and autophagy of
endothelial progenitor cells by targeting GABARAPL1. Biosci Rep.
36(pii): e003962016. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Wagner EF and Nebreda AR: Signal
integration by JNK and p38 MAPK pathways in cancer development. Nat
Rev Cancer. 9:537–549. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Eswaran J, Horvath A, Godbole S, Reddy SD,
Mudvari P, Ohshiro K, Cyanam D, Nair S, Fuqua SA, Polyak K, et al:
RNA sequencing of cancer reveals novel splicing alterations. Sci
Rep. 3:16892013. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Suo C, Hrydziuszko O, Lee D, Pramana S,
Saputra D, Joshi H, Calza S and Pawitan Y: Integration of somatic
mutation, expression and functional data reveals potential driver
genes predictive of breast cancer survival. Bioinformatics.
31:2607–2613. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Yosudjai J, Inpad C, Chomwong S, Dana P,
Sawanyawisuth K, Phimsen S, Wongkham S, Jirawatnotai S and Kaewkong
W: An aberrantly spliced isoform of anterior gradient-2, AGR2vH
promotes migration and invasion of cholangiocarcinoma cell. Biomed
Pharmacother. 107:109–116. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Mcneely S, Beckmann R and Bence Lin AK:
CHEK again: Revisiting the development of CHK1 inhibitors for
cancer therapy. Pharmacol Ther. 142:1–10. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Zhang H, Yang T, Wu M and Shen F:
Intrahepatic cholangiocarcinoma: Epidemiology, risk factors,
diagnosis and surgical management. Cancer Lett. 379:198–205. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Goere D, Wagholikar GD, Pessaux P, Carrère
N, Sibert A, Vilgrain V, Sauvanet A and Belghiti J: Utility of
staging laparoscopy in subsets of biliary cancers: Laparoscopy is a
powerful diagnostic tool in patients with intrahepatic and
gallbladder carcinoma. Surg Endosc. 20:721–725. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
He RQ, Zhou XG, Yi QY, Deng CW, Gao JM,
Chen G and Wang QY: Prognostic signature of alternative splicing
events in bladder urothelial carcinoma based on spliceseq data from
317 cases. Cell Physiol Biochem. 48:1355–1368. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Lin P, He RQ, Huang ZG, Zhang R, Wu HY,
Shi L, Li XJ, Li Q, Chen G, Yang H and He Y: Role of global
aberrant alternative splicing events in papillary thyroid cancer
prognosis. Aging (Albany NY). 11:2082–2097. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Zhang D, Duan Y, Cun J and Yang Q:
Identification of prognostic alternative splicing signature in
breast carcinoma. Front Genet. 10:2782019. View Article : Google Scholar : PubMed/NCBI
|