Introduction
Lung cancer, as a major cause of human mortality, is
considered one of the most common types of cancer. Every year, 1.6
million individuals succumb to lung cancer, accounting for
one-third of all cancer-associated mortalities. It is the most
common cause of cancer-associated mortalities in males and the
second most common in females (1,2).
Non-small-cell lung cancer (NSCLC) accounts for 85% of all cases of
lung cancer; within this group, adenocarcinoma (Ade) and squamous
cell carcinoma (SCC) are the major histopathological types
(3). Once diagnosed with lung
cancer, the 5-year survival rate of patients is only 17.8%
(4,5). In the previous decade, although
promising treatments have emerged, for the majority of patients
with distant metastases, surgery is not a viable option and there
are no radical treatments available (6,7).
Therefore, it is important to explore the potential mechanism of
tumorigenesis and tumor progression in NSCLC, and to identify
underlying biomarkers for prognosis that may be targeted by lung
cancer-specific chemotherapeutic drugs. Furthermore, the results of
these investigations may be utilized for early personalized
methods, individualized precise treatment strategies and improved
survival cycles in patients with lung cancer.
Ubiquitylation is a widespread post-translational
modification of proteins that occurs in eukaryotic cells, and
constitutes an important topic of research in the post-genome era.
This type of modification is considered an important complement to
the regulation of the genetic central dogma (8). Ubiquitylation is achieved by the
generation of an isopeptide bond, which is formed by covalent
bonding between the C-terminal carboxyl groups of ubiquitin in the
lysine side chain of the substrate protein. The ubiquitination
level of proteins in eukaryotic cells is regulated by the E1-E2-E3
enzyme synthesis system and the deubiquitinating enzymes (DUBs)
system (9). Ubiquitinating enzymes
and DUBs function together to form a unique ubiquitin network
through multi-level modification of the substrate (including
monoubiquitin, 8 ubiquitin chains, mixed ubiquitin chains and
bifurcated ubiquitin chains), to regulate cellular processes. In
contrast to the ubiquitination process, DUBs modulate
ubiquitin-associated processes by reverse-modifying monoubiquitin
and ubiquitin chain modifications on substrate proteins (10,11).
Ubiquitination modification is involved in protein
degradation, autophagy, DNA damage repair, cell cycle, signal
transduction, gene expression, inflammation, immunity and other
vital life processes (12,13). The dysfunction of ubiquitinating
enzymes is associated with a variety of severe diseases, including
cancer, and cardiovascular and neurodegenerative diseases (14–17). In
particular, in malignant tumors, previous studies have demonstrated
that several types of DUBs are involved in the development and
progression of cancer (18),
including follicular lymphoma (19),
and prostate (20), colon and breast
cancer (21).
A total of ~90 DUBs are encoded in the human genome
(22). Ovarian tumor-associated
proteases (OTUs) are a subtype of DUBs that may be classified into
4 subfamilies according to sequence similarities. OTU
domain-containing proteins (OTUDs) are a subfamily of OTUs
comprising eight members: OTUD1; OTUD2; OTUD3; OTUD4; OTUD5/DUBA;
OTUD6A; OTUD6B; and putative bifunctional UDP-N-acetylglucosamine
transferase and deubiquitinase ALG13 (ALG13) (23).
Previous studies have focused on the function of
OTUDs in tumor development, invasion and metastasis; numerous
studies have demonstrated that OTUD1 serves a key role in the
metastasis of thyroid and breast cancer (24–26). In
addition, OTUD3 (27), OTUD4
(28), OTUD5 (29) and OTUD6B (30) also serve roles in the pathogenesis of
tumors. However, the knowledge concerning the roles of different
OTUDs in the development of lung cancer is limited. Therefore, the
present study explored large sample-based databases to determine
the expression and prognostic value of each isoenzyme of the OTUD
subfamily in NSCLC.
Materials and methods
Oncomine analysis
Data entries from February 2018 to August 2018 in
the Oncomine database (http://www.oncomine.org/) (31,32) were
searched to determine the individual mRNA expression levels of the
OTUD subfamily in different cancer types. The Oncomine 4.5 Research
Edition is a web-based data mining database with cancer microarray
information, aimed at promoting the expression of whole genome
analysis and comparative transcriptome data analysis for the major
types of cancer and in normal tissues. It currently contains 715
datasets and 86,733 samples. The present study compared the mRNA
levels in datasets of patients with NSCLC and normal individuals.
P=0.05, fold-change value ‘all’ and the top 10% gene rank were
selected as thresholds to obtain the highest number of genes in the
datasets.
Kaplan-Meier survival analysis
The prognostic values of OTUD sub-members (OTUD1,
OTUD2, OTUD3, OTUD4, OTUD5, OTUD6B and ALG13) specifically
expressed in NSCLC samples were evaluated by overall survival (OS)
using the Kaplan-Meier plotter resource (33–35).
Hazard ratios (HR) with 95% confidence intervals (CIs) and log-rank
P-values were calculated subsequently. To evaluate the prognostic
value of each member, the patient samples were divided into two
groups (high vs. low expression group) based on the median gene
expression value. Subsequently, GraphPad Prism 7 software (GraphPad
Software, Inc.) was used to produce Kaplan-Meier survival curves
according to these setting conditions. The Affymetrix identity of
each gene in NSCLC was validated and summarized in Table I. In the present study, the ‘array
quality control’ option was selected to ‘exclude biased arrays’ and
the results of the figures were obtained by multivariate Cox
regression analysis.
 | Table I.Desired Affymetrix ID of OTUD family
genes in the Kaplan-Meier plotter resource. |
Table I.
Desired Affymetrix ID of OTUD family
genes in the Kaplan-Meier plotter resource.
| OTUD | Affymetrix ID |
|---|
| OTUD1 | 226140_s_at |
| OTUD2 | 215150_at |
| OTUD3 | 213216_at |
| OTUD4 | 203480_s_at |
| OTUD5 | 233933_s_at |
| OTUD6B | 222825_at |
| ALG13 | 205583_s_at |
Results
Basic characteristics of 8 OTUD
isoenzymes
To date, 8 OTUD isoenzymes have been identified in
the human genome. Their characteristics are associated with protein
data bank identification [RCSB Protein Data Bank (https://www.rcsb.org)], physiological processes and
various types of cancer (23,27–30,36,37),
as demonstrated in Table II.
 | Table II.Basic characteristics of 8 OTUD
isoenzymes. |
Table II.
Basic characteristics of 8 OTUD
isoenzymes.
| Isoenzyme | PDB-ID | Physiological
process | Associated
diseases |
|---|
| OTUD1 | 4bop | N/A | Thyroid cancer |
| OTUD2 | 4bop | Endoplasmic
reticulum degradation | Cervical
cancer |
| OTUD3 | 4bop | PI3K/AKT | Breast cancer |
| OTUD4 | N/A | DNA alkylation
Damage repair | N/A |
| OTUD5 | 3pfy | p53 | N/A |
| OTUD6A | N/A | N/A | N/A |
| OTUD6B | N/A | B-cells within the
lymphatic system | NSCLC |
| ALG13 | N/A | N/A | N/A |
Different expression of the OTUD
subfamily in NSCLC
Analysis of the Oncomine database revealed the
patterns of OTUD family genes expression in tissues from patients
with NSCLC compared with normal tissues; the data are summarized in
Table III. The analysis
demonstrated that the mRNA expression levels of these OTUD
subfamily members were different significantly over-expression or
under-expression in patients with NSCLC compared with normal
samples in different datasets.
 | Table III.OTUD family genes expression in lung
cancer (Oncomine database). |
Table III.
OTUD family genes expression in lung
cancer (Oncomine database).
| A, Lung
adenocarcinoma vs. normal |
|---|
|
|---|
|
|
|
| Sample number |
|
|
|---|
|
|
|
|
|
|
|
|---|
| Gene | Fold change | Dataset | Normal | Cancer | Total | P-value |
|---|
| OTUD1 | −2.678 | Hou et
al | 65 | 45 | 110 |
2.79×10−19 |
|
| −3.761 | Garber et
al | 5 | 40 | 45 |
2.99×10−4 |
| OTUD2 | 1.161 | TCGA | 390 | 261 | 651 |
6.30×10−43 |
|
| 1.093 | Weiss et
al | 59 | 77 | 136 |
3.73×10−11 |
| OTUD5 | 1.302 | Hou et
al | 65 | 45 | 110 |
3.15×10−7 |
| OTUD6B | 1.135 | TGCA | 390 | 261 | 651 |
1.72×10−24 |
|
| 1.108 | Weiss et
al | 59 | 77 | 136 |
6.99×10−10 |
| ALG13 | −1.465 | Yamagata et
al | 3 | 9 | 12 | 0.002 |
|
| B, Squamous cell
lung carcinoma vs. normal |
|
|
|
|
| Sample
number |
|
|
|
|
|
|
|
|
|
| Gene | Fold
change | Dataset | Normal | Cancer | Total | P-value |
|
| OTUD1 | −2.921 | Hou et
al | 65 | 27 | 92 |
4.39×10−16 |
|
| −3,206 | Garber et
al | 5 | 12 | 17 |
4.55×10−4 |
| OTUD2 | 1.109 | TGCA | 390 | 348 | 738 |
5.77×10−14 |
| OTUD4 | −1.350 | Yamagata et
al | 3 | 11 | 14 | 0.007 |
|
| −1.128 | TGCA | 390 | 348 | 738 |
1.83×10−40 |
|
| −1.078 | Weiss et
al | 59 | 155 | 214 |
3.18×10−15 |
| OTUD6B | 1.100 | Weiss et
al | 59 | 155 | 214 |
1.36×10−16 |
| ALG13 | −1.672 | Yamagata et
al | 3 | 11 | 14 |
2.23×10−5 |
|
| C, Large cell
lung carcinoma vs. normal |
|
|
|
|
| Sample
number |
|
|
|
|
|
|
|
|
|
| Gene | Fold
change | Dataset | Normal | Cancer | Total | P-value |
|
| OTUD1 | −4.192 | Hou et
al | 65 | 19 | 84 |
1.57×10−13 |
| ALG13 | −1.548 | Yamagata et
al | 3 | 5 | 8 | 0.004 |
|
| D, Small cell
lung carcinoma vs. normal |
|
|
|
|
| Sample
number |
|
|
|
|
|
|
|
|
|
| Gene | Fold
change | Dataset | Normal | Cancer | Total | P-value |
|
| OTUD1 | −3.625 | Garber et
al | 5 | 4 | 9 | 0.002 |
| OTUD3 | 2.205 | Bhattacharjee et
al | 17 | 6 | 23 | 0.005 |
Distinct prognostic value of the OTUD
subfamily in NSCLC
The prognostic value of the mRNA expression of OTUDs
was examined with Kaplan-Meier plotter (Kaplan Meier-plotter [Lung
Cancer] 2015 version). Among all the OTUD subfamily members, only
OTUD6A was not detected. The datasets are presented in Table IV. Firstly, the prognostic value of
OTUD1 mRNA expression was determined. Survival curves were plotted
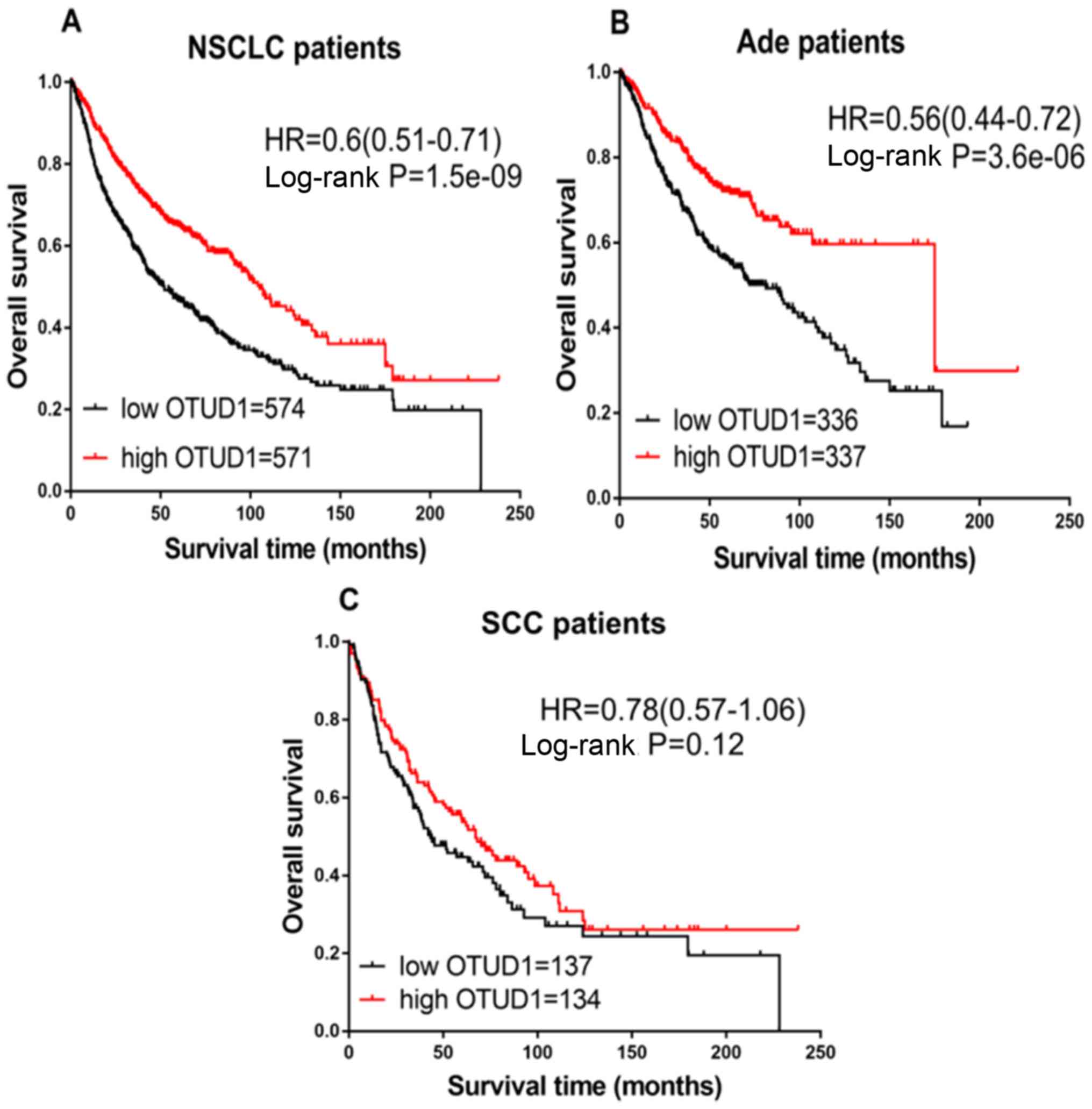
for all patients with NSCLC (Fig.
1A), Ade (Fig. 1B) and SCC
(Fig. 1C). High expression of OTUD1
was associated with significantly increased OS in all NSCLC
(HR=0.60; CI, 0.51–0.71; P=1.5×10−9) and Ade cases
(HR=0.56; CI, 0.44–0.72; P=3.6×10−6), but not in
patients with SCC (HR=0.78; CI, 0.57–1.06; P=0.12).
 | Table IV.Association between OTUD isoforms and
pathology subtype in patients with NSCLC. |
Table IV.
Association between OTUD isoforms and
pathology subtype in patients with NSCLC.
|
|
| Expression group
(n) |
|
|
|---|
|
|
|
|
|
|
|---|
| Gene | Pathology
subtype | Low | High | HR (95% CI) | P-value |
|---|
| OTUD1 | NSCLC | 574 | 571 | 0.60
(0.51–0.71) |
1.5×10−9 |
|
| Ade | 336 | 337 | 0.56
(0.44–0.72) |
3.6×10−6 |
|
| SCC | 137 | 134 | 0.78
(0.57–1.06) | 0.120 |
| OTUD2 | NSCLC | 966 | 960 | 1.27
(1.12–1.45) |
1.7×10−4 |
|
| Ade | 366 | 354 | 1.13
(0.9–1.43) | 0.290 |
|
| SCC | 263 | 261 | 0.88
(0.69–1.11) | 0.270 |
| OTUD3 | NSCLC | 964 | 962 | 0.84
(0.74–0.96) | 0.009 |
|
| Ade | 361 | 359 | 0.65
(0.51–0.82) |
3.5×10−4 |
|
| SCC | 262 | 262 | 0.93
(0.73–1.18) | 0.550 |
| OTUD4 | NSCLC | 963 | 963 | 0.78
(0.69–0.88) |
9.6×10−5 |
|
| Ade | 360 | 360 | 0.47
(0.37–0.6) |
6.6×10−10 |
|
| SCC | 263 | 261 | 1.01
(0.8–1.28) | 0.940 |
| OTUD5 | NSCLC | 578 | 567 | 1.02
(0.87–1.2) | 0.810 |
|
| Ade | 339 | 334 |
1.22(0.96–1.55) | 0.110 |
|
| SCC | 136 | 135 | 1.09
(0.8–1.49) | 0.590 |
| OTUD6B | All | 574 | 571 | 1.05
(0.89–1.24) | 0.560 |
|
| Ade | 336 | 337 | 0.62
(0.48–0.79) |
9.2×10−5 |
|
| SCC | 136 | 135 | 1.41
(1.03–1.93) | 0.029 |
| ALG13 | All | 965 | 961 | 0.76 (0.67-
0.87) |
3.2×10−5 |
|
| Ade | 360 | 360 | 0.47
(0.37–0.6) |
7.2×10−10 |
|
| SCC | 262 | 262 | 0.8
(0.63–1.02) | 0.068 |
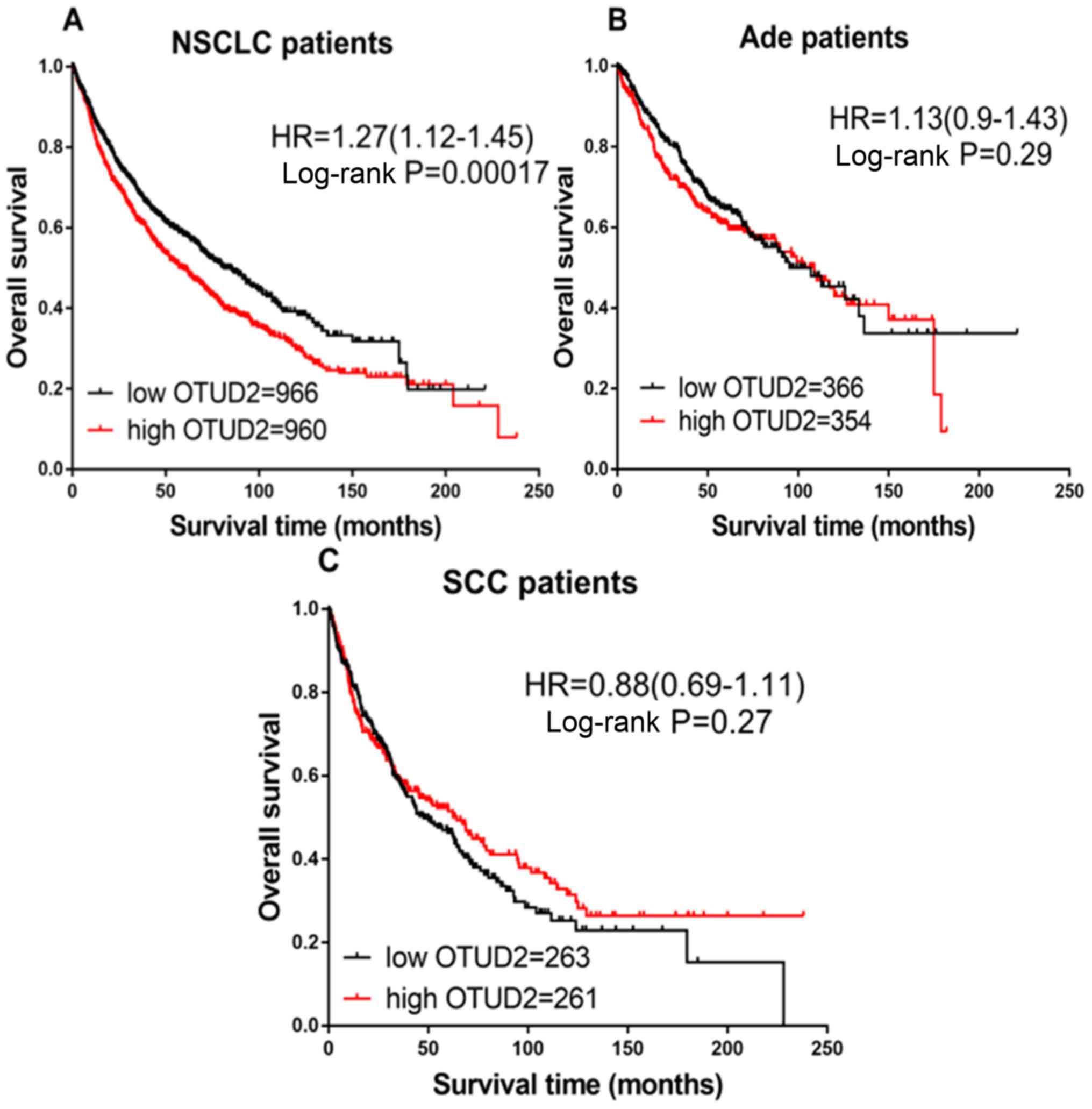
Then, the prognostic value of OTUD2 mRNA expression
was analyzed. Increased expression levels of OTUD2 mRNA were
associated with decreased OS in patients with NSCLC (HR=1.27; CI,
1.12–1.45; P=0.00017; Fig. 2A).
However, high expression of OTUD2 mRNA was not associated with OS
in patients with Ade (HR=1.13; CI, 0.90–1.43; P=0.29; Fig. 2B) or SCC (HR=0.88; CI, 0.69–1.11;
P=0.27; Fig. 2C).
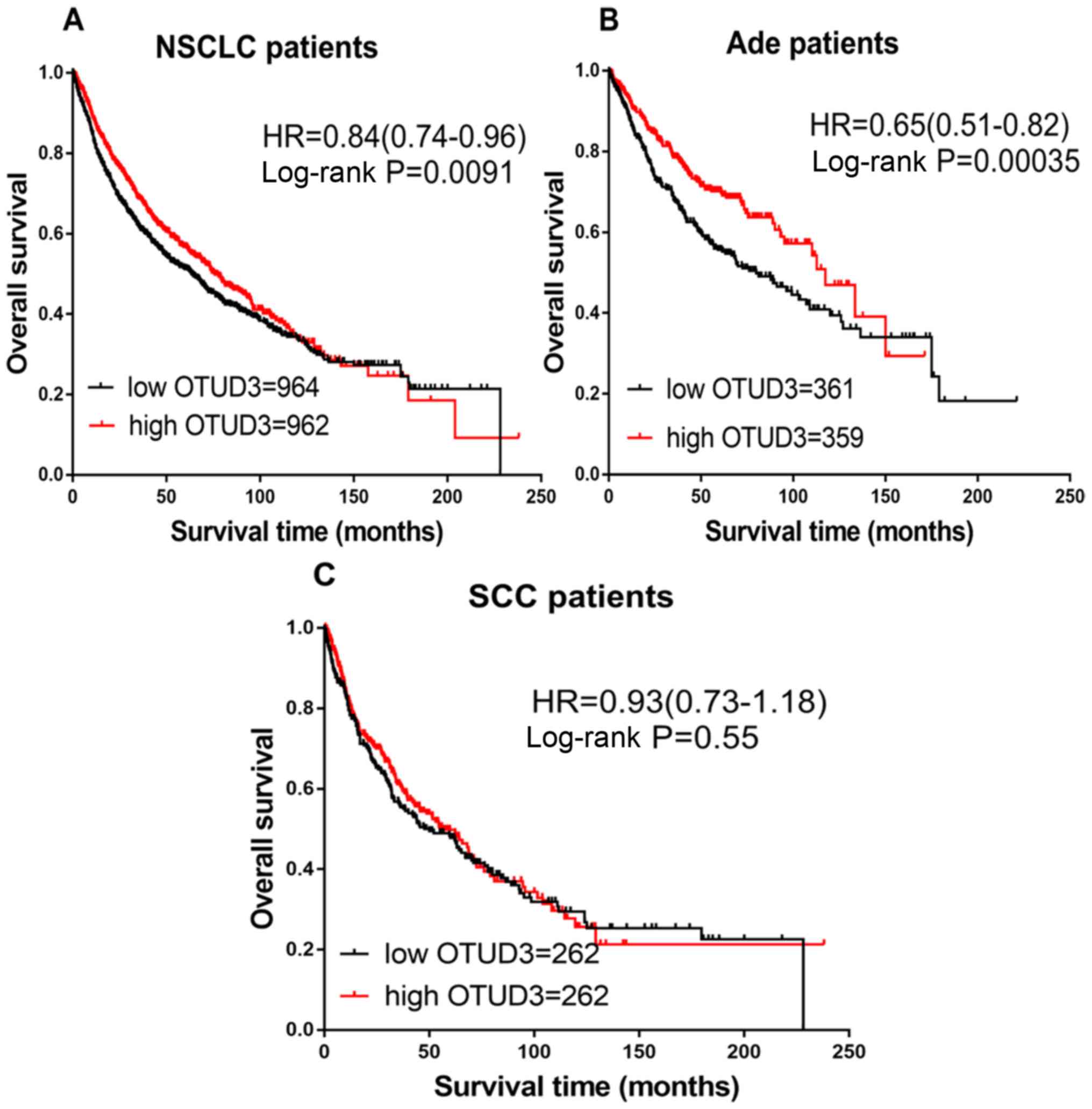
As indicated in Fig.
3, the prognostic significance of OTUD3 expression was also
evaluated. Increased expression of OTUD3 mRNA was associated with
good OS in all patients with NSCLC (HR=0.84; CI, 0.74–0.96;
P=0.0091; Fig. 3A) and Ade (HR=0.65;
CI, 0.51–0.82; P=0.00035; Fig. 3B),
but not with SCC (HR=0.93; CI, 0.73–1.18; P=0.55; Fig. 3C).
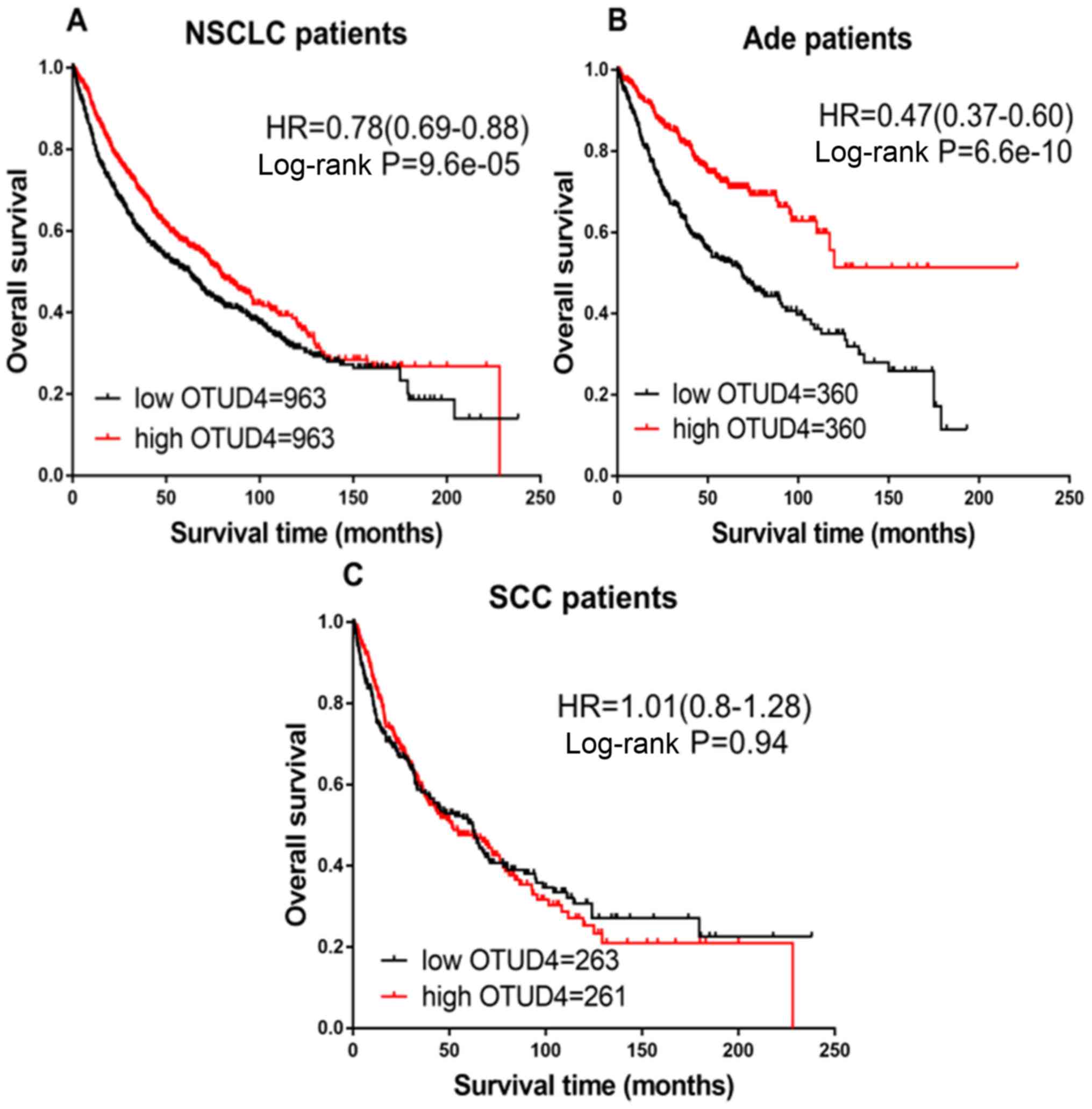
For OTUD4, high mRNA expression was associated with
favorable OS in all patients with NSCLC (HR=0.78; CI, 0.69–0.88;
P=9.6×10−5; (Fig. 4A) and
Ade (HR=0.47; CI, 0.37–0.60; P=6.6×10−10; Fig. 4B), but not with SCC (HR=1.01; CI,
0.80–1.28; P=0.94; Fig. 4C).
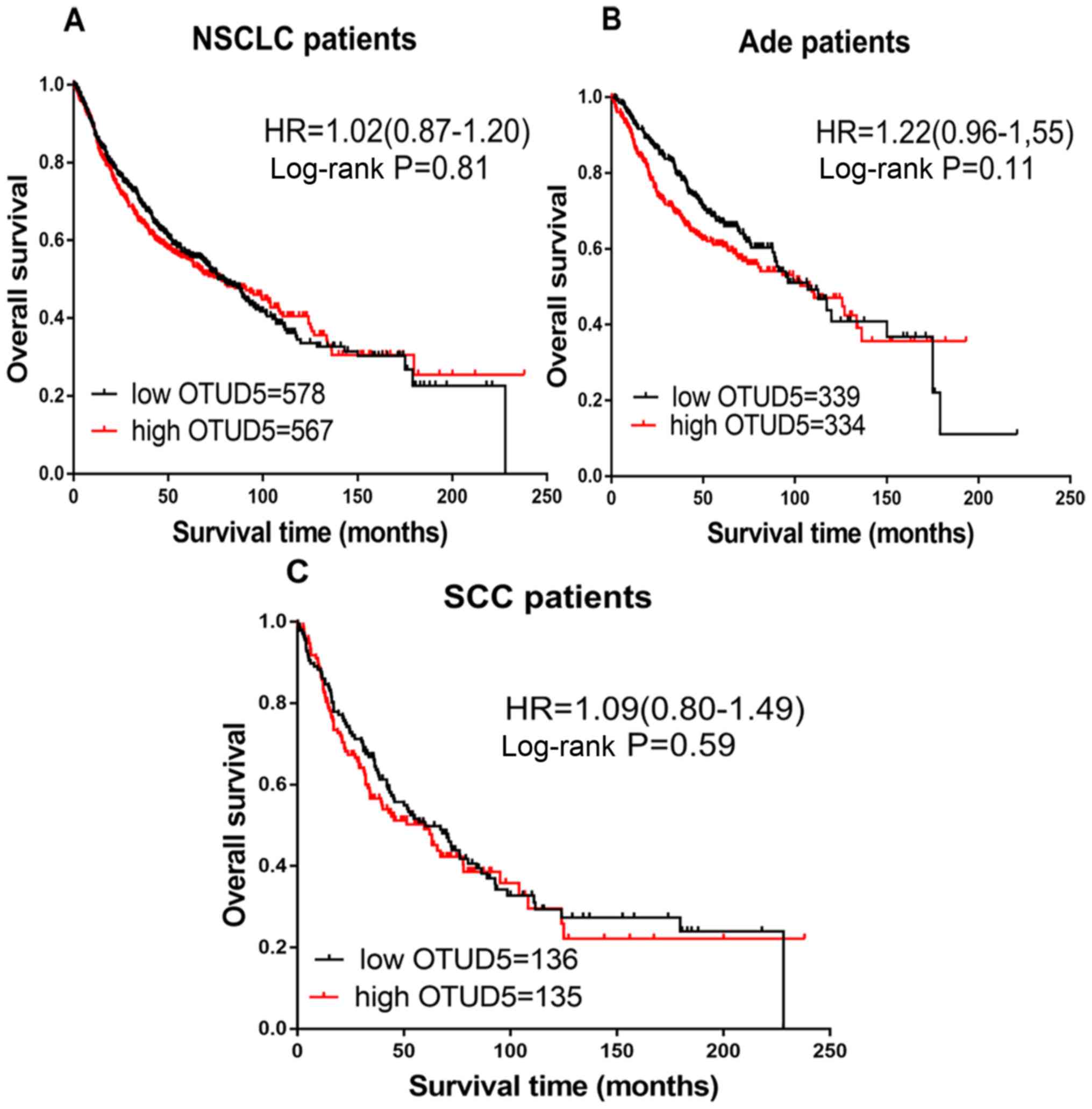
Fig. 5 demonstrates
the prognostic effect of OTUD5 mRNA expression. High or low
expression of OTUD5 did not elicit an effect on the prognosis of
patients with NSCLC (HR=1.02; CI, 0.87–1.20; P=0.81; Fig. 5A), Ade (HR=1.22; CI, 0.96–1.55;
P=0.11; Fig. 5B) or SCC (HR=1.09;
CI, 0.80–1.49; P=0.59; Fig. 5C).
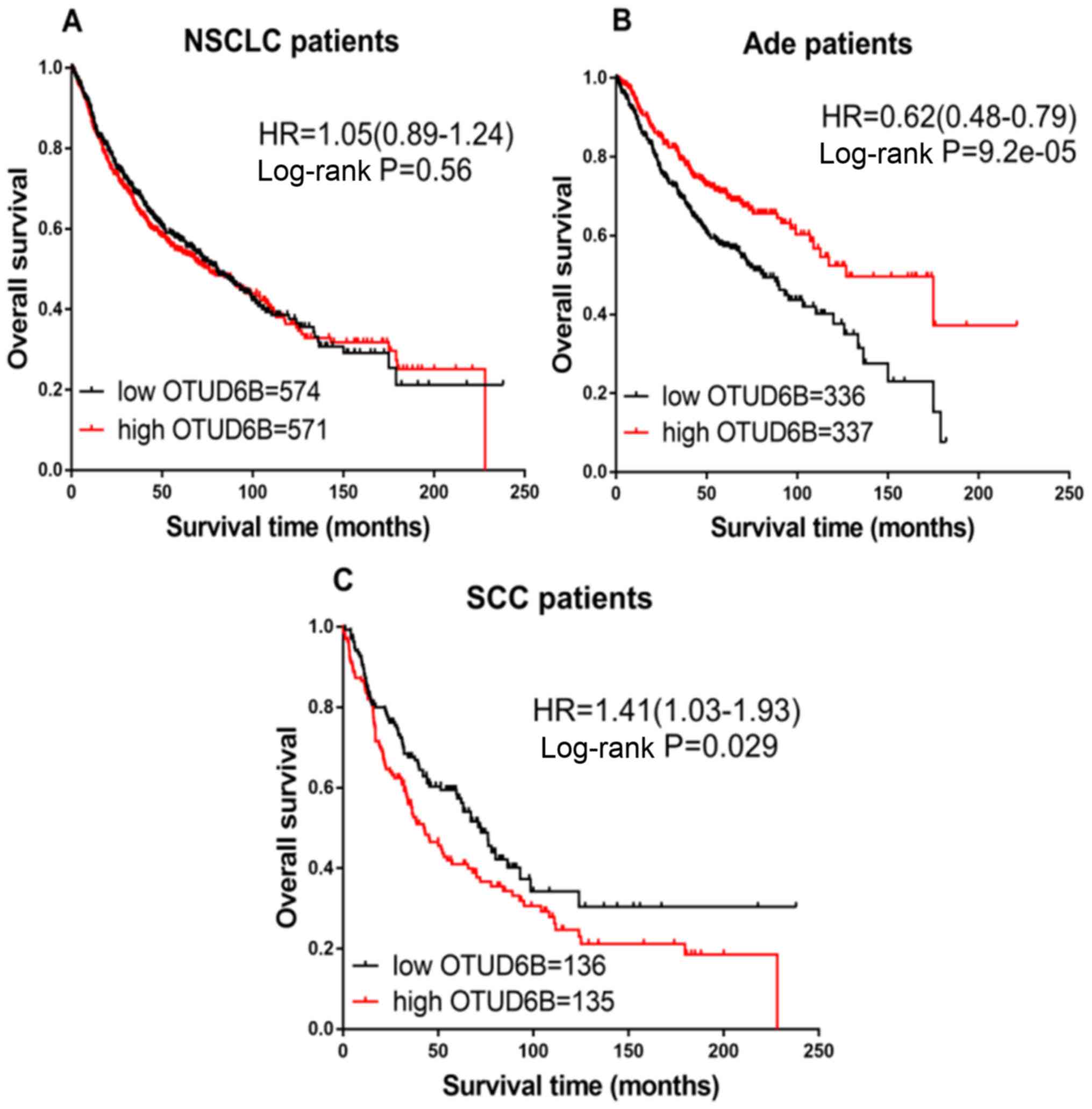
Next, the prognostic value of OTUD6B expression was
examined. In patients with NSCLC, no significant differences in
prognoses were observed between the high or low OTUD6B expression
groups (HR=1.05; CI, 0.89–1.24; P=0.56; Fig. 6A). However, the increased
transcriptional expression of OTUD5 was associated with favorable
OS in patients with Ade (HR=0.62; CI, 0.48–0.79;
P=9.2×10−5; Fig. 6B),
while patients with SCC exhibited worse OS (HR=1.41; CI, 1.03–1.93;
P=0.029; Fig. 6C).
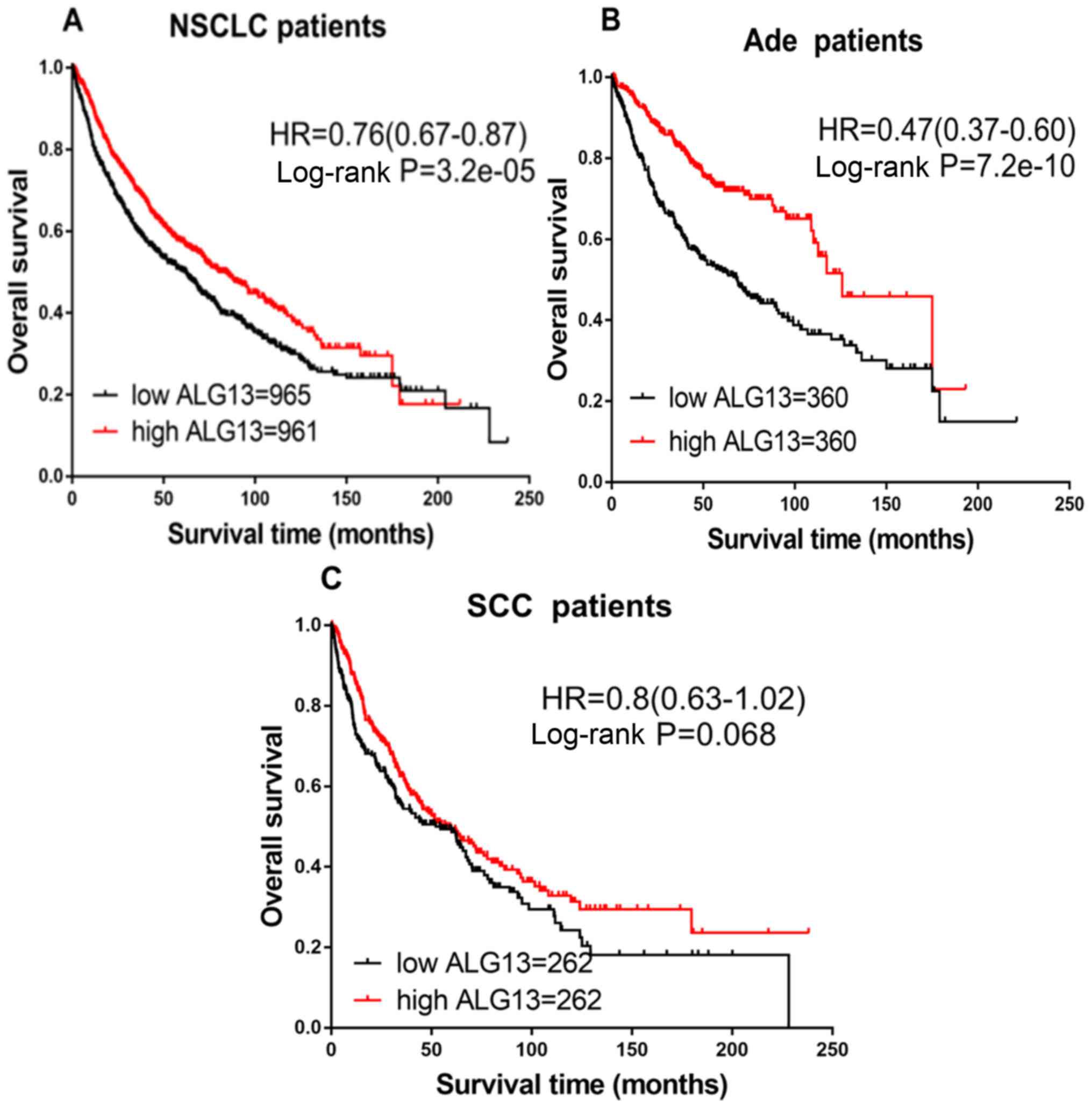
Finally, the prognostic effect of the expression of
ALG13 was explored. The prognoses in the high and low ALG13
expression groups were different in patients with NSCLC (HR=0.76;
CI, 0.67–0.87; P=3.2×10−5; Fig. 7A) and Ade (HR=0.47; CI, 0.37–0.60;
P=7.2×10−10; Fig. 7B).
Patients with high expression of ALG13 exhibited longer OS.
However, no difference was observed in patients with SCC (HR=0.80;
CI, 0.63–1.02; P=0.068; Fig.
7C).
The associations between OTUDs and
clinicopathological features in the NSCLC patients, including tumor
stages (NCCN Non-Small Cell Lung Cancer, Version 5.2017) (38), lymph node status (NCCN Non-Small Cell
Lung Cancer, Version 5.2017), smoking status, sex and chemotherapy,
were also examined. Tumor stage, lymph node status and chemotherapy
were not demonstrated to be associated with OTUD expression (data
not shown). As demonstrated in Table
SI, OTUD1 expression was identified to be associated with a
significantly poorer OS in all patients with stage I and II NSCLC,
while high expression of OTUD5 was associated with significantly
improved OS in patients with stage II NSCLC. As indicated in
Table SII, OTUD1, OTUD6B and ALG13
were significantly associated with lymph node status of patients
with NSCLC. All OTUDs, with the exception of OTUD3 and OTUD5, were
significantly associated with smoking status of NSCLC patients
(Table SIII). Meanwhile, all OTUDs
with the exception of OTUD5 and OTUD6B were significantly
associated with gender of NSCLC patients (Table SIV). Only OTUD3 expression was
significantly associated with chemotherapy treatment in patients
with NSCLC (Table SV).
Discussion
Among the cancer markers identified to date, OTUDs
have been extensively studied. However, few studies have analyzed
the expression of OTUDs in lung cancer, in particular the different
OTUD isoforms. Therefore, to the best of knowledge, the present
study was the first to analyze and discuss the different roles of
OTUD isoenzymes in the prognosis of NSCLC.
Screening of the Oncomine database identified that
OTUD1, OTUD2, OTUD6B and ALG13 met the filter criteria of the
present study. The analysis revealed that different OTUD subfamily
mRNA expression levels were significantly different in NSCLC
compared with normal samples. Hou et al (39) and Garber et al (40) have demonstrated that OTUD1 exhibits
lower mRNA levels in Ade and SCC compared with normal tissue. In
addition, the datasets analyzed in the studies by Yamagata et
al (41) and Selamat et
al (42) demonstrated that ALG13
mRNA was also expressed at decreased levels in Ade and SCC.
Conversely, OTUD2 and OTUD6B mRNA were expressed at increased
levels in Ade and SCC.
OTUD1 is a DUB, which belongs to the OTU family. It
is an important enzyme that controls the activity or abundance of
substrates by removing covalently linked ubiquitin from proteins
(43). However, its substrates and
its role in cells are unknown. OTUD1 directly suppresses the
ubiquitination of p53 in cells to increase apoptosis and decrease
cell proliferation, and a previous study by Piao et al
(26) indicated that OTUD1 exerted a
pivotal role in regulating p53 stability and activity. These
results suggest that OTUD1 is a novel regulator of p53. In
addition, OTUD1 has a role in the occurrence and development of
thyroid carcinogenesis (24). OTUD1
is a metastasis suppressor, and its high expression may inhibit
cancer stem cell (CSC) self-renewal and unlimited proliferation,
and prevent metastasis. A study by Zhang et al (25) concluded that the absence of OTUD1
allowed breast cancer cells to undergo epithelial-mesenchymal
transition and acquire CSC traits that promoted metastasis to
distant organs including lungs and bone. However, the
downregulation of OTUD1 expression was associated with an improved
OS compared with that of patients with high OTUD1 expression in
urothelial bladder carcinoma (44).
In addition, the present study revealed that mRNA expression of
OTUD1 had a prognostic value, as OTUD1 was downregulated in NSCLC
and high mRNA expression of OTUD1 predicted a longer prognosis.
OTUD2, also termed YOD1 deubiquitinase, is a member
of the OTU DUB family and contains the K11-specific OTU domain
(23,45). Originally, OTUD2 was identified as a
cofactor for protein processing (46). Then, Rumpf et al (46) suggested that it was released from
tumor necrosis factor receptor-associated factor 6 upon
interleukin-1 stimulation and that its depletion enhanced the
canonical activation of NF-κB. Various studies have suggested that
NF-κB was responsible for the malignant metastasis of lung cancer
(47–50). Based on these observations, we
hypothesized that OTUD2 was associated with the metastasis and
prognosis of lung cancer. The results of the present study also
demonstrated that high OTUD2 mRNA expression was significantly
associated with poorer OS in patients with NSCLC.
OTUD3 belongs to the DUB family and contains an OTU
domain with a priority hydrolyzed K6- and K11-linked distal
ubiquitin (45). A Toxoplasma
gondii deubiquitinase within the OTU family, TgOTUD3A, is the
most similar ortholog of human OTUD3 in terms of structure
(51). TgOTUD3A mRNA serves a key
role in cell cycle regulation and exhibits decreased expression in
the G1 phase of the cell cycle (52). Furthermore, as suggested by Yuan
et al (27), OTUD3 may
exhibit a tumor-suppressive role. The present study also revealed
that increased expression levels of OTUD3 mRNA were significantly
associated with favorable OS in patients with NSCLC. Therefore,
OTUD3 may inhibit the growth of NSCLC cells.
OTUD4 is an additional novel deubiquitinase that is
considered to serve a role in DNA alkylation repair (28). However, its role in cancer has not
yet been explored. In the present study, it was observed that OTUD4
was highly expressed in patients with NSCLC and Ade, where it was a
good prognostic marker according to Kaplan-Meier analysis, but not
in SCC.
OTUD5 or DUBA is a 571-amino-acid protein (53). It is a deubiquitinase that regulates
the production of type I interferon to regulate tissue factor R3
signaling (54). Park et al
(55) identified the programmed cell
death 5-OTUD5 network as a central hub for regulating p53-mediated
apoptosis. p53 is an important gene in tumor growth and metastasis.
However, the results of the present did not identify a significant
association between OTUD5 and NSCLC prognosis.
To the best of our knowledge, OTUD6A has not been
described in the relevant literature to date. OTUD6B is also a DUB.
It is a cleavable ubiquitin-linked protease that has recently been
demonstrated to be involved in regulating B-cell proliferation
following cytokine stimulation. Santiago-Sim et al (55) observed that OTUD6B was associated
with a severe intellectual disability syndrome. However, there is
no relevant information on the role of this molecule in cancer,
particularly in lung cancer. The results from the present study
demonstrated that high mRNA expression of OTUD6B in Ade is
associated with a good prognosis, whereas in SCC it exhibited an
inverse association, as patients with increased mRNA expression of
OTUD6B had poorer prognosis.
ALG13 is a highly conserved protein in the majority
of eukaryotes and also belongs to the OTU family (56). De Antonellis et al (57) observed that ALG13 has been identified
as an early target of microRNA-34a, with relevance to neuroblastoma
tumorigenesis. However, to the best of our knowledge, expression of
ALG13 in NSCLC has not been detected to date. In the previously
described results of the present study, the mRNA expression of
ALG13 was decreased in patients with Ade and SCC compared with that
in normal tissues. High mRNA expression of ALG13 did not exhibit a
significant association with the prognosis of patients with SCC,
but did predict an improved OS in patients with NSCLC and Ade.
In conclusion, the present study evaluated the
differential expression of OTUDs in NSCLC and normal tissues, and
the results revealed that the expression levels of OTUD1 and ALG13
were decreased in NSCLC compared with normal lung tissue according
to Oncomine analysis. In the Kapan-Meier analysis, the effect of 7
OTUDs on the prognoses of patients with NSCLC was analyzed, and it
was demonstrated that increased expression levels of OTUD1, OTUD3,
OTUD4 and ALG13 were associated with increased OS in patients with
NSCLC and Ade, but not in patients with SCC. Similarly, increased
expression of OTUD2 mRNA indicated poorer prognosis in all NSCLC
cases, but no association was observed in the Ade or SCC patient
cohorts. Increased mRNA expression of OTUD5 was not associated with
OS in NSCLC, Ade or SCC. In addition, the increased expression of
OTUD6B in patients with NSCLC was not associated with survival.
These data reveal the complexity and heterogeneity of the molecular
biology of lung cancer, and may provide novel avenues for prognosis
prediction, although the mechanism of its carcinogenicity and the
investigation of novel drug treatment targets requires additional
analysis in future studies.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Funds from
The Key Discipline of Jiaxing Respiratory, Medicine Construction
Project (grant no. 04-Z-11); ‘Early Diagnosis and Comprehensive
Treatment of Lung Cancer Innovation Team Building’ Project of
Zhejiang.
Availability of data and materials
The datasets analyzed in the present study are
available in the Oncomine database (http://www.oncomine.org/) and the Kaplan-Meier plotter
[Lung Cancer] (http://kmplot.com/analysis/index.php?p=service&cancer=lung).
Authors' contributions
JJD, JLL and XDL conceived the study and wrote and
revised the manuscript. JJD and XDL reviewed, collected and
analyzed the data. JJD, GXH and ZXF designed the study and acquired
the data. All authors contributed to the writing of the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre L, Bray F, Siegel R, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Goldstraw P, Ball D, Jett J, Le Chevalier
T, Lim E, Nicholson AG and Shepherd FA: Non-small-cell lung cancer.
Lancet. 378:1727–1740. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ramalingam S, Owonikoko T and Khuri F:
Lung cancer: New biological insights and recent therapeutic
advances. CA Cancer J Clin. 61:91–112. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chaffer C and Weinberg R: A perspective on
cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ramalingam S and Belani C: Systemic
chemotherapy for advanced non-small cell lung cancer: Recent
advances and future directions. Oncologist. 13 (Suppl 1):S5–S13.
2008. View Article : Google Scholar
|
|
7
|
Detterbeck F, Boffa D and Tanoue L: The
new lung cancer staging system. Chest. 136:260–271. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lothrop AP, Torres MP and Fuchs SM:
Deciphering post-translational modification codes. FEBS Lett.
587:1247–1257. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Husnjak K and Dikic I: Ubiquitin-binding
proteins: Decoders of ubiquitin-mediated cellular functions. Annu
Rev Biochem. 81:291–322. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Komander D, Clague M and Urbé S: Breaking
the chains: Structure and function of the deubiquitinases. Nat Rev
Mol Cell Biol. 10:550–563. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Clague M, Barsukov I, Coulson J, Liu H,
Rigden D and Urbé S: Deubiquitylases from genes to organism.
Physiol Rev. 93:1289–1315. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Swatek KN and Komander D: Ubiquitin
modifications. Cell Res. 26:399–422. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen ZJ and Sun LJ: Nonproteolytic
functions of ubiquitin in cell signaling. Molecular cell.
33:275–286. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sippl W, Collura V and Colland F:
Ubiquitin-specific proteases as cancer drug targets. Future Oncol.
7:619–632. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
McClurg UL and Robson CN: Deubiquitinating
enzymes as oncotargets. Oncotarget. 6:9657–9668. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kee Y and Huang TT: Role of
Deubiquitinating enzymes in DNA repair. Mol Cell Biol. 36:524–544.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Clague MJ, Coulson JM and Urbé S: Cellular
functions of the DUBs. J Cell Sci. 125:277–286. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang JM: Emerging roles of
deubiquitinating enzymes in human cancer. Acta Pharmacol Sin.
28:1325–1330. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schwickart M, Huang X, Lill JR, Liu J,
Ferrando R, French DM, Maecker H, O'Rourke K, Bazan F,
Eastham-Anderson J, et al: Deubiquitinase USP9X stabilizes MCL1 and
promotes tumour cell survival. Nature. 463:103–107. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Priolo C, Tang D, Brahamandan M, Benassi
B, Sicinska E, Ogino S, Farsetti A, Porrello A, Finn S, Zimmermann
J, et al: The isopeptidase USP2a protects human prostate cancer
from apoptosis. Cancer Res. 66:8625–8632. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Popov N, Wanzel M, Madiredjo M, Zhang D,
Beijersbergen R, Bernards R, Moll R, Elledge SJ and Eilers M: The
ubiquitin-specific protease USP28 is required for MYC stability.
Nat Cell Biol. 9:765–774. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Coyne ES and Wing SS: The business of
deubiquitination-location, location, location. F1000Res. 5:F1000
Faculty Rev-163. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mevissen TE, Hospenthal MK, Geurink PP,
Elliott PR, Akutsu M, Arnaudo N, Ekkebus R, Kulathu Y, Wauer T, El
Oualid F, et al: OTU deubiquitinases reveal mechanisms of linkage
specificity and enable ubiquitin chain restriction analysis. Cell.
154:169–184. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Carneiro AP, Reis CF, Morari EC, Maia YC,
Nascimento R, Bonatto JM, de Souza MA, Goulart LR and Ward LS: A
putative OTU domain-containing protein 1 deubiquitinating enzyme is
differentially expressed in thyroid cancer and identifies
less-aggressive tumours. Br J Cancer. 111:551–558. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Z, Fan Y, Xie F, Zhou H, Jin K, Shao
L, Shi W, Fang P, Yang B, van Dam H, et al: Breast cancer
metastasis suppressor OTUD1 deubiquitinates SMAD7. Nat Commun.
8:21162017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Piao S, Pei HZ, Huang B and Baek SH:
Ovarian tumor domain-containing protein 1 deubiquitinates and
stabilizes p53. Cell Signal. 33:22–29. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yuan L, Lv Y, Li H, Gao H, Song S, Zhang
Y, Xing G, Kong X, Wang L, Li Y, et al: Deubiquitylase OTUD3
regulates PTEN stability and suppresses tumorigenesis. Nat Cell
Biol. 17:1169–1181. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao Y, Majid MC, Soll JM, Brickner JR,
Dango S and Mosammaparast N: Noncanonical regulation of alkylation
damage resistance by the OTUD4 deubiquitinase. EMBO J.
34:1687–1703. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Luo J, Lu Z, Lu X, Chen L, Cao J, Zhang S,
Ling Y and Zhou X: OTUD5 regulates p53 stability by
deubiquitinating p53. PLoS One. 8:e776822013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sobol A, Askonas C, Alani S, Weber MJ,
Ananthanarayanan V, Osipo C and Bocchetta M: Deubiquitinase OTUD6B
isoforms are important regulators of growth and proliferation. Mol
Cancer Res. 15:117–127. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rhodes D, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rhodes D, Kalyana-Sundaram S, Mahavisno V,
Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal
C, Kulkarni P, et al: Oncomine 3.0: Genes, pathways, and networks
in a collection of 18,000 cancer gene expression profiles.
Neoplasia. 9:166–180. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Györffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Győrffy B, Surowiak P, Budczies J and
Lánczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8:e822412013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ivanova L, Zandberga E, Siliņa K, Kalniņa
Z, Ābols A, Endzeliņš E, Vendina I, Romanchikova N, Hegmane A,
Trapencieris P, et al: Prognostic relevance of carbonic anhydrase
IX expression is distinct in various subtypes of breast cancer and
its silencing suppresses self-renewal capacity of breast cancer
cells. Cancer Chemother Pharmacol. 75:235–246. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wertz IE, Newton K, Seshasayee D¸, Kusam
S, Lam C, Zhang J, Popovych N, Helgason E, Schoeffler A, Jeet S, et
al: Phosphorylation and linear ubiquitin direct A20 inhibition of
inflammation. Nature. 528:370–375. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xu Z, Zheng Y, Zhu Y, Kong X and Hu L:
Evidence for OTUD-6B participation in B lymphocytes cell cycle
after cytokine stimulation. PLoS One. 6:e145142011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ettinger DS, Wood DE, Aisner DL, Akerley
W, Bauman J, Chirieac LR, D'Amico TA, DeCamp MM, Dilling TJ,
Dobelbower M, et al: Non-small cell lung cancer, version 5.2017,
NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc
Netw. 15:504–535. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hou J, Aerts J, den Hamer B, van Ijcken W,
den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens
JA, Hoogsteden HC, et al: Gene expression-based classification of
non-small cell lung carcinomas and survival prediction. PLoS One.
5:e103122010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Garber ME, Troyanskaya OG, Schluens K,
Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen
GD, Perou CM, Whyte RI, et al: Diversity of gene expression in
adenocarcinoma of the lung. Proc Natl Acad Sci USA. 98:13784–13789.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yamagata N, Shyr Y, Yanagisawa K, Edgerton
M, Dang TP, Gonzalez A, Nadaf S, Larsen P, Roberts JR, Nesbitt JC,
et al: A training-testing approach to the molecular classification
of resected non-small cell lung cancer. Clin Cancer Res.
9:4695–4704. 2003.PubMed/NCBI
|
|
42
|
Selamat SA, Chung BS, Girard L, Zhang W,
Zhang Y, Campan M, Siegmund KD, Koss MN, Hagen JA, Lam WL, et al:
Genome-scale analysis of DNA methylation in lung adenocarcinoma and
integration with mRNA expression. Genome Res. 22:1197–1211. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Makarova KS, Aravind L and Koonin EV: A
novel superfamily of predicted cysteine proteases from eukaryotes,
viruses and Chlamydia pneumoniae. Trends Biochem Sci. 25:50–52.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cancer Genome Atlas Research Network, .
Comprehensive molecular characterization of urothelial bladder
carcinoma. Nature. 507:315–322. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Flierman D, van der Heden van Noort GJ,
Ekkebus R, Geurink PP, Mevissen TE, Hospenthal MK, Komander D and
Ovaa H: Non-hydrolyzable diubiquitin probes reveal linkage-specific
reactivity of deubiquitylating enzymes mediated by S2 pockets. Cell
Chem Biol. 23:472–482. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Rumpf S and Jentsch S: Functional division
of substrate processing cofactors of the ubiquitin-selective Cdc48
chaperone. Mol Cell. 21:261–269. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li X, Wang S, Zhu R, Li H, Han Q and Zhao
RC: Lung tumor exosomes induce a pro-inflammatory phenotype in
mesenchymal stem cells via NFκB-TLR signaling pathway. J Hematol
Oncol. 9:422016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lu Z, Li Y, Wang J, Che Y, Sun S, Huang J,
Chen Z and He J: Long non-coding RNA NKILA inhibits migration and
invasion of non-small cell lung cancer via NF-κB/Snail pathway. J
Exp Clin Cancer Res. 36:542017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Richardson JSM, Aminudin N and Abd Malek
SN: Chalepin: A compound from Ruta angustifolia L. pers
exhibits cell cycle arrest at S phase, suppresses nuclear
factor-kappa B (NF-κB) pathway, signal transducer and activation of
transcription 3 (STAT3) phosphorylation and extrinsic apoptotic
pathway in non-small cell lung cancer carcinoma (A549). Pharmacogn
Mag. 13 (Suppl 3):S489–S498. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Tang X, Sun L, Wang G, Chen B and Luo F:
RUNX1: A regulator of NF-kB signaling in pulmonary diseases. Curr
Protein Pept Sci. 19:172–178. 2018.PubMed/NCBI
|
|
51
|
Dhara A and Sinai AP: A cell
cycle-regulated toxoplasma deubiquitinase, TgOTUD3A, targets
polyubiquitins with specific lysine linkages. mSphere. 1:e00085–16.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Behnke MS, Wootton JC, Lehmann MM, Radke
JB, Lucas O, Nawas J, Sibley LD and White MW: Coordinated
progression through two subtranscriptomes underlies the tachyzoite
cycle of Toxoplasma gondii. PLoS One. 5:e123542010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Huang OW, Ma X, Yin J, Flinders J, Maurer
T, Kayagaki N, Phung Q, Bosanac I, Arnott D, Dixit VM, et al:
Phosphorylation-dependent activity of the deubiquitinase DUBA. Nat
Struct Mol Biol. 19:171–175. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kayagaki N, Phung Q, Chan S, Chaudhari R,
Quan C, O'Rourke KM, Eby M, Pietras E, Cheng G, Bazan JF, et al:
DUBA: A deubiquitinase that regulates type I interferon production.
Science. 318:1628–1632. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Santiago-Sim T, Burrage LC, Ebstein F,
Tokita MJ, Miller M, Bi W, Braxton AA, Rosenfeld JA, Shahrour M,
Lehmann A, et al: Biallelic variants in OTUD6B cause an
intellectual disability syndrome associated with seizures and
dysmorphic features. Am J Hum Genet. 100:676–688. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang X, Weldeghiorghis T, Zhang G,
Imperiali B and Prestegard JH: Solution structure of Alg13: The
sugar donor subunit of a yeast N-acetylglucosamine transferase.
Structure. 16:965–975. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
De Antonellis P, Carotenuto M,
Vandenbussche J, De Vita G, Ferrucci V, Medaglia C, Boffa I,
Galiero A, Di Somma S, Magliulo D, et al: Early targets of miR-34a
in neuroblastoma. Mol Cell Proteomics. 13:2114–2131. 2014.
View Article : Google Scholar : PubMed/NCBI
|