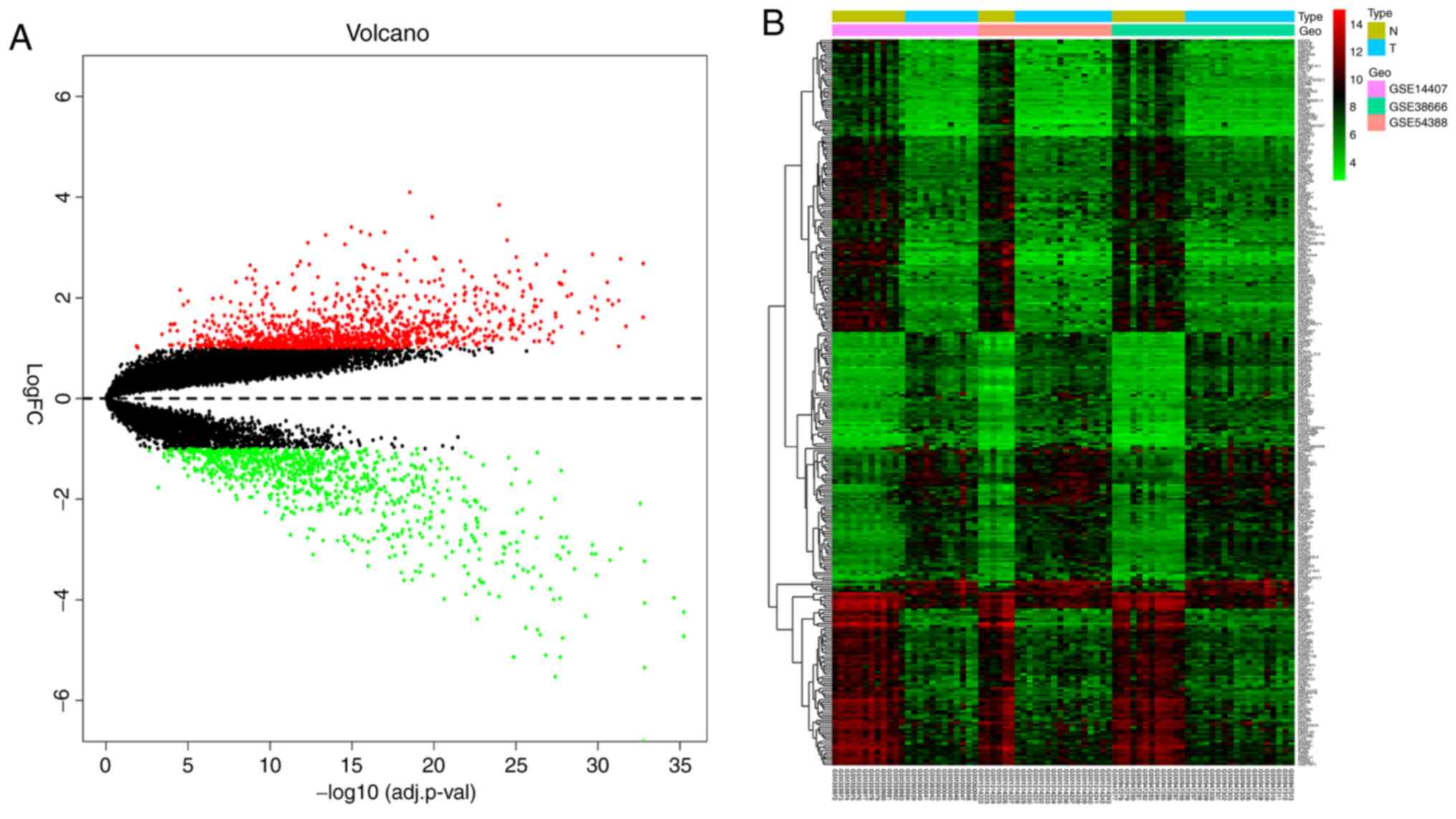
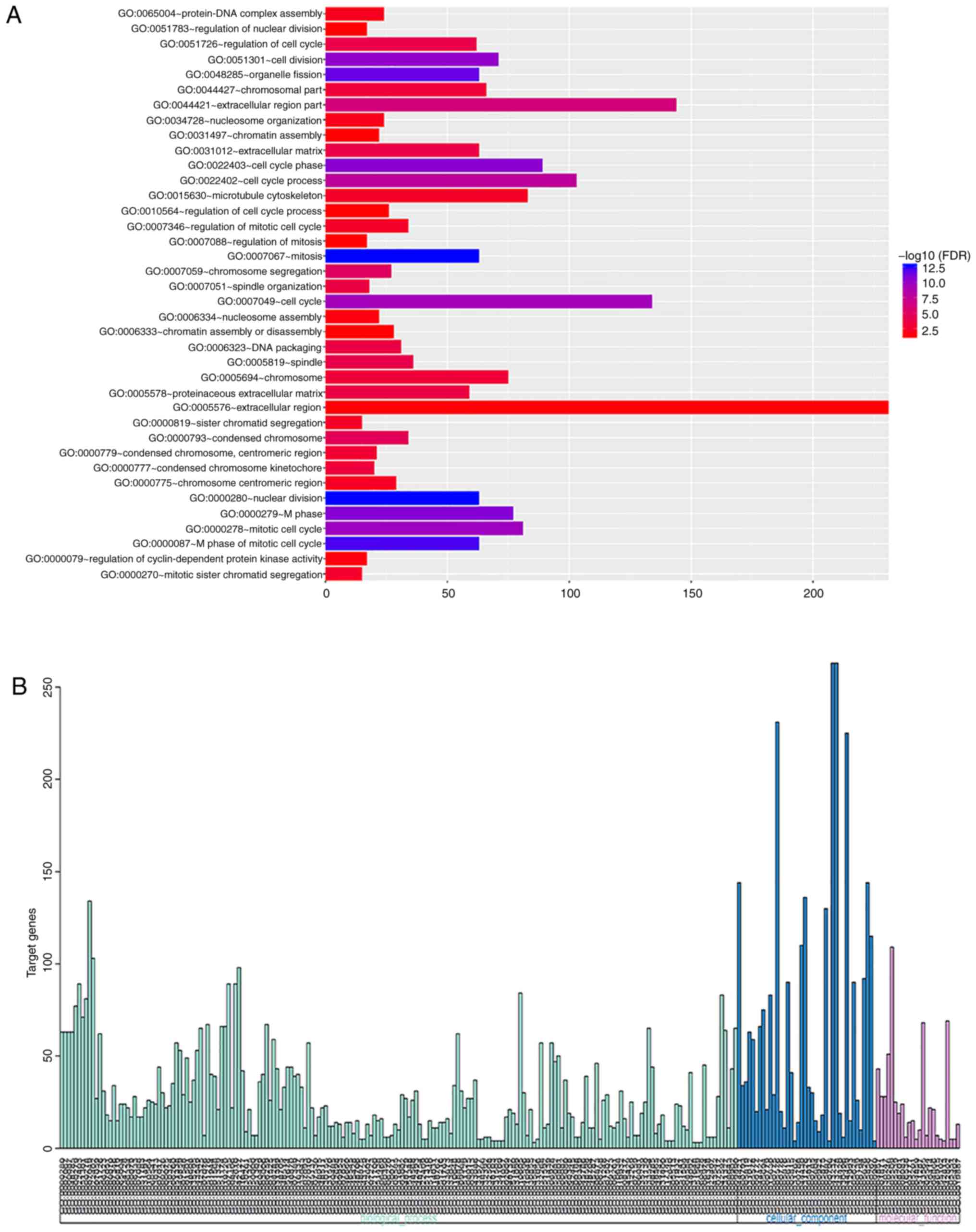
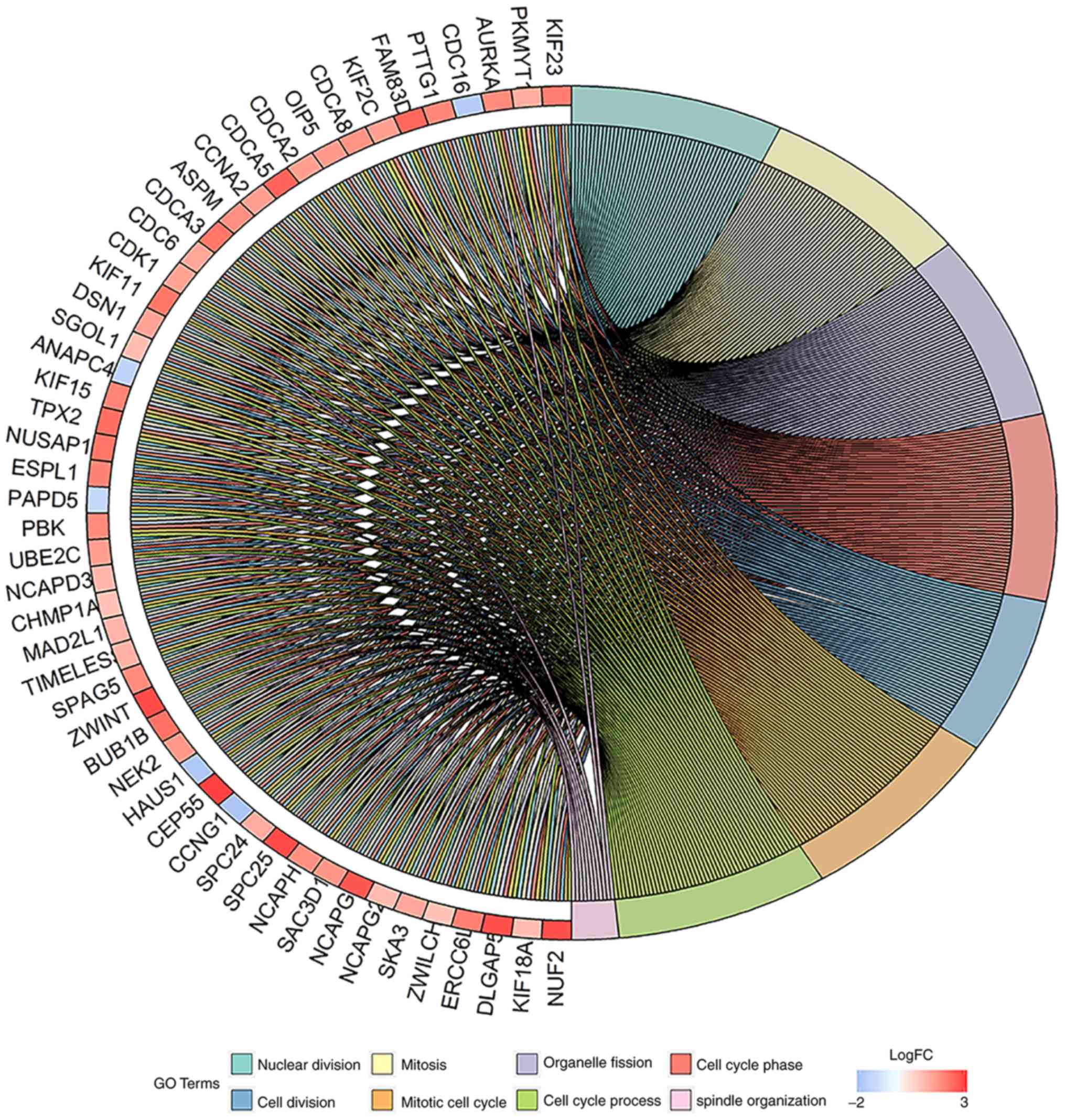
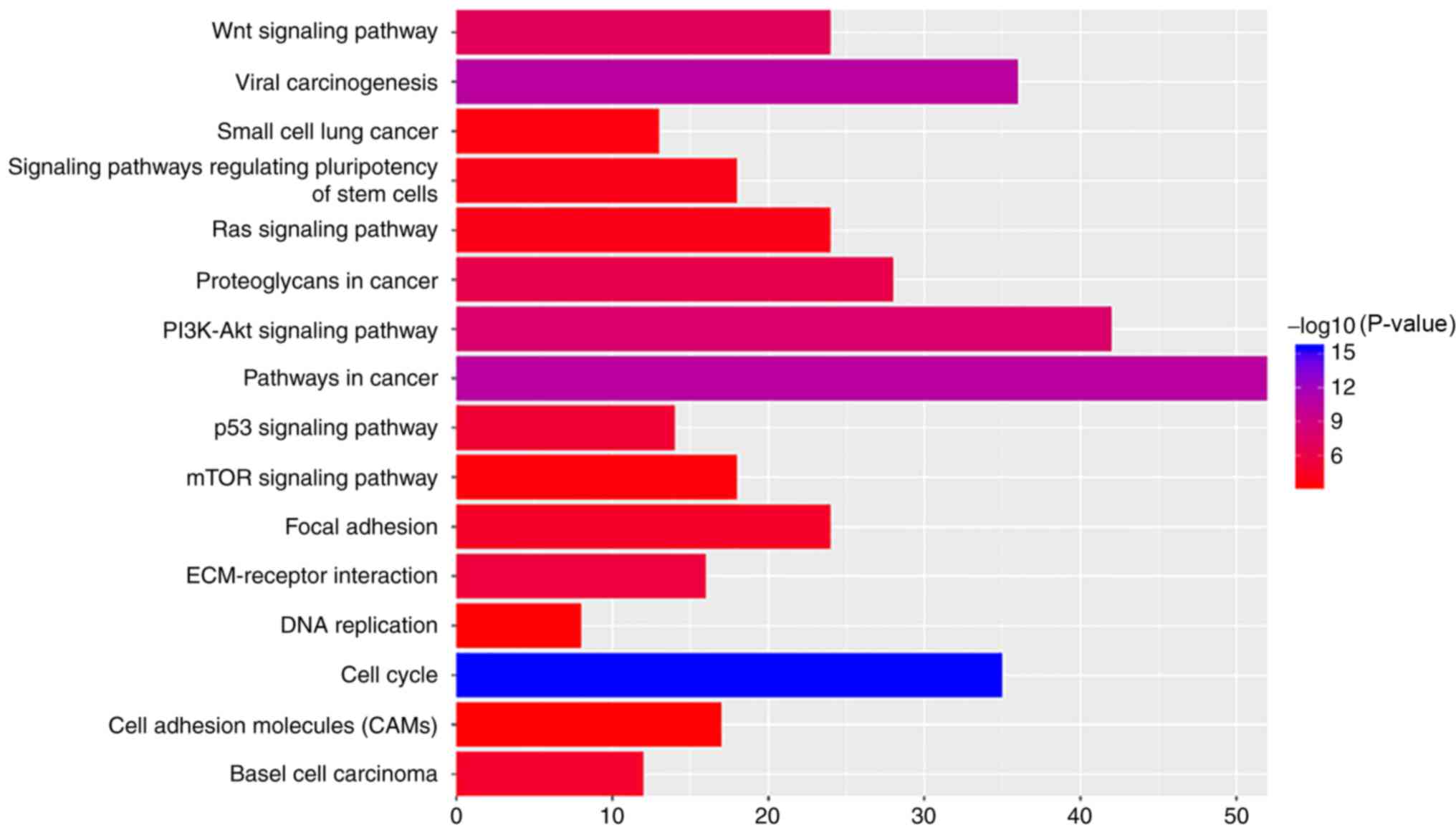
|
1
|
Emmings E, Mullany S, Chang Z, Landen CN
Jr, Linder S and Bazzaro M: Targeting mitochondria for treatment of
chemoresistant ovarian cancer. Int J Mol Sci. 20(pii): E2292019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhou Y, Layton O and Hong L:
Identification of genes and pathways involved in ovarian epithelial
cancer by bioinformatics analysis. J Cancer. 9:3016–3022. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cortez AJ, Tudrej P, Kujawa KA and
Lisowska KM: Advances in ovarian cancer therapy. Cancer Chemother
Pharmacol. 81:17–38. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Szabova L, Bupp S, Kamal M, Householder
DB, Hernandez L, Schlomer JJ, Baran ML, Yi M, Stephens RM,
Annunziata CM, et al: Pathway-specific engineered mouse allograft
models functionally recapitulate human serous epithelial ovarian
cancer. PLoS One. 9:e956492014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rojas V, Hirshfield KM, Ganesan S and
Rodriguez-Rodriguez L: Molecular characterization of epithelial
ovarian cancer: Implications for diagnosis and treatment. Int J Mol
Sci. 17(pii): E21132016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lengauer C, Kinzler KW and Vogelstein B:
Genetic instabilities in human cancer. Nature. 396:643–649. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Previs RA, Sood AK, Mills GB and Westin
SN: The rise of genomic profiling in ovarian cancer. Expert Rev Mol
Diagn. 16:1337–1351. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sanchez-Pena ML, Isaza CE, Perez-Morales
J, Rodriguez-Padilla C, Castro JM and Cabrera-Rios M:
Identification of potential biomarkers from microarray experiments
using multiple criteria optimization. Cancer Med. 2:253–265. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Su LJ, Hsu SL, Yang JS, Tseng HH, Huang SF
and Huang CY: Global gene expression profiling of
dimethylnitrosamine-induced liver fibrosis: From pathological and
biochemical data to microarray analysis. Gene Expr. 13:107–132.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Muller C, Schillert A, Rothemeier C,
Trégouët DA, Proust C, Binder H, Pfeiffer N, Beutel M, Lackner KJ,
Schnabel RB, et al: Removing batch effects from longitudinal gene
Expression-Quantile normalization plus combat as best approach for
microarray transcriptome data. PLoS One. 11:e01565942016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Clough E and Barrett T: The gene
expression omnibus database. Methods Mol Biol. 1418:93–110. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bowen NJ, Walker LD, Matyunina LV, Logani
S, Totten KA, Benigno BB and McDonald JF: Gene expression profiling
supports the hypothesis that human ovarian surface epithelia are
multipotent and capable of serving as ovarian cancer initiating
cells. BMC Med Genomics. 2:712009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yeung TL, Leung CS, Wong KK,
Gutierrez-Hartmann A, Kwong J, Gershenson DM and Mok SC: ELF3 is a
negative regulator of epithelial-mesenchymal transition in ovarian
cancer cells. Oncotarget. 8:16951–16963. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lili LN, Matyunina LV, Walker LD, Benigno
BB and McDonald JF: Molecular profiling predicts the existence of
two functionally distinct classes of ovarian cancer stroma. Biomed
Res Int. 2013:8463872013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fasold M and Binder H: Estimating
RNA-quality using GeneChip microarrays. BMC Genomics. 13:1862012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Leek JT, Johnson WE, Parker HS, Jaffe AE
and Storey JD: The Sva package for removing batch effects and other
unwanted variation in high-throughput experiments. Bioinformatics.
28:882–883. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drug. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Braun P and Gingras AC: History of
protein-protein interactions: From egg-white to complex networks.
Proteomics. 12:1478–1498. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang K, Kong X, Feng G, Xiang W, Chen L,
Yang F, Cao C, Ding Y, Chen H, Chu M, et al: Investigation of
hypoxia networks in ovarian cancer via bioinformatics analysis. J
Ovarian Res. 11:162018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rhodes DR, Kalyana-Sundaram S, Mahavisno
V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ,
Kincead-Beal C, Kulkarni P, et al: Oncomine 3.0: Genes, pathways,
and networks in a collection of 18,000 cancer gene expression
profiles. Neoplasia. 9:166–180. 2017. View Article : Google Scholar
|
|
24
|
Gyorffy B, Lánczky A and Szállási Z:
Implementing anonline tool for genome-wide validation of
survival-associated biomarkers in ovarian-cancer using microarray
data from 1287 patients. Endocr Relat Cancer. 19:197–208. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Drakes ML, Mehrotra S, Aldulescu M, Potkul
RK, Liu Y, Grisoli A, Joyce C, O'Brien TE, Stack MS and Stiff PJ:
Stratification of ovarian tumor pathology by expression of
programmed cell Death-1 (Pd-1) and Pd-Ligand-1 (Pd-L1) in ovarian
cancer. J Ovarian Res. 11:432018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Moufarrij S, Dandapani M, Arthofer E,
Gomez S, Srivastava A, Lopez-Acevedo M, Villagra A and Chiappinelli
KB: Epigenetic therapy for ovarian cancer: Promise and progress.
Clin Epigenetics. 11:72019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hennessy BT, Coleman RL and Markman M:
Ovarian cancer. Lancet. 374:1371–1382. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Baron JA: Screening for cancer with
molecular markers: Progress comes with potential problem. Nat Rev
Cancer. 12:368–371. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Soon WW, Hariharan M and Snyder MP:
High-throughput sequencing for biology and medicine. Mol Syst Biol.
9:6402013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Duffy MJ: Use of biomarkers in screening
for cancer. Adv Exp Med Biol. 867:27–39. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Vlasova-St Louis I and Bohjanen PR:
Post-transcriptional regulation of cytokine and growth factor
signaling in cancer. Cytokine Growth Factor Rev. 33:83–93. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Peyressatre M, Prevel C, Pellerano M and
Morris MC: Targeting cyclin-dependent kinases in human cancers:
From small molecules to Peptide inhibitors. Cancers (Basel).
7:179–237. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Garcia-Reyes B, Kretz AL, Ruff JP, von
Karstedt S, Hillenbrand A, Knippschild U, Henne-Bruns D and Lemke
J: The emerging role of Cyclin-dependent kinases (CDKs) in
pancreatic ductal adenocarcinoma. Int J Mol Sci. 19(pii):
E32192018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mohammed MK, Shao C, Wang J, Wei Q, Wang
X, Collier Z, Tang S, Liu H, Zhang F, Huang J, et al: Wnt/β-Catenin
signaling plays an ever-expanding role in stem cell self-renewal,
tumorigenesis and cancer chemoresistance. Genes Dis. 3:11–40. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhan T, Rindtorff N and Boutros M: Wnt
signaling in cancer. Oncogene. 36:1461–1473. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Clements WM, Wang J, Sarnaik A, Kim OJ,
MacDonald J, Fenoglio-Preiser C, Groden J and Lowy AM: Beta-Catenin
mutation is a frequent cause of Wnt pathway activation in gastric
cancer. Cancer Res. 62:3503–3506. 2002.PubMed/NCBI
|
|
37
|
Parker AL, Kavallaris M and McCarroll JA:
Microtubules and their role in cellular stress in cancer. Front
Oncol. 4:1532014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Burgess RJ and Zhang Z: Histone chaperones
in nucleosome assembly and human disease. Nat Struct Mol Biol.
20:14–22. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Polo SE, Theocharis SE, Grandin L,
Gambotti L, Antoni G, Savignoni A, Asselain B, Patsouris E and
Almouzni G: Clinical significance and prognostic value of chromatin
assembly factor-1 overexpression in human solid tumours.
Histopathology. 57:716–724. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Huang J, Zhang L, Greshock J, Colligon TA,
Wang Y, Ward R, Katsaros D, Lassus H, Butzow R, Godwin AK, et al:
Frequent genetic abnormalities of the PI3K/AKT pathway in primary
ovarian cancer predict patient outcome. Genes Chromosomes Cancer.
50:606–618. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chien AJ, Conrad WH and Moon RT: A Wnt
survival guide: From flies to human disease. J Invest Dermatol.
129:1614–1627. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bitler BG, Nicodemus JP, Li H, Cai Q, Wu
H, Hua X, Li T, Birrer MJ, Godwin AK, Cairns P and Zhang R: Wnt5a
suppresses epithelial ovarian cancer by promoting cellular
senescence. Cancer Res. 71:6184–6194. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bodnar L, Stanczak A, Cierniak S, Smoter
M, Cichowicz M, Kozlowski W, Szczylik C, Wieczorek M and
Lamparska-Przybysz M: Wnt/β-catenin pathway as a potential
prognostic and predictive marker in patients with advanced ovarian
cancer. J Ovarian Res. 7:162014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Conciatori F, Ciuffreda L, Bazzichetto C,
Falcone I, Pilotto S, Bria E, Cognetti F and Milella M: mTOR
cross-talk in cancer and potential for combination therapy. Cancers
(Basel). 10:E232018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Saxton RA and Sabatini DM: mTOR signaling
in growth, metabolism, and disease. Cell. 169:361–371. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Evan GI and Vousden KH: Proliferation,
cell cycle and apoptosis in cancer. Nature. 411:342–348. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang R, Shi H, Ren F, Zhang M, Ji P, Wang
W and Liu C: The aberrant upstream pathway regulations of CDK1
protein were implicated in the proliferation and apoptosis of
ovarian cancer cells. J Ovarian Res. 10:602017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sung WW, Lin YM, Wu PR, Yen HH, Lai HW, Su
TC, Huang RH, Wen CK, Chen CY, Chen CJ and Yeh KT: High
nuclear/cytoplasmic ratio of Cdk1 expression predicts poor
prognosis in colorectal cancer patients. BMC Cancer. 14:9512014.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yang W, Cho H, Shin HY, Chung JY, Kang ES,
Lee EJ and Kim JH: Accumulation of cytoplasmic Cdk1 is associated
with cancer growth and survival rate in epithelial ovarian cancer.
Oncotarget. 7:49481–49497. 2016.PubMed/NCBI
|
|
50
|
Ou Y, Ma L, Huang Z, Zhou W, Zhao C, Zhang
B, Song Y, Yu C and Zhan Q: Overexpression of cyclin B1 antagonizes
chemotherapeutic-induced apoptosis through PTEN/Akt pathway in
human esophageal squamous cell carcinoma cells. Cancer Biol Ther.
14:45–55. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bonnet ME, Gossart JB, Benoit E, Messmer
M, Zounib O, Moreau V, Behr JP, Lenne-Samuel N, Kedinger V, Meulle
A, et al: Systemic delivery of sticky siRNAs targeting the cell
cycle for lung tumor metastasis inhibition. J Control Release.
170:183–190. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kedinger V, Meulle A, Zounib O, Bonnet ME,
Gossart JB, Benoit E, Messmer M, Shankaranarayanan P, Behr JP,
Erbacher P and Bolcato-Bellemin AL: Sticky siRNAs targeting
survivin and cyclin B1 exert an antitumoral effect on melanoma
subcutaneous xenografts and lung metastases. BMC Cancer.
13:3382013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Matthess Y, Raab M, Sanhaji M, Lavrik IN
and Strebhardt K: Cdk1/cyclin B1 controls Fas-mediated apoptosis by
regulating caspase-8 activity. Mol Cell Biol. 30:5726–5740. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Soria JC, Jang SJ, Khuri FR, Hassan K, Liu
D, Hong WK and Mao L: Overexpression of cyclin B1 in early-stage
non-small cell lung cancer and its clinical implication. Cancer
Res. 60:4000–4004. 2000.PubMed/NCBI
|
|
55
|
Nozoe T, Korenaga D, Kabashima A, Ohga T,
Saeki H and Sugimachi K: Significance of cyclin B1 expression as an
independent prognostic indicator of patients with squamous cell
carcinoma of the esophagus. Clin Cancer Res. 8:817–822.
2002.PubMed/NCBI
|
|
56
|
Ding K, Li W, Zou Z, Zou X and Wang C:
CCNB1 is a prognostic biomarker for ER+ breast cancer. Med
Hypotheses. 83:359–364. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Weng L, Du J, Zhou Q, Cheng B, Li J, Zhang
D and Ling C: Identification of cyclin B1 and Sec 62 as biomarkers
for recurrence in patients with HBV-related hepatocellular
carcinoma after surgical resection. Mol Cancer. 11:392012.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lee KS, Burke TR Jr, Park JE, Bang JK and
Lee E: Recent Advances and new strategies in targeting Plk1 for
anticancer therapy. Trends Pharmacol Sci. 36:858–877. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Iyer RS, Nicol SM, Quinlan PR, Thompson
AM, Meek DW and Fuller-Pace FV: The RNA Helicase/Transcriptional
Co-regulator, P68 (DDX5), stimulates expression of oncogenic
protein kinase, Polo-like Kinase-1 (PLK1), and is associated with
elevated PLK1 levels in human breast cancers. Cell Cycle.
13:1413–1423. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Liu Z, Sun Q and Wang X: PLK1, A potential
target for cancer therapy. Transl Oncol. 10:22–32. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Weichert W, Denkert C, Schmidt M, Gekeler
V, Wolf G, Köbel M, Dietel M and Hauptmann S: Polo-like kinase
isoform expression is a prognostic factor in ovarian carcinoma. Br
J Cancer. 90:815–821. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
de Carcer G, Venkateswaran SV, Salgueiro
L, El Bakkali A, Somogyi K, Rowald K, Montañés P, Sanclemente M,
Escobar B, de Martino A, et al: Plk1 overexpression induces
chromosomal instability and suppresses tumor development. Nat
Commun. 9:30122018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Yasuda H, Lindorfer MA, Myung CS and
Garrison JC: Phosphorylation of the G protein gamma12 subunit
regulates effector specificity. J Biol Chem. 273:21958–21965. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Larson K.C, Lipko M, Dabrowski M and
Draper MP: Gng12 is a novel negative regulator of LPS-induced
inflammation in the microglial cell line BV-2. Inflamm Res.
59:15–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Luo C, Zhao S, Dai W, Zheng N and Wang J:
Proteomic analyses reveal GNG12 regulates cell growth and casein
synthesis by activating the Leu-mediated mTORC1 signaling pathway.
Biochim Biophys Acta Proteins Proteom. 1866:1092–1101. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Hao X and Qu T: Expression of CENPE and
its prognostic role in non-small cell lung cancer. Open Med (Wars).
14:497–502. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Balamuth NJ, Wood A, Wang Q, Jagannathan
J, Mayes P, Zhang Z, Chen Z, Rappaport E, Courtright J, Pawel B, et
al: Serial transcriptome analysis and cross-species integration
identifies centromere-associated protein E as a novel neuroblastoma
target. Cancer Res. 70:2749–2758. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Bieche I, Vacher S, Lallemand F,
Tozlu-Kara S, Bennani H, Beuzelin M, Driouch K, Rouleau E,
Lerebours F, Ripoche H, et al: Expression analysis of mitotic
spindle checkpoint genes in breast carcinoma: Role of NDC80/HEC1 in
early breast tumorigenicity, and a two-gene signature for
aneuploidy. Mol Cancer. 10:232011. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Bolanos-Garcia VM and Blundell TL: BUB1
and BUBR1: Multifaceted kinases of the cell cycle. Trends Biochem
Sci. 36:141–150. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Karess RE, Wassmann K and Rahmani Z: New
insights into the role of BubR1 in mitosis and beyond. Int Rev Cell
Mol Biol. 306:223–273. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Elowe S: Bub1 and BubR1: At the interface
between chromosome attachment and the spindle checkpoint. Mol Cell
Biol. 31:3085–3093. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Hudler P, Britovsek NK, Grazio SF and
Komel R: Association between polymorphisms in segregation genes
BUB1B and TTK and gastric cancer risk. Radiol Oncol. 50:297–307.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Fu X, Chen G, Cai ZD, Wang C, Liu ZZ, Lin
ZY, Wu YD, Liang YX, Han ZD, Liu JC and Zhong WD: Overexpression of
BUB1B contributes to progression of prostate cancer and predicts
poor outcome in patients with prostate cancer. Onco Targets Ther.
9:2211–2220. 2016.PubMed/NCBI
|
|
74
|
Hahn MM, Vreede L, Bemelmans SA, van der
Looij E, van Kessel AG, Schackert HK, Ligtenberg MJ, Hoogerbrugge
N, Kuiper RP and de Voer RM: Prevalence of germline mutations in
the spindle assembly checkpoint gene BUB1B in individuals with
early-onset colorectal cancer. Genes Chromosomes Cancer.
55:855–863. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Mansouri N, Movafagh A, Sayad A, Heidary
Pour A, Taheri M, Soleimani S, Mirzaei HR, Alizadeh Shargh S,
Azargashb E, Bazmi H, et al: Targeting of BUB1b gene expression in
sentinel lymph node biopsies of invasive breast cancer in iranian
female patients. Asian Pac J Cancer Prev. 17:317–321. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Do TV, Xiao F, Bickel LE, Klein-Szanto AJ,
Pathak HB, Hua X, Howe C, O'Brien SW, Maglaty M, Ecsedy JA, et al:
Aurora kinase A mediates epithelial ovarian cancer cell migration
and adhesion. Oncogene. 33:539–549. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Zou Z, Yuan Z, Zhang Q, Long Z, Chen J,
Tang Z, Zhu Y, Chen S, Xu J, Yan M, et al: Aurora kinase a
inhibition-induced autophagy triggers drug resistance in breast
cancer cells. Autophagy. 8:1798–1810. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Reiter R, Gais P, Jutting U, Steuer-Vogt
MK, Pickhard A, Bink K, Rauser S, Lassmann S, Höfler H, Werner M
and Walch A: Aurora kinase a messenger RNA overexpression is
correlated with tumor progression and shortened survival in head
and neck squamous cell carcinoma. Clin Cancer Res. 12:5136–5141.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wan XB, Long ZJ, Yan M, Xu J, Xia LP, Liu
L, Zhao Y, Huang XF, Wang XR, Zhu XF, et al: Inhibition of Aurora-A
suppresses epithelial-mesenchymal transition and invasion by
downregulating MAPK in nasopharyngeal carcinoma cells.
Carcinogenesis. 29:1930–1937. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Mignogna C, Staropoli N, Botta C, De Marco
C, Rizzuto A, Morelli M, Di Cello A, Franco R, Camastra C, Presta
I, et al: Aurora Kinase A expression predicts platinum-resistance
and adverse outcome in high-grade serous ovarian carcinoma
patients. J Ovarian Res. 9:312016. View Article : Google Scholar : PubMed/NCBI
|