Introduction
Hepatocellular carcinoma (HCC) accounts for ~95% of
all primary liver cancers. HCC is one of the solid tumors with the
poorest prognosis, with the 6th highest incidence and 3rd highest
mortality rates worldwide (1).
Although various new treatments have been applied in the clinic,
the incidence and mortality rates of HCC have not adequately
improved (2). Patients with HCC have
a higher recurrence rate even after curative therapy, and a
five-year survival rate of only 11% (3). The poor prognosis of patients with HCC
highlights the need to explore the mechanisms of disease
progression. Understanding the molecular pathogenesis of HCC,
particularly the mechanisms of tumorigenesis, is important for
developing novel biomarkers and treatment strategies.
MicroRNAs (miRNAs) are endogenous, small
single-stranded, non-coding RNAs of ~20–25 nucleotides in length.
miRNAs regulate translation by binding to specific sequences in the
3′-untranslated region (3′-UTR) of target mRNAs (4,5).
Accumulating evidence suggests that miRNA deregulation contributes
to a wide range of human diseases, including cancer (6). In terms of the onset and prognosis of
human cancers, miRNAs are able to regulate oncogenes or tumor
suppressor genes during tumorigenesis, in a target-dependent manner
(7). It is important to clarify the
function of miRNAs in tumor pathogenesis and progression, as miRNAs
may regulate a variety of critical biological processes, including
cell differentiation, proliferation, cell cycle distribution,
apoptosis, migration, invasion and tumor cell drug resistance
(8–10).
Studies have indicated a preference for the
detection of miRNA (miR)-107 over a-fetoprotein (AFP) for the early
diagnosis of HCC (11), and the
combination of serum AFP and ultrasound surveillance is the most
widely used strategy for screening and detecting HCC in high-risk
groups (12). miR-107 is located on
chromosome 10 and has been found to be abnormally expressed in
various tumors (13–15). Evidence has supported the hypothesis
that reduced expression levels of miR-107 are associated with the
growth, migration and invasion of various cancer types, including
HCC (16,17). Furthermore, previous research has
revealed that miR-107 enhances the viability, migration and
invasion of U2OS cells, which may be associated with the activation
of the MEK/extracellular signal-regulated kinase and nuclear
factor-κB signaling pathways via targeting of the tumor suppressor
gene tropomyosin 1 (18). It was
also reported that miR-107 may inhibit glioma cell proliferation by
targeting sal-like protein 4 (19),
and that miR-107 overexpression inhibited colon cancer cell
proliferation by targeting hypoxia inducible factor-β (20). These reports demonstrate the complex
nature of miR-107 activity. Therefore, it was speculated that
depleted plasma expression levels of miR-107 may be associated with
tumor progression and poor outcome in patients with HCC.
Regulator of G-protein signaling (RGS) proteins are
expressed in the majority of cell types, tissues and organ systems.
RGS proteins have been implicated in a variety of physiologies and
pathologies (21), including
hematopoiesis (22), cancer
migration and invasion (23), and
synaptic signaling plasticity in brain-anxiety disorder (24). RGS4 belongs to the B/R4 subfamily
(25) of the RGS protein family,
which is characterized by a conserved 120-amino acid RGS region
flanking the short amino and carboxyl termini (26). RGS4 is an intracellular protein
primarily recognized for its GTPase activating function, which by
stimulating Gα-bound GTP hydrolysis, inactivates the Gα subunits of
the heterotrimeric G protein, and subsequently inhibits G-protein
coupled receptor (GPCR) signaling (27). Data has shown that RGS4 induces
G2/M arrest in the breast cancer cell cycle, and
therefore, the signal transduction pathway initiated by RGS4 may be
associated with breast cancer cell proliferation (28). RGS4 has been extensively studied in
the central nervous and circulatory systems (29), and its involvement in cancer is also
increasingly being investigated. Given that RGS proteins serve
important roles in tumorigenesis, it was hypothesized that
optimizing the function or overexpression of RGS proteins in tumor
tissues may be an effective strategy for tumor therapy.
Additionally, as a tumor suppressor, RGS4 inhibited
tumor invasiveness and metastasis by regulating matrix
metalloproteinases (MMPs) and epithelial mesenchymal transition
(EMT)-associated markers (30).
Previous studies have also demonstrated that MMP-2 and MMP-9 are
involved in the development and metastasis of cutaneous melanoma
(29,31,32), and
have shown that epidermal growth factor receptor (EGFR) serves a
key role in the occurrence and development of primary liver cancer
(33). CXC chemokine receptor type 4
(CXCR4) is one of the most widely expressed chemokine receptors in
tissues, serving an important role in cell growth and invasion, and
the metastasis of various malignant tumors. There is evidence to
indicate the activation of CXCR4/stromal cell-derived factor 1
promoted prostate cancer cell invasion through MMP-9, and that the
overexpression of RGS4 can block the increased invasive and
metastatic influences of EGFR and CXCR4 (30). It has also been shown that RGS4
overexpression can completely reverse CXCR4-mediated, and partially
reverse EGFR-mediated invasion and metastasis. This suggests that
CXCR4, as a GPCR, may be directly negatively regulated by RGS4.
In the present study, it was discovered that the
RGS4 3′-UTR contained a potential binding site for miR-107 in the
putative target sequence, and a previous study has confirmed that
the expression level of miR-107 was lower in 30 HCC samples
relative to their corresponding adjacent liver tissues (17). However, the role of miR-107 in the
progression of HCC, and the direct targets of miR-107 in the
regulation of HCC remain undefined. Therefore, the present study
investigated the potential role of miR-107 in RGS4 expression and
the tumor characteristics of HCC.
Materials and methods
Cell lines and culture
The HCC cell line SK-HEP-1 was purchased from the
American Type Culture Collection (ATCC), and maintained in 25
cm2 flasks containing Dulbecco's modified eagle's medium
(DMEM; ATCC) supplemented with 10% fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc.) at 37°C in a humidified incubator
(5% CO2 and 95% humidity).
Lentivirus transduction of
miR-107
miR-107 mimics and control mimics were transduced
into SK-HEP-1 cells. Lentiviruses expressing miR-107 mimics
(5′-AGCAGCAUUGUACAGGGCUAUCA-3′) and control mimics
(5′-UUCUCCGAACGUGUCACGUTT-3′) were purchased from Shanghai Sun
Biotechnology Co., Ltd. In total, 100 pmol lentivirus and 8 µg/ml
polybrene were added to the culture medium at a multiplicity of
infection of 10–15. After incubation overnight, the medium was
replaced, and the cells were maintained for another 5–7 days to
stabilize the lentiviral transduction. Subsequently, the
transduction efficiency was verified by reverse
transcription-quantitative (RT-q) PCR. The groups were designated a
as the negative control (NC), miR-107 mimics and control mimics
groups.
RT-qPCR analysis
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to isolate the total RNA from the
miR-107-SK-HEP-1 cells, and cDNA was synthesized using Moloney
murine leukemia virus reverse transcriptase (New England BioLabs,
Inc.) with an oligo(dT)18 primer. The RT reaction was
performed by incubating a reaction mixture containing 0.5 µg RNA,
100 pmol random hexamer primer (Applied Biosystems; Thermo Fisher
Scientific, Inc.), 50 units reverse transcriptase (Applied
Biosystems; Themro Fisher Scientific, Inc.), 20 units RNase
inhibitor (Promega Corporation), and 1 mM dNTP (Thermo Fisher
Scientific, Inc.) in a total 20 µl volume. The RT conditions were
as follows: 25°C for 15 min, 45°C for 30 min and 94°C for 5 min.
Then, qPCR was performed using a Light Cycler system with
LightCycler FastStart DNA Master PLUS SYBR®-Green I
(Roche Diagnostics) under the following conditions: Intial
denaturation at 94°C for 10 min, followed by 40 cycles at 94°C for
10 sec and 60°C for 1 min, and 72°C 30 sec for elongation. The qPCR
primers used were as follows: miR-107 forward,
5′-ATACCGCTCGAGTGCCATGTGTCCACTGAAT-3′ and reverse,
5′-ATACCGCTCGAGTTCCATGCCTCAACTCCT-3′; U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. U6
was used as a reference gene for miR-107 quantification, and the
relative quantitative 2−ΔΔCq method (34) was used for data analysis. The
experiments were repeated three times.
MTT assay
HCC cell proliferation was assessed using an MTT
assay. SK-HEP-1 cells (1×104 cells/well) in the
exponential growth phase were seeded into 96-well plates. Then, 1
mg/ml MTT solution was added to each well and further incubated for
4 h at 37°C. To dissolve the formazan crystals, 100 µl dimethyl
sulfoxide was added to each well and the absorbance at 570 nm was
measured using a microplate reader.
Dual-luciferase reporter assay
SK-HEP-1 cells (1×104 cells/well) were
seeded into 96-well plates, and a pGL3 firefly luciferase reporter
gene vector (Promega Corporation) with the 3′-UTR-wild-type (WT) or
3′-UTR-mutant (MUT) fragment of human RGS4 cDNA, containing a
putative target site for miR-107, was co-transfected into SK-HEP-1
cells with the miR-107 mimics and negative controls at a 100 nM
final concentration using lentiviral transfection. Lipofectamine™
3000 transfection reagent (Thermo Fisher Scientific, Inc.) was used
for all transfections following manufacturer's instructions. At 48
h post-transfection, the luciferase activity of the cell extracts,
which was normalized with Renilla luciferase, was measured
using the Dual-Luciferase Reporter Assay System (Promega
Corporation) and a fluorescence microscope, following the
manufacturer's protocol. Experiments were independently repeated ≥3
times.
Colony formation assays
At 24 h post-transfection, SK-HEP-1 cells were
resuspended in serum-free DMEM containing 1% N2, 2% B27, 20 ng/ml
human fibroblast growth factor-2 and 20 ng/ml EGF (Invitrogen;
Thermo Fisher Scientific, Inc.). Subsequently, cells were seeded in
6-well ultra-low attachment plates (300 cells/well). Following 9
days of incubation in 5% CO2 at 37°C with fresh medium
replaced every 3 days, the supplement was discarded and the cells
were then washed with phosphate-buffered saline (PBS). The cells
were then fixed with 4% paraformaldehyde for 15 min at room
temperature and stained with Giemsa stain (Beijing Solarbio Science
& Technology, Co., Ltd.) for 20 min. Colonies were counted
under a light inverted microscope (TS100; Nikon Corporation) and
each assay was performed in triplicate.
Wound-healing migration assay
A wound-healing assay was employed to assess the
migrational capacity of SK-HEP-1 cells following transfection. The
SK-HEP-1 cells were plated into 24-well plates (2 ml,
2.5×104 cells/well) and cultured in serum-free medium
for 24 h to obtain a monolayer. When the cell confluence had
reached 80%, a sterile pipette tip held perpendicular to the bottom
of the well, was used to scratch the cell surface to create a
wound. Following removal of the debris with the tip of the pipette,
the culture was replenished with fresh medium and cells were
incubated at 5% CO2 and 37°C for 24 h. Images of the
migrated cells were captured at 24 h under a light inverted
microscope (TS100; Nikon Corporation) and analyzed using ImageJ
(v1.8.0; National Institutes of Health).
Transwell invasion assay
A Transwell assay was performed to identify the
invasion ability of SK-HEP-1 cells after transfection with miR-107
mimics. Transwell culture inserts pre-coated with Matrigel (8-mm
pore size; BD Biosciences) were placed into upper chambers at 37°C
for 30 min. A total of 150 µl cell suspension (2.5×104
cells) suspended into serum-free medium was added to the upper
chamber and 500 µl RPMI-1640 medium containing 10% FBS was placed
into lower chambers of 24-wells culture plates. Then, the
non-invaded cells on the upper surface of the membrane were removed
with a cotton swab after incubation at 37°C for 24 h. After
fixation with methanol, cells on the lower surface of membrane were
stained with 0.005% crystal violet at room temperature in PBS for 1
h, and the number of migrated or invaded cells in 10 random fields
was counted under a light inverted microscope (TS100; Nikon
Corporation; magnification, ×200).
Flow cytometry analysis
After 48 h of transfection with miR-107 mimics,
SK-HEP-1 cells were centrifuged at 200 × g for 10 min and fixed
with 70% ice-cold ethanol for 24 h at 4°C. Next, the cells were
harvested and washed twice with cold PBS and then stained with
Annexin V and 7-aminoactinomycin D (7-AAD; BD Biosciences).
Following the addition of 5 µl Annexin V and 5 µl 7-AAD with RNaseA
(Sigma-Aldrich; Merck KGaA), the cells were incubated in the dark
for 30 min at 4°C. The apoptotic cells and the cell cycle
distribution were detected using a FACS Calibur flow cytometer (BD
Biosciences) and Cell Quest Software (v3.1; BioMedica, Diagnostics,
Inc.) following the manufacturer's instructions. All experiments
were performed three times.
Western blotting
RIPA lysis buffer (Gibco; Thermo Fisher Scientific,
Inc.) was used to extract the total protein from the cells, and the
protein concentration was determined using the bicinchoninic acid
method. Proteins (40 µg) were separated by 8–15% SDS-PAGE and
transferred onto PVDF membranes (Thermo Fisher Scientific, Inc.).
GAPDH was used as the internal control. Anti-RGS4 (ab97307;
1:1,000), -EGFR (ab52894; 1:1,000), -CXCR4 (ab1670; 1:500), -MMP-2
(ab37150; 1:500), and -MMP-9 (ab119906; 1:500) were purchased from
Abcam. The PVDF membranes were then blocked with 5% non-fat milk in
20 mM Tris-HCl, 137 mM NaCl and 1% Tween-20 (pH 7.6; Invitrogen;
Thermo Fisher Scientific, Inc.) for 2 h at room temperature. After
that, the membranes were incubated with the primary antibodies
overnight at 4°C, followed by incubation with horseradish
peroxidase (HRP)-conjugated secondary antibodies for 2 h at room
temperature. Subsequently, the bands were developed with Immobile
Western Chemiluminescence HRP Substrates (EMD Millipore). The
images were captured using a Luminescence/Fluorescence Imaging
System (GE Healthcare), and the signal intensities were quantified
using ImageJ analysis software version 1.51j8 (National Institutes
of Health). This experiment was performed in triplicate.
Statistical analysis
All data are presented as the mean ± standard
deviation obtained from at least three independent experiments. The
expression levels of miRNA and mRNA were quantified using the
2−ΔΔCq method. Statistical analysis was performed using
SPSS software version 21.0 (IBM Corp.) and GraphPad Prism version
5.01 (Graph-Pad Software, Inc.). Significant differences between
two groups were determined using the Student's t-test, while
differences among several groups were assessed by one-way analysis
of variance followed by Dunnett's post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
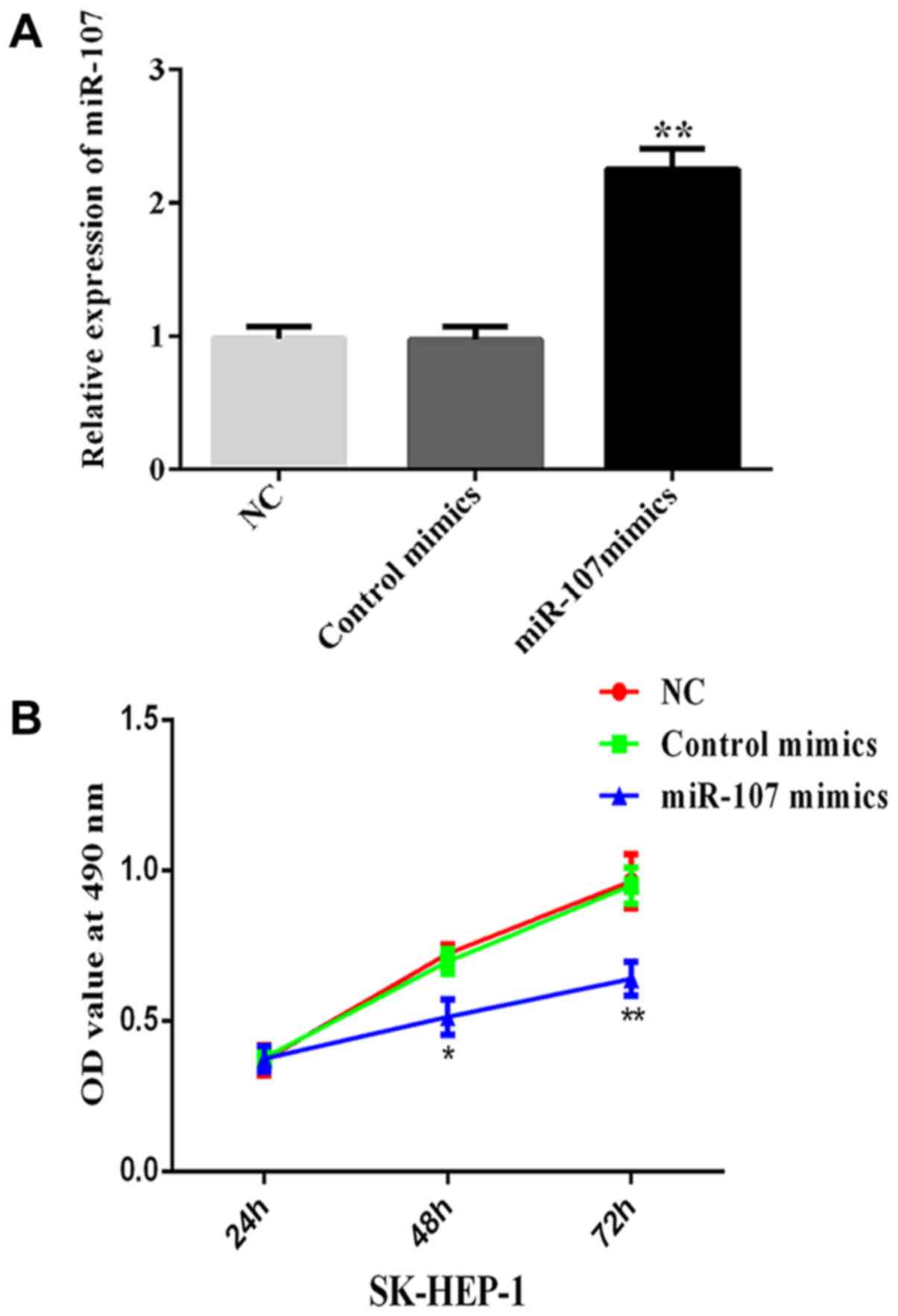
miR-107 expression level is increased
in SK-HEP-1 cells after transfection
After transfecting with miR-107 mimics and control
mimics, the expression level of miR-107 was determined in SK-HEP-1
cells using RT-qPCR. The results indicated a marked rise in miR-107
expression levels in the miR-107 mimics group compared with the NC
and control mimics groups (**P< 0.01; Fig. 1A). The result also confirmed the
successful transfection of the miR-107 mimics plasmid.
Overexpression of miR-107 represses
the growth, migration, invasion and colony formation of HCC
cells
To evaluate the biological functions of miR-107 in
HCC cells, SK-HEP-1 cells were infected with miR-107-expressing
adenoviruses and the effects on cell growth, colony formation,
migration and invasion were analyzed. The results of the MTT assay
showed that the overexpression of miR-107 significantly reduced
cell proliferation, compared with the NC and control mimics groups
(P<0.05, at 48 h; P<0.01, at 72 h; Fig. 1B). In addition, the number of
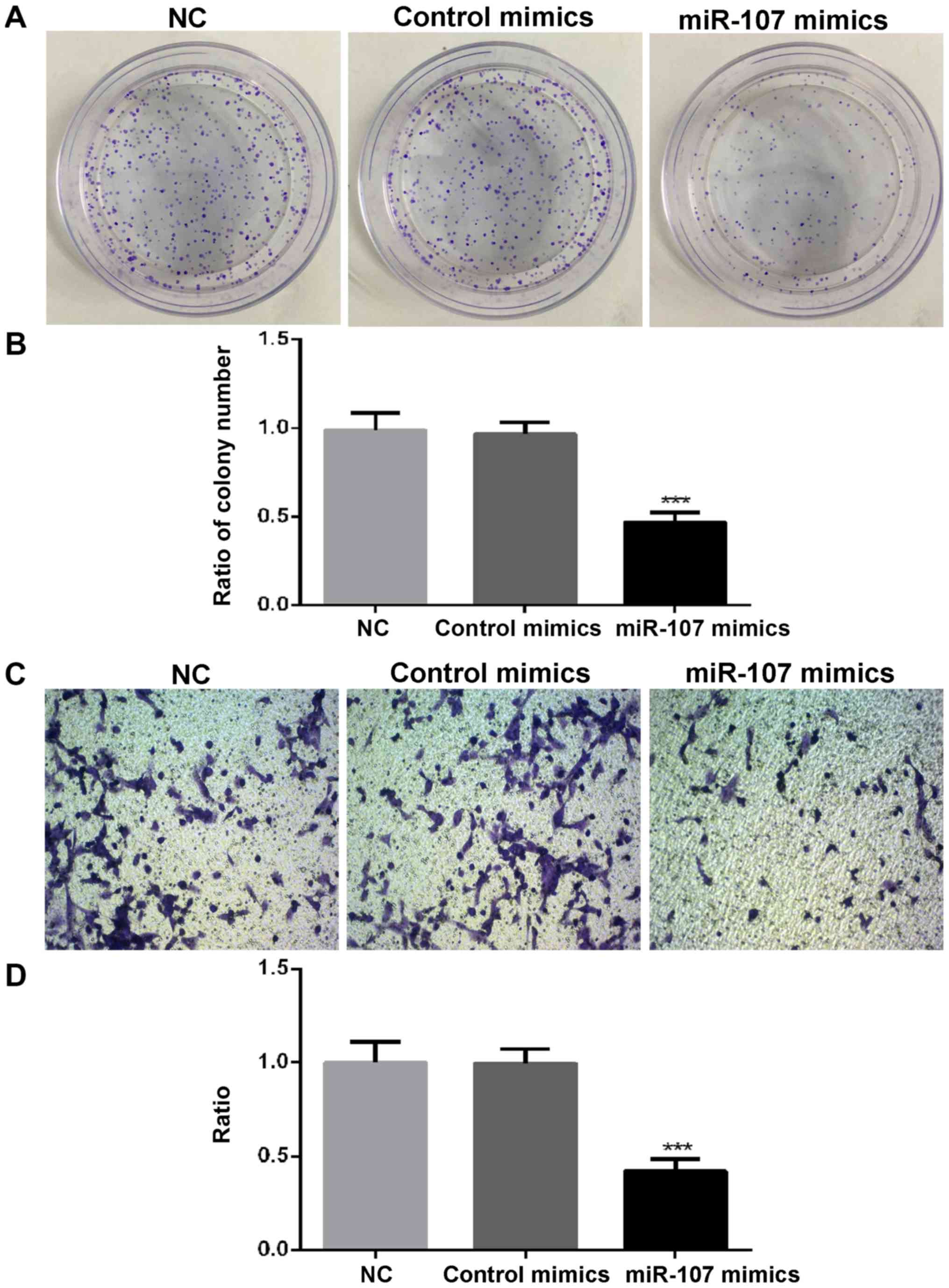
SK-HEP-1 cell colonies formed was significantly reduced by miR-107
overexpression (P<0.001; Fig. 2A and
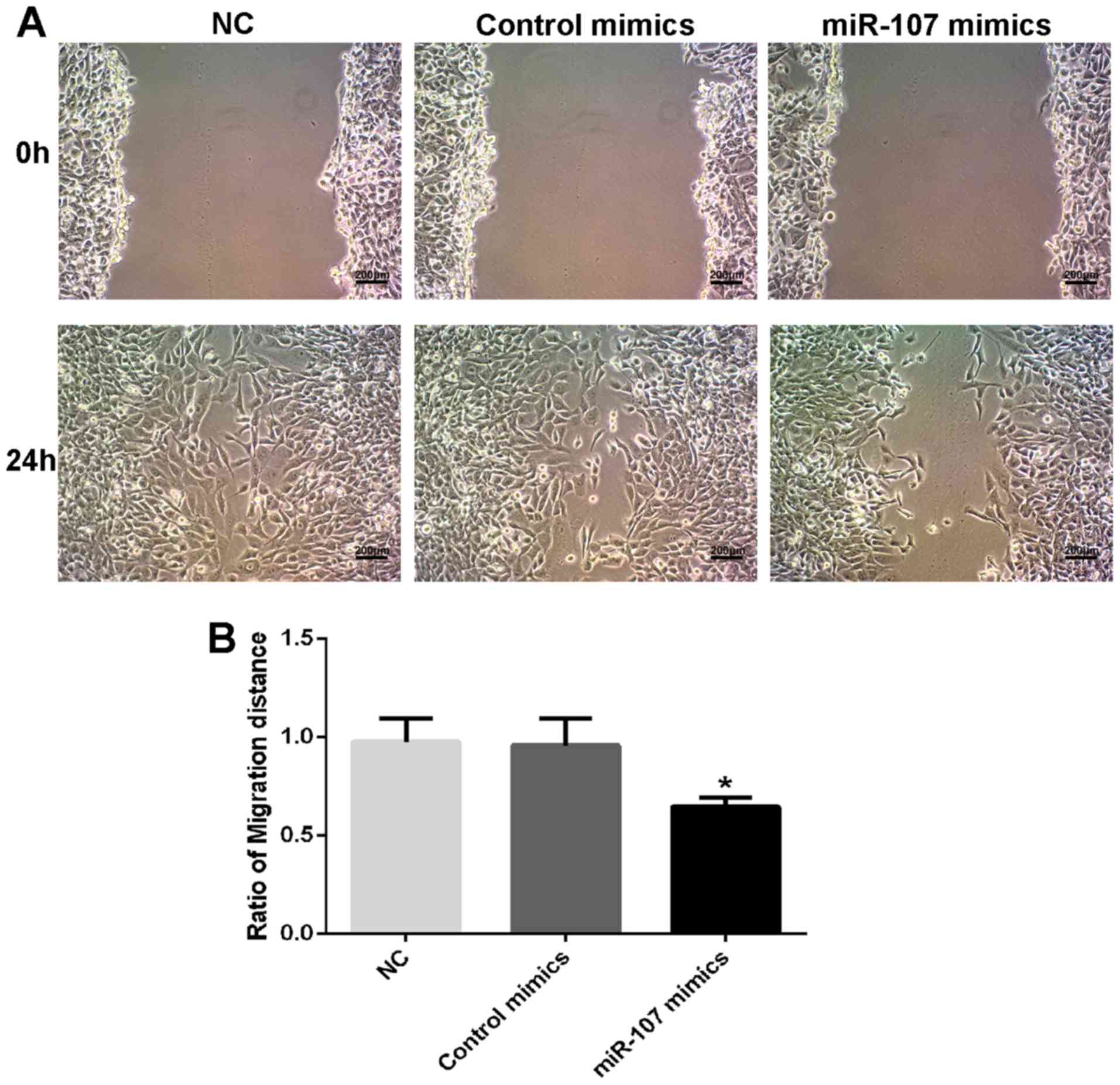
B). Transwell and wound-healing assays indicated that both the
invasion (P<0.001; Fig. 2C and D)
and migration (P<0.05; Fig. 3A and
B) capacities of SK-HEP-1 cells were significantly repressed by
miR-107 overexpression. The results indicated that miR-107
regulated the proliferation, migration, invasion and colony
formation of HCC cells.
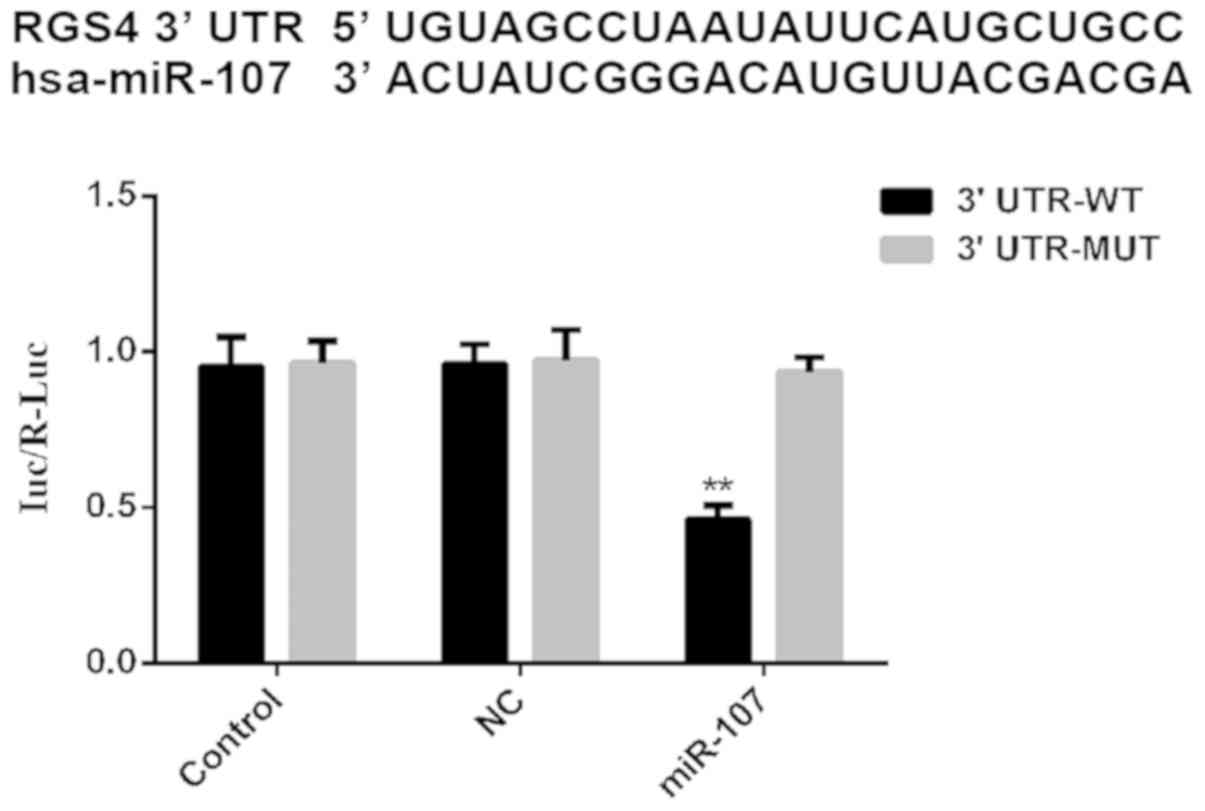
The RGS4 3′UTR contains a putative
miR-107 binding site that is required for the miR-107-mediated
regulation of RGS4 expression
To verify whether the miR-107 binding site is
required for regulating RGS4 expression, two luciferase reporter
plasmids were constructed, a wild-type 3′-UTR (RGS4-3′-UTR-WT) and
a mutant (RGS4-3′-UTR-MUT). To further investigate whether the
predicted binding site of miR-107 to the RGS4 3′-UTR was
responsible for this regulation, the 3′-UTR of RGS4 was cloned
downstream of a luciferase reporter gene (WT-RGS4). It was observed
that the overexpression of miR-107 remarkably reduced the
luciferase activity of RGS4-3′-UTR-WT (P<0.01; Fig. 4), but not RGS4-3′-UTR-MUT when
compared with the NC and control mimics groups. From these results,
it may be concluded that the sequence 5′-AUGCUGC-3′ within the RGS4
3′-UTR is required for miR-107-mediated regulation of RGS4
expression in HCC cells.
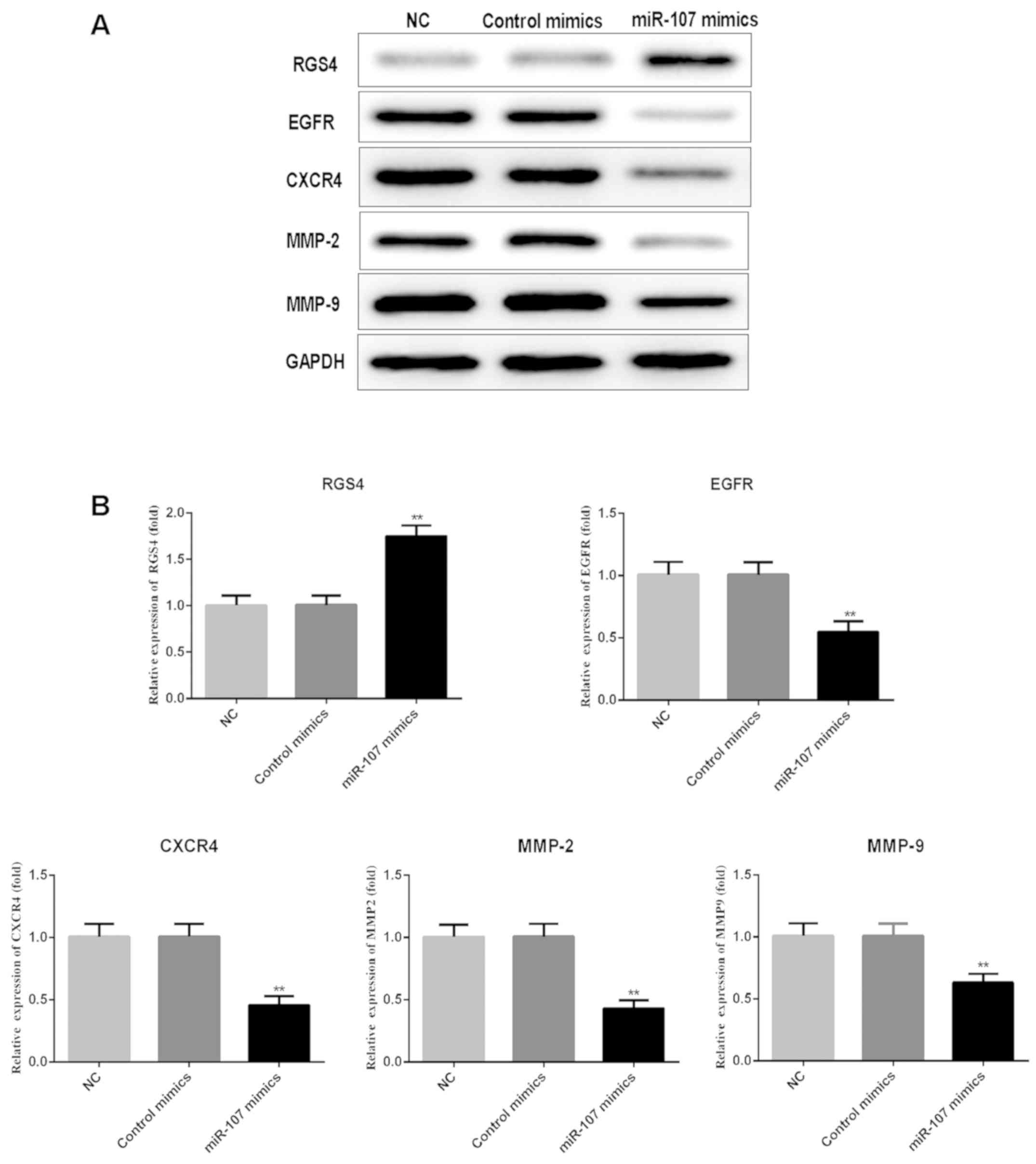
Overexpression of miR-107 enhances the
expression of RGS4, and reduces that of EGFR, CXCR4 and MMP-2 and
−9
To further confirm that miR-107 regulates the
expression of RGS4, EGFR, CXCR4, MMP-2 and MMP-9, SK-HEP-1 cells
were transfected with miR-107 mimics and control mimics, and the
protein expression levels were detected by western blot analysis.
The results showed that enhancing miR-107 expression significantly
promoted GRS4 expression, compared with the NC and control mimics
groups, whereas enhanced miR-107 significantly repressed EGFR,
CXCR4, MMP-2 and MMP-9 expression (P<0.01; Fig. 5).
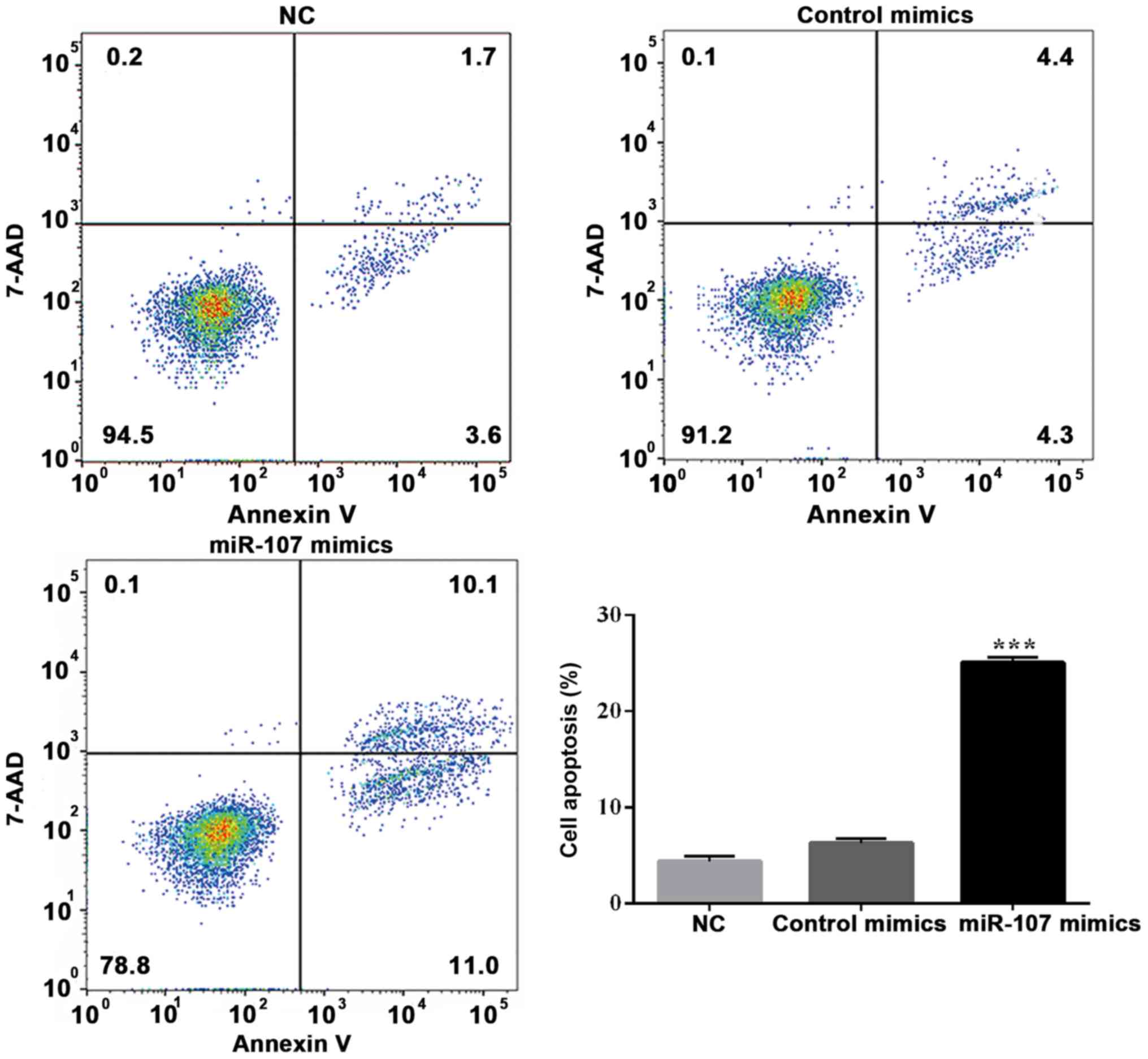
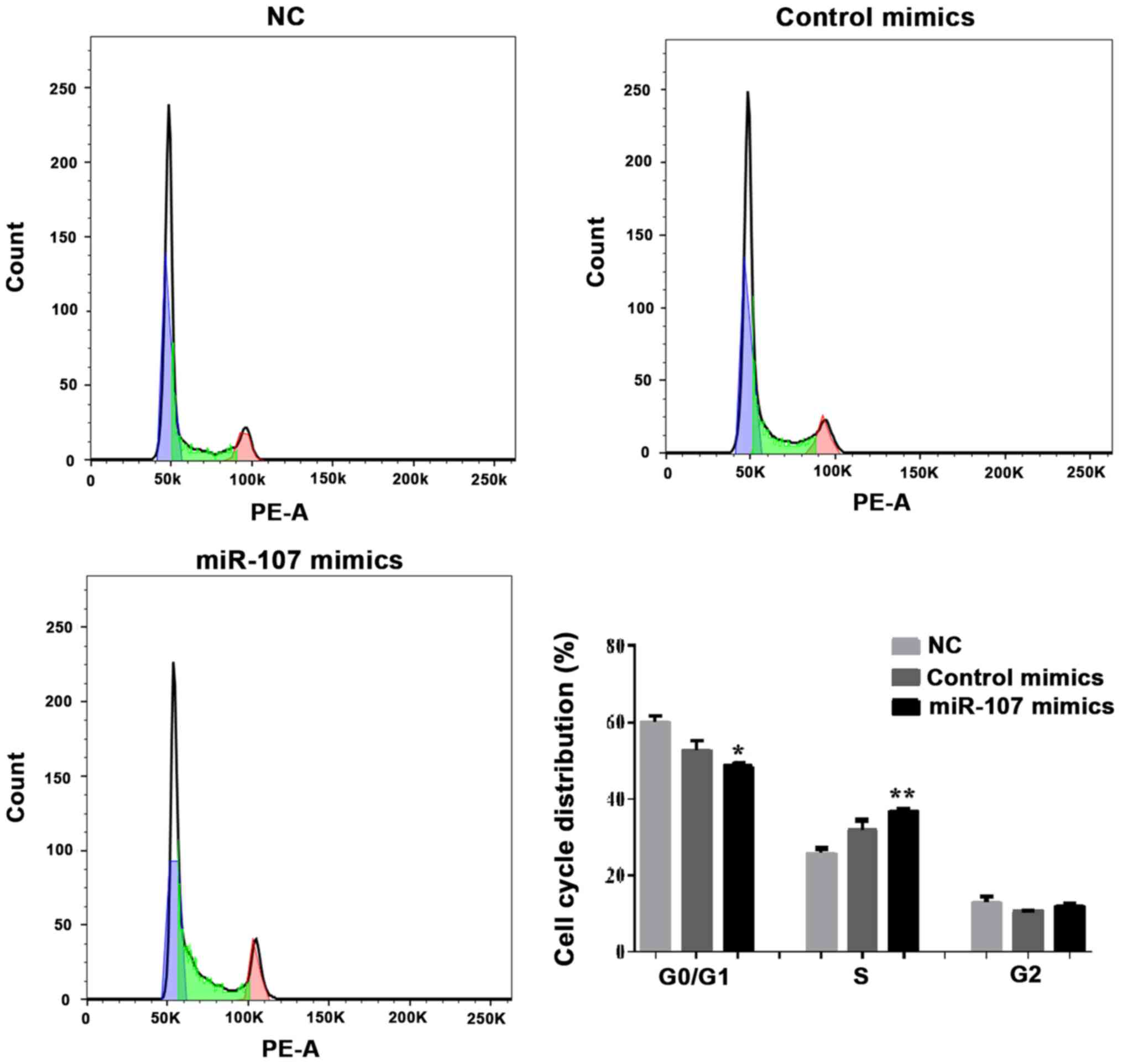
Effects of miR-107 overexpression on
SK-HEP-1 apoptosis and cell cycle distribution
To explore the possible mechanisms of miR-107
overexpression in SK-HEP-1 cells, the effect of miR-107 mimics on
the apoptotic rate and cell cycle distribution was determined.
7-Amino-actinomycin D (7-AAD) has a high DNA binding constant and
is efficiently excluded by cells in early apoptosis, while it can
enter cells in late apoptosis and necrotic cells to stain their
nuclei. Therefore, the matching use of Annexin V and 7-AAD can be
used to distinguish cells between early and late apoptosis and dead
cells. As shown in Fig. 6, flow
cytometry indicated that the early apoptotic rate was notably
increased following miR-107 overexpression (P<0.001; Fig. 6). Furthermore, the overall level of
apoptosis was increased. Cells treated with miR-107 mimics were
arrested in S phase, and S phase injury repair was not completed.
(P<0.05 and P<0.01; Fig. 7).
There was no significant difference in the G2 phase
population size among the miR-107 mimics, mimics control and NC
groups. These results suggested that the overexpression of miR-107
promoted cell proliferation by inhibiting the cell cycle and
inducing apoptosis.
Discussion
Globally, HCC is one of the most common human
malignancies, and its incidence in China is the highest among the
Asian countries (35). The number of
patients diagnosed with HCC each year is >700,000, and
>600,000 mortalities are associated with malignant HCC (36). In previous years, several studies
have shown that miRNAs are involved in the progression and
metastasis of tumors (37).
miRNA-107 is regarded as a tumor suppressor and may be
downregulated in breast cancer, possibly through the regulation of
its downstream target, brain-derived neurotrophic factor (38). Imamura et al (39) identified downregulated tumor
suppressor miRNAs in the plasma (including miR-107) for the
treatment of pancreatic cancer, using a comprehensive miRNA
array-based approach. Similarly, other studies have shown that
miR-107 acted as a tumor suppressor in several different tumor
types, including breast cancer, non-small cell lung cancer and head
and neck squamous cell carcinoma (40–42). In
the present study, the overexpression of miR-107 suppressed cell
proliferation, invasion, migration and colony-forming ability,
promoted apoptosis and caused G1 arrest, indicating that
miR-107 acts as a tumor suppressor in hepatic carcinoma cells.
Moreover, in vitro experiments revealed that
the overexpression of miR-107 enhanced RGS4 expression. The G
protein-signaling pathway serves a key role in the development and
regeneration of normal liver tissue, and an important role in the
development of HCC. Reportedly, altered levels of RGS4 expression
are associated with several human diseases, including cancer
(43). A number of studies have
shown that specific members of the RGS family are also involved in
the occurrence of various tumors types. RGS2 inhibition contributes
to the development of bladder cancer (44). Furthermore, RGS5 and RGS10 are
involved in the development of ovarian cancer (45), and RGS6 and RGS16 are both associated
with the EGF-mediated apoptosis and proliferation of breast cancer
cells (46). RGS17 can promote the
proliferation of lung cancer cells through the cyclic adenosine
monophosphate (cAMP)-protein kinase-cAMP response element-binding
protein signal transduction pathway (47). Additionally, RGS22 can also inhibit
the migration of pancreatic cancer cells (48), and numerous studies have demonstrated
that RGS4 may regulate GPCR signaling during breast cancer cell
proliferation (49).
In addition, RGS4 may inhibit the signal pathway by
downregulating the Gi-coupled receptors protease-activated receptor
1 and CXCR4 signal transduction, which were associated with the
migration and invasion of breast cancer cells (23). RGS4 is also present in NSCLC cells,
and its overexpression reduces the invasion and migration of tumor
cells by inhibiting MMP-2 and-9 and reversing EMT (30). In addition, MMP-2 and −9 were
confirmed to be involved in the development and metastasis of
cutaneous melanoma (31,32). In human cancer cells, the function of
kinase-independent EGFR is to prevent autophagic cell death
(50). Activated EGFR is involved in
the regulation of cell survival, proliferation and differentiation.
The present study showed that an increase in the expression level
of RGS4 protein resulted in a corresponding reduction in migration,
invasion and colony-forming ability of HCC cells. Therefore, it was
hypothesized that miR-107 serves as a tumor suppressor and
negatively regulates RGS4, which in turn alters the expression
levels of EGFR, CXCR4 and MMP-2 and −9. These findings suggest that
miR-107 may be a promising therapeutic target, and support its
involvement in the regulation of RGS4 overexpression in HCC.
In conclusion, the present study indicated that
miR-107 directly regulates RGS4 expression by targeting the RGS4
3′-UTR, thereby suppressing the migration and invasion of human HCC
cells, and promoting apoptosis by reducing the expression levels of
EGFR, CXCR4 and MMP-2 and −9. To the best of our knowledge, this is
the first report to demonstrate that miR-107 is depleted in HCC
cells, and that it may act as a biomarker and therapeutic target
for HCC. These findings support the accumulating evidence that
miR-107 participates in the progression of hepatic carcinoma by
modulating RGS4 expression.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors contributions
DX designed the research, wrote the manuscript, and
performed cell culture and molecular biology experiments; HXG
designed the research and critically revised the manuscript for
intellectual content. Both authors have read and approved the final
manuscript.
Ethics approval and consent for
publication
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Diaz-Gonzalez A, Forner A, Rodriguez de
Lope C and Varela M: New challenges in clinical research on
hepatocellular carcinoma. Rev Esp Enferm Dig. 108:485–493.
2016.PubMed/NCBI
|
|
2
|
Singal AG, Pillai A and Tiro J: Early
detection, curative treatment, and survival rates for
hepatocellular carcinoma surveillance in patients with cirrhosis: A
metaanalysis. PLoS Med. 11:e10016242014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mittal S, Kanwal F, Ying J, Chung R, Sada
YH, Temple S, Davila JA and El-Serag HB: Effectiveness of
surveillance for hepatocellular carcinoma in clinical practice: A
United States cohort. J Hepatol. 65:1148–1154. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Iorio MV and Croce CM: microRNA
involvement in human cancer. Carcinogenesis. 33:1126–1133. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ryan BM, Robles AI and Harris CC: Genetic
variation in microRNA networks: The implications for cancer
research. Nat Rev Cancer. 10:389–402. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Y, Li T, Qiu Y, Zhang T, Guo P, Ma
X, Wei Q and Han L: Serum microRNA panel for early diagnosis of the
onset of hepatocellular carcinoma. Medicine (Baltimore).
96:e56422017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
El-Serag HB, Marrero JA, Rudolph L and
Reddy R: Diagnosis and treatment of hepatocellular carcinoma.
Gastroenterology. 134:1752–1763. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ayremlou N, Mozdarani H, Mowla SJ and
Delavari A: Increased levels of serum and tissue miR-107 in human
gastric cancer: Correlation with tumor hypoxia. Cancer Biomark.
15:851–860. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Takahashi Y, Forrest AR, Maeno E,
Hashimoto T, Daub CO and Yasuda J: MiR-107 and MiR-185 can induce
cell cycle arrest in human non-small cell lung cancer cell lines.
PLoS One. 4:e66772009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee KH, Lotterman C, Karikari C, Omura N,
Feldmann G, Habbe N, Goggins MG, Mendell JT and Maitra A:
Epigenetic silencing of MicroRNA miR-107 regulates cyclin-dependent
kinase 6 expression in pancreatic cancer. Pancreatology. 9:293–301.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang S, Ma G, Zhu H, Lv C, Chu H, Tong N,
Wu D, Qiang F, Gong W, Zhao Q, et al: miR-107 regulates tumor
progression by targeting NF1 in gastric cancer. Sci Rep.
6:365312016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Y, Chen F, Zhao M, Yang Z, Zhang S,
Ye L, Gao H and Zhang X: MiR-107 suppresses proliferation of
hepatoma cells through targeting HMGA2 mRNA 3′UTR. Biochem Biophys
Res Commun. 480:455–460. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang R, Zhang C, Liu G, Gu R and Wu H:
MicroRNA-107 promotes proliferation, migration, and invasion of
osteosarcoma cells by targeting tropomyosin 1. Oncol Res.
25:1409–1419. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
He J, Zhang W, Zhou Q, Zhao T, Song Y,
Chai L and Li Y: Low-expression of microRNA-107 inhibits cell
apoptosis in glioma by upregulation of SALL4. Int J Biochem Cell
Biol. 45:1962–1973. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yamakuchi M, Lotterman CD, Bao C, Hruban
RH, Karim B, Mendell JT, Huso D and Lowenstein CJ: P53-induced
microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad
Sci USA. 107:6334–6339. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hollinger S and Hepler JR: Cellular
regulation of RGS proteins: Modulators and integrators of G protein
signaling. Pharmacol Rev. 54:527–559. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gerber KJ, Squires KE and Hepler JR: Roles
for regulator of G protein signaling proteins in synaptic signaling
and plasticity. Mol Pharmacol. 89:273–286. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xie Y, Wolff DW, Wei T, Wang B, Deng C,
Kirui JK, Jiang H, Qin J, Abel PW and Tu Y: Breast cancer migration
and invasion depend on proteasome degradation of regulator of
G-protein signaling 4. Cancer Res. 69:5743–5751. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Louwette S, Van Geet C and Freson K:
Regulators of G protein signaling: role in hematopoiesis,
megakaryopoiesis and platelet function. J Thromb Haemost.
10:2215–2222. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xie Z, Chan EC and Druey KM: R4 regulator
of G protein signaling (RGS) proteins in inflammation and immunity.
AAPS J. 18:294–304. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Siderovski DP and Willard FS: The GAPs,
GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int J
Biol Sci. 1:51–66. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ross EM and Wilkie TM: GTPase-activating
proteins for heterotrimeric G proteins: Regulators of G protein
signaling (RGS) and RGS-like proteins. Annu Rev Biochem.
69:795–827. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Park HJ, Kim SH and Moon DO: Growth
inhibition of human breast carcinoma cells by overexpression of
regulator of G-protein signaling 4. Oncol Lett. 13:4357–4363. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bansal G, Druey KM and Xie Z: R4RGS
proteins: Regulation of G-protein signaling and beyond. Pharmacol
Ther. 116:473–495. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cheng C, Yue W, Li L, Li S, Gao C, Si L
and Tian H: Regulator of G-protein signaling 4: A novel tumor
suppressor with prognostic significance in non-small cell lung
cancer. Biochem Biophys Res Commun. 469:384–391. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Falzone L, Salemi R, Travali S, Scalisi A,
McCubrey JA, Candido S and Libra M: MMP-9 overexpression is
associated with intragenic hypermethylation of MMP9 gene in
melanoma. Aging (Albany NY). 8:933–944. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Marusak C, Bayles I, Ma J, Gooyit M, Gao
M, Chang M and Bedogni B: The thiirane-based selective MT1-MMP/
MMP2 inhibitor ND-322 reduces melanoma tumor growth and delays
metastatic dissemination. Pharmacol Res. 113:515–520. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sooro MA, Zhang N and Zhang P: Targeting
EGFR-mediated autophagy as a potential strategy for cancer therapy.
Int J Cancer. 143:2116–2125. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C (T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cai L and Cai X: Up-regulation of miR-9
expression predicate advanced clinicopathological features and poor
prognosis in patients with hepatocellular carcinoma. Diagn Pathol.
9:10002014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ram Kumar RM, Boro A and Fuchs B:
Involvement and clinical aspects of microRNA in osteosarcoma. Int J
Mol Sci. 17:E8772016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gao B, Hao S, Tian W, Jiang Y, Zhang S,
Guo L, Zhao J, Zhang G, Yan J and Luo D: MicroRNA-107 is
downregulated and having tumor suppressive effect in breast cancer
by negatively regulating BDNF. J Gene Med. Dec 19–2017.(Epub ahead
of print). doi: 10.1002/jgm.2932. View Article : Google Scholar
|
|
39
|
Imamura T, Komatsu S, Ichikawa D, Miyamae
M, Okajima W, Ohashi T, Kiuchi J, Nishibeppu K, Konishi H, Shiozaki
A, et al: Depleted tumor suppressor miR-107 in plasma relates to
tumor progression and is a novel therapeutic target in pancreatic
cancer. Sci Rep. 7:57082017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang L, Ma P, Sun LM, Han YC, Li BL, Mi
XY, Wang EH and Song M: MiR-107 down-regulates SIAH1 expression in
human breast cancer cells and silencing of miR-107 inhibits tumor
growth in a nude mouse model of triple-negative breast cancer. Mol
Carcinog. 55:768–777. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xia H, Li Y and Lv X: MicroRNA-107
inhibits tumor growth and metastasis by targeting the BDNF-mediated
PI3K/AKT pathway in human nonsmall lung cancer. Int J Oncol.
49:1325–1333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Datta J, Smith A, Lang JC, Islam M, Dutt
D, Teknos TN and Pan Q: microRNA-107 functions as a candidate
tumor-suppressor gene in head and neck squamous cell carcinoma by
downregulation of protein kinase cvarepsilon. Oncogene.
31:4045–4053. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hao J, Michalek C, Zhang W, Zhu M, Xu X
and Mende U: Reguation of cardiomyocyte signaling by RGS proteins:
Differential selectivity towards G proteins and susceptibility to
regulation. J Mol Cell Cardiol. 41:51–61. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cao X, Qin J, Xie Y, Khan O, Dowd F,
Scofield M, Lin MF and Tu Y: Regulator of G-protein signaling 2
(RGS2) inhibits androgen-independent activation of androgen
receptor in prostate cancer cells. Oncogene. 25:3719–3734. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xu Z, Zuo Y, Wang J, Yu Z, Peng F, Chen Y,
Dong Y, Hu X, Zhou Q, Ma H, et al: Overexpression of the regulator
of G-protein signaling 5 reduces the survival rate and enhances the
radiation response of human lung cancer cells. Oncol Rep.
33:2899–2907. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liang G, Bansal G, Xie Z and Druey KM:
RGS16 inhibits breast cancer cell growth by mitigating
phosphatidylinositol 3-kinase signaling. J Biol Chem.
284:21719–21727. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
James MA, Lu Y, Liu Y, Vikis HG and You M:
RGS17, an overexpressed gene in human lung and prostate cancer,
induces tumor cell proliferation through the cyclic AMP-PKA-CREB
pathway. Cancer Res. 69:2108–2116. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kelly P, Moeller BJ, Juneja J, Booden MA,
Der CJ, Daaka Y, Dewhirst MW, Fields TA and Casey PJ: The G12
family of heterotrimeric G proteins promotes breast cancer invasion
and metastasis. Proc Natl Acad Sci USA. 103:8173–8178. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hu Y, Xing J, Chen L, Zheng Y and Zhou Z:
RGS22 inhibits pancreatic adenocarcinoma cell migration through the
G12/13 α subunit/F-actin pathway. Oncol Rep. 34:2507–2514. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Tan X, Thapa N, Sun Y and Anderson RA: A
kinase-independent role for EGF receptor in autophagy initiation.
Cell. 160:145–160. 2015. View Article : Google Scholar : PubMed/NCBI
|