Introduction
Originating from adult brain tissue, glioblastoma
(GBM) is a highly malignant primary tumor with numerous
characteristics, including complicated pathological diagnosis, poor
prognosis and easy relapse (1).
Tumor cells have a strong ability for proliferation and invasion,
and can invade the central nervous system (2). In 2018, patients with GBM had a poor
prognosis, and the median survival time of patients was ~17 months
(3). In 2019, following standard
treatment, the 6-month progression-free survival rate was 25.2% in
patients with recurrent glioblastoma (rGBM) (4). Traditional treatment for GBM includes
surgical resection, radiation and chemotherapy with temozolomide
(5). Following surgical resection of
GBM, substantial invading tumor cells are located in the near
resection edge of the tumor or within 2–3 cm from the resection
cavity, and these are the most common recurrence sites (6).
In previous years, with the development of medicine
and bioinformatics, gene expression information has been generated
and stored in the Gene Expression Omnibus (GEO) database.
Bioinformatics analysis allows an improved understanding of the
alterations in essential genes in the process of tumor development.
Furthermore, it can provide a basis for target treatment and tumor
diagnosis (7). A previous study
identified that both the peritumoral brain zone (PBZ) and tumor
core (TC) are heterogeneous in the same patients, and tumor cells
in the PBZ exhibit increased invasiveness compared with cells in
the TC (8). Van Meter et al
(9) demonstrated that there were a
number of different gene expression products between the PBZ and
TC. Even in the absence of tumor cells in the PBZ, it is possible
to detect the expression of genes associated with invasion and
neo-angiogenesis (10). Therefore,
identifying differentially expressed genes (DEGs) between the PBZ
and TC may reveal the causes of GBM recurrence, and may provide a
target for tumor treatment.
In the present study, the gene expression profiles
of the PBZ were compared with the gene expression profiles of the
TC to identify DEGs. The DEGs of the PBZ and TC were screened by
GEO2R online analysis, and the Database for Annotation,
Visualization, and Integrated Discovery (DAVID) was used for Gene
Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)
enrichment analyses. A protein-protein interaction (PPI) network
was constructed using Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING) and Cytoscape software tools, and the hub
genes were obtained using the Cytoscape plug-in Cytohubba.
Furthermore, the Gene Expression Profiling Interactive Analysis
(GEPIA) database was used to analyze the survival rate of patients
with aberrant expression levels of the hub genes.
Materials and methods
Microarray data analysis
GEO (https://www.ncbi.nlm.nih.gov/geo/) is a database that
stores a large amount of genetic information. For analysis,
‘glioblastoma’ was selected as a key word, the tissue type was set
as ‘Homo sapiens’ and the study type was set as ‘expression
profiling by array’. The screening criteria were: i) Samples of
peritumoral brain area and core of GBM; and ii) both samples should
be obtained from the same patient. The two datasets that met the
criteria were GSE13276 and GSE116520. The GSE13276 dataset, which
was submitted by Mangiola et al (2), is based on the GPL96 platform and
contains 5 cases of GBM core samples and 5 cases of GBM surrounding
tissue (no replicate samples included). The GSE116520 dataset,
which was submitted by Kruthika et al (11), is based on the GPL10558 platform and
contains 17 cases of GBM core samples and 17 cases of GBM
surrounding tissue (Table I).
 | Table I.Details of the GEO datasets. |
Table I.
Details of the GEO datasets.
| Author, year | Samples | GEO accession | Country | Platform | PBZ, n | TC, n | Total, n | (Refs.) |
|---|
| Mangiola et
al, 2013 | GBM WHO grade IV
tumor tissue | GSE13276 | Italy | GLP96 | 5 | 5 | 10 | (2) |
| Kruthika et
al, 2019 | GBM WHO grade IV
tumor tissue | GSE116520 | India | GPL10558 | 17 | 17 | 34 | (11) |
Screening of DEGs
DEGs were screened using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/), which
allowed a comparison of the differences between two different sets
of samples in a series. In the GSE13276 dataset, |log fold change
(FC)| ≥2 and P<0.05 were defined as statistically significant
for the DEGs. Similarly, in the GSE116520 dataset, |log FC| ≥1 (and
P<0.05 were defined as statistically significant for the DEGs).
If the same criteria in the two datasets were selected, the
resulting genes would be limited, therefore in order to obtain a
larger number of target genes different criteria were selected.
Excel software (Office 2016; Microsoft Corporation) was used to
delete all genes without gene symbols and duplicate genes
corresponding to multiple probe IDs in the GSE13276 and GSE116520
datasets. A Venn diagram of the DEGs obtained from the
bioinformatics and evolutionary genomics online tool ‘Calculate and
draw custom Venn diagrams’ (http://bioinformatics.psb.ugent.be/webtools/Venn/) was
constructed, and the common DEGs were identified.
GO and KEGG enrichment analysis
GO (http://www.geneontology.org) is a database used to
describe the characteristics of genes and gene products. KEGG
(https://www.kegg.jp/) is a database containing
information on genome, chemistry and system functions (12). DAVID (version 6.8; http://david.ncifcrf.gov/) is an online database, and
was used to perform GO term and KEGG pathway enrichment analyses
(13,14). P<0.05 was considered to indicate a
statistically significant difference.
Interaction network analysis and hub
gene identification
The STRING (https://string-db.org/) database is a tool for
analyzing the interrelationships between genes, including direct
(physical) and indirect (functional) links, and constructing PPI
networks. STRING version 11.0 contains 24,584,628 proteins from
5,090 organisms (15). The PPI
network association among DEGs was analyzed using the STRING
database and the default minimum required interaction score was set
as >0.4. Subsequently, the Cytoscape (version 3.7.1; http://www.cytoscape.org/) plug-in Cytohubba (version
was 0.1; http://apps.cytoscape.org/apps/cytohubba) was used to
screen the PPI network, and the top10 genes, which were the hub
genes in the PBZ, were obtained using the Degree algorithm.
Survival analysis
GEPIA (http://gepia.cancer-pku.cn/detail.php) is a database
used to analyze RNA sequencing expression data, and data of the
tumors (GBM) and normal samples included in The Cancer Genome Atlas
(TCGA: http://portal.gdc.cancer.gov/) and
the Genotype-Tissue Expression (GTEx: http://gtexportal.org/home/) databases were analyzed
(16). All the parameters were set
to the default value, and the cut-off value was median=50%. The GBM
sample was selected as the dataset and the hazard ratio (HR) was
calculated based on the Cox Proportional-Hazards Model. The 95% CI
was not calculated in the present study. For the HR, P<0.05 was
considered to indicate a statistically significant difference.
Results
Screening of DEGs
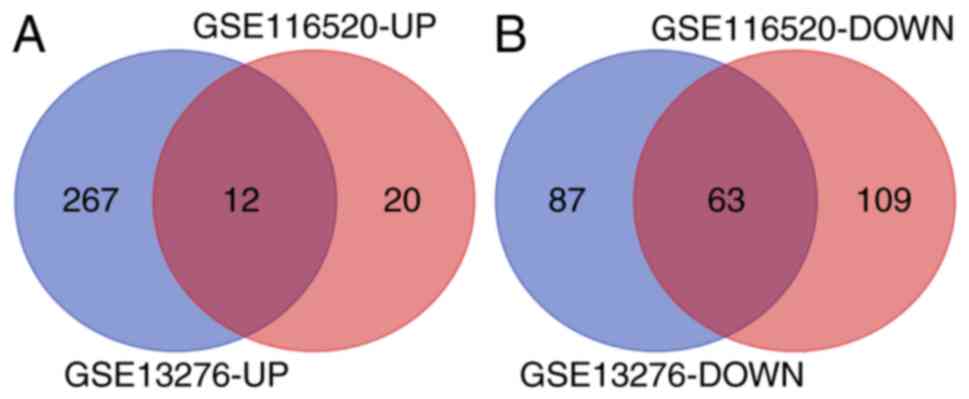
For the GSE13276 dataset, 429 DEGs were identified,
including 279 upregulated (UP) genes and 150 downregulated (DOWN)
genes. For the GSE116520 dataset, 204 DEGs were identified,
including 32 UP genes and 172 DOWN genes. The bioinformatics and
evolutionary genomics webpage tool, Venn, was used to construct the
Venn diagram of the two groups of DEGs, and through the Venn
diagram, 75 common DEGs were identified (Table II), including 12 common UP and 63
common DOWN genes (Fig. 1).
 | Table II.Common differentially expressed
genes, including 12 upregulated and 63 downregulated genes, of the
GSE13276 and GSE116520 datasets. |
Table II.
Common differentially expressed
genes, including 12 upregulated and 63 downregulated genes, of the
GSE13276 and GSE116520 datasets.
| A, Common
upregulated genes |
|---|
|
|---|
| Number of
genes | Gene symbols |
|---|
| 12 | CD163, LOX, ABCC3,
NAMPT, HILPDA, IGFBP3, TREM1, C8orf4, VEGFA, TGFBI, PI3, CXCL8 |
|
| B, Common
downregulated genes |
|
| Number of
genes | Gene
symbols |
|
| 63 | KLK6, GRM3,
RAPGEF5, FA2H, PLLP, CAPN3, PPP1R16B, SH3GL3, RAB40B, PTPRD, GPR37,
CLCA4, ASPA, PTGDS, SOX10, MBP, HSPA2, ABCA2, MAP7, MYRF, SEC14L5,
MAP6D1, PKP4, SHTN1, CHN2, STXBP6, PIP4K2A, PAQR6, NINJ2, MAG,
PLEKHB1, SLCO1A2, KCNK1, TSPAN8, ZNF536, SLCO3A1, PLP1, EFHD1,
PEX5L, TF, ENPP2, SLC31A2, LHPP, ALDH1A1, UGT8, CYP2J2, MOG, MAL,
CNTN2, MOBP, BCAS1, CNTNAP2, RASGRP3, ADARB2, ADAP1, NIPAL3, SEPT4,
APLP1, TMEM144, ST18, MVB12B, DAAM2, BIN1 |
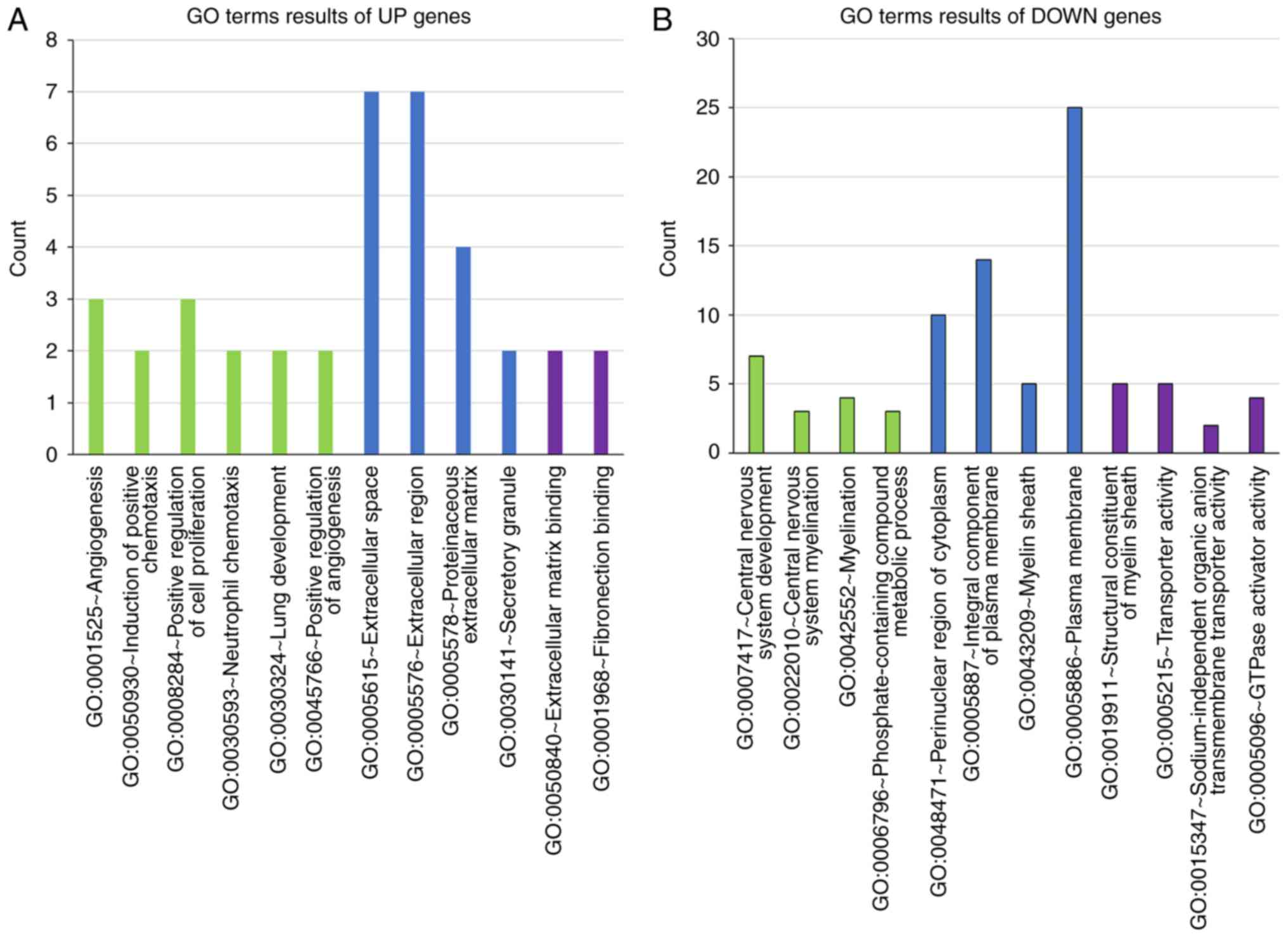
GO term enrichment analysis
The DAVID online tool was used to analyze the GO
terms and KEGG pathways of the DEGs. The GO term enrichment
analysis results demonstrated that the UP genes were significantly
enriched in ‘regulation of angiogenesis’ (P<0.01; Fig. 2A). DOWN genes were significantly
enriched in ‘central nervous system development’ (P<0.01;
Fig. 2B).
KEGG pathway analysis
By analyzing the DEGs carefully, the most
significantly enriched KEGG pathways were identified (Table III). The UP genes were mainly
enriched in ‘bladder cancer’ and the DOWN genes were mainly
enriched in ‘endocytosis’ (Table
III).
 | Table III.Kyoto Encyclopedia of Genes and
Genomes pathway analysis of differentially expressed genes. |
Table III.
Kyoto Encyclopedia of Genes and
Genomes pathway analysis of differentially expressed genes.
| Term | Status | Count, n | P-value | Genes |
|---|
| hsa05219: Bladder
cancer | Upregulated | 2 | 0.023633 | VEGFA, CXCL8 |
| hsa04144:
Endocytosis | Downregulated | 4 | 0.049978 | SH3GL3, HSPA2,
BIN1, MVB12B |
PPI network and hub gene analysis
STRING online software was used to analyze the DEGs
and then a PPI network was constructed to predict the hub genes.
Following analysis of the PPI network, it was identified that 52
nodes and 101 interactions were involved in the PPI network. The
top 10 hub genes were calculated using the Degree algorithm of the
Cytohubba plug-in, as shown in Fig.
3. The 10 hub genes included proteolipid protein 1 (PLP1),
myelin associated oligodendrocyte basic protein (MOBP), contactin 2
(CNTN2), C-X-C motif chemokine ligand 8 (CXCL8), myelin
oligodendrocyte glycoprotein (MOG), vascular endothelial growth
factor A (VEGFA), myelin basic protein (MBP), myelin associated
glycoprotein (MAG), SRY-box transcription factor 10 (SOX10) and
plasmolipin (PLLP). Compared with TC, significant changes in all
these hub genes occurred in the PBZ. Additionally, the DAVID
database was used for GO enrichment and KEGG pathway analyses of
these 10 hub genes. Through GO enrichment analysis, it was
determined that the 10 hub genes were mainly enriched in ‘cell
maturation’, ‘substantia nigra development’ and ‘central nervous
system development’ for biological processes, ‘compact myelin’ and
‘myelin sheath’ for cellular components, and ‘structural
constituent of myelin sheath’ for molecular functions (Table IV). According to the KEGG pathway
analysis, the 10 genes were enriched in ‘bladder cancer’ and
‘rheumatoid arthritis’ (Table
V).
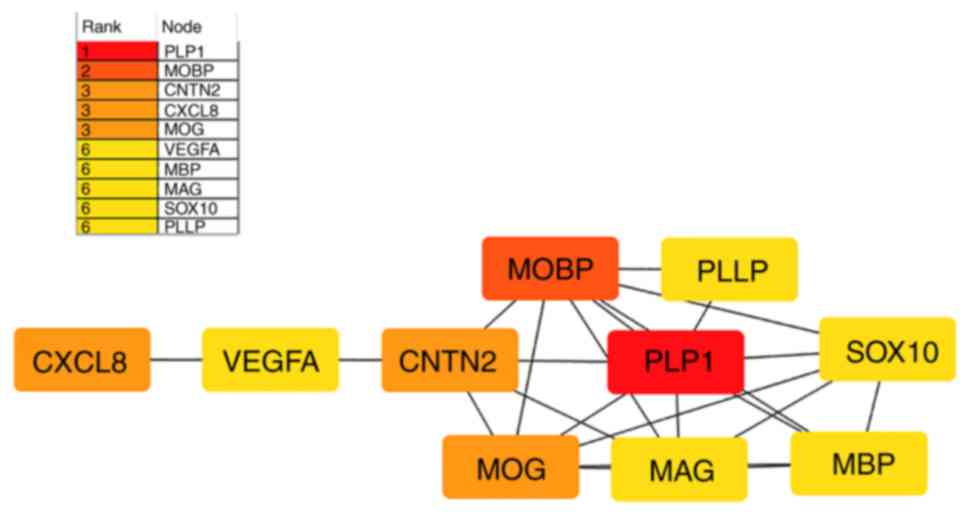 | Figure 3.Top 10 hub genes obtained from the
analysis of the protein-protein interaction networks, constructed
using Cytohubba, a plug-in of Cytoscape. Different colors represent
the different importance of the hub genes, in terms of their degree
of connectivity. The redder the color block is, the more important
it is to other genes. PLP1, proteolipid protein 1; MOBP, myelin
associated oligodendrocyte basic protein; CNTN2, contactin 2;
CXCL8, C-X-C motif chemokine ligand 8; MOG, myelin oligodendrocyte
glycoprotein; VEGFA, vascular endothelial growth factor A; MBP,
myelin basic protein; MAG, myelin associated glycoprotein; SOX10,
SRY-box transcription factor 10; PLLP, plasmolipin. |
 | Table IV.GO enrichment analysis of 10 hub
genes. |
Table IV.
GO enrichment analysis of 10 hub
genes.
| A,
GOTERM_BP_DIRECT |
|---|
|
|---|
| Term | Count, n | P-value | Genes |
|---|
| GO:0048469~cell
maturation | 3 |
1.59×10−4 | SOX10, PLP1,
VEGFA |
|
GO:0021762~substantia nigra
development | 3 |
2.96×10−4 | MAG, PLP1, MBP |
| GO:0007417~central
nervous system development | 3 | 0.001764 | CNTN2, MOG,
MBP |
| GO:0008366~axon
ensheathment | 2 | 0.003746 | PLP1, MBP |
| GO:0022010~central
nervous system myelination | 2 | 0.003746 | PLP1, CNTN2 |
|
GO:0050930~induction of positive
chemotaxis | 2 | 0.008013 | VEGFA, CXCL8 |
| GO:0002052~positive
regulation of neuroblast proliferation | 2 | 0.010671 | SOX10, VEGFA |
| GO:0031623~receptor
internalization | 2 | 0.022817 | CNTN2, CXCL8 |
| GO:0007155~cell
adhesion | 3 | 0.023633 | MAG, CNTN2,
MOG |
|
GO:0042552~myelination | 2 | 0.024392 | PLLP, MBP |
|
| B,
GOTERM_CC_DIRECT |
|
| Term | Count,
n | P-value | Genes |
|
| GO:0043218~compact
myelin | 2 | 0.001974 | PLLP, MBP |
| GO:0043209~myelin
sheath | 3 | 0.002394 | PLP1, MOBP,
CNTN2 |
|
| C,
GOTERM_MF_DIRECT |
|
| Term | Count,
n | P-value | Genes |
|
|
GO:0019911~structural constituent of
myelin sheath | 4 |
1.26×10−8 | PLP1, MOBP, PLLP,
MBP |
 | Table V.Kyoto Encyclopedia of Genes and
Genomes pathway analysis of 10 hub genes. |
Table V.
Kyoto Encyclopedia of Genes and
Genomes pathway analysis of 10 hub genes.
| Term | Count, n | P-value | Genes |
|---|
| hsa05219: Bladder
cancer | 2 | 0.017777 | VEGFA, CXCL8 |
| hsa05323:
Rheumatoid arthritis | 2 | 0.037894 | VEGFA, CXCL8 |
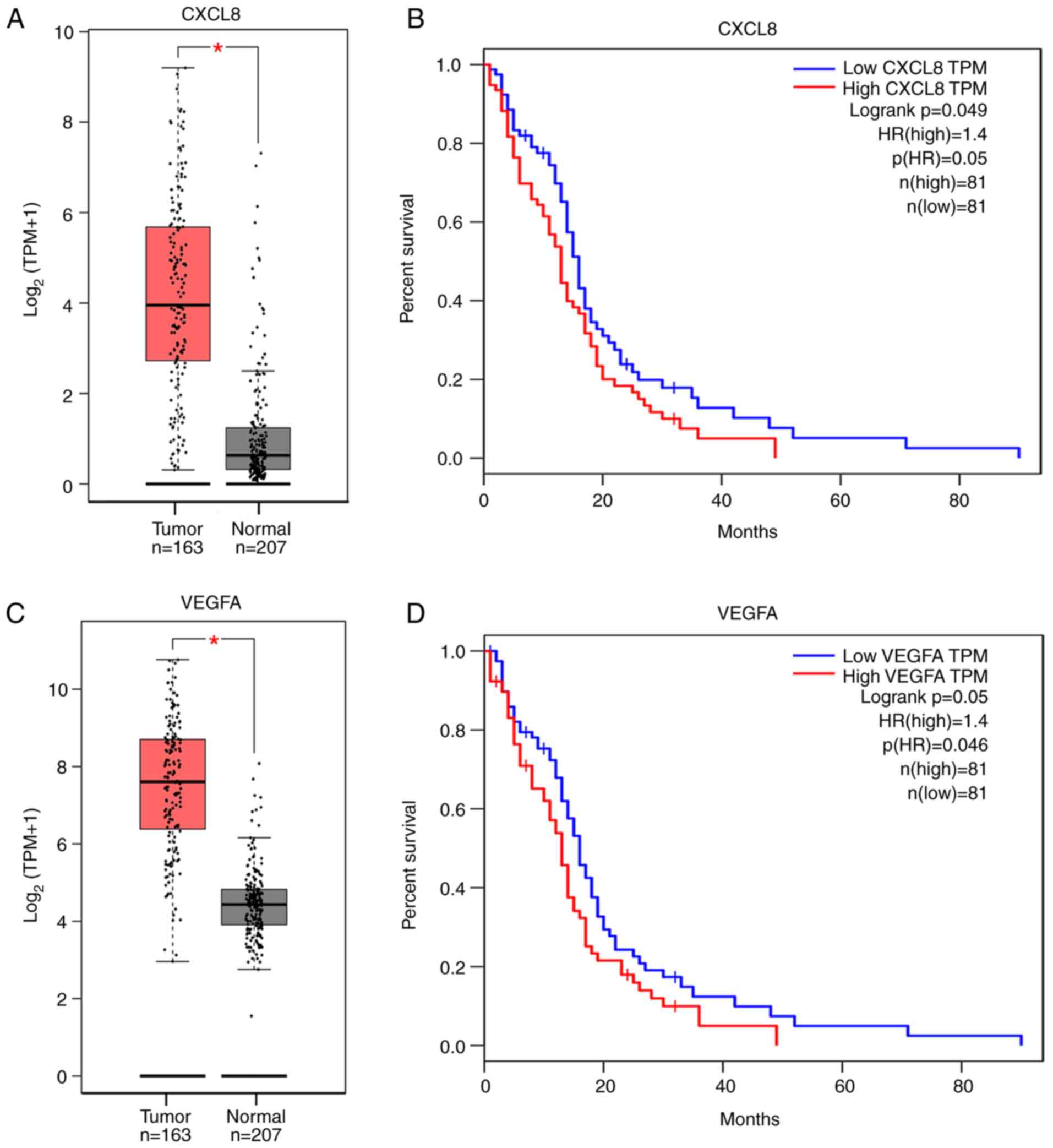
Survival analysis
The prognostic value of the 10 hub genes was
evaluated using the GEPIA online tool and survival curves were
obtained. The GEPIA online tool is based on the TCGA and GTEx
databases. Survival analysis of the 10 hub genes revealed that
PLP1, MOBP, CNTN2, MOG, MBP, MAG, SOX10 and PLLP were not
associated with the overall survival rate of patients with GBM.
However, CXCL8 and VEGFA were significantly associated with a
reduction in the overall survival rate of patients with GBM
(Fig. 4B and D). The expression
levels of the 10 hub genes in the normal and tumor groups in the
GEPIA database were also compared. The results of the present study
demonstrated that the VEGFA and CXCL8 genes expression were
differentially expressed in tumor tissues and normal tissues
(Fig. 4A and C), and both genes were
highly expressed in the PBZ region.
Discussion
In the present study, to investigate the causes of
GBM recurrence, the DEGs between the PBZ and TC were evaluated by
bioinformatics analysis. The GSE13276 and GSE116520 datasets were
analyzed and 75 common DEGs were identified, including 12 common UP
genes and 63 common DOWN genes. DAVID was used to perform a
functional annotation analysis of the 75 DEGs. In the GO analysis,
the UP genes were enriched in ‘regulation of angiogenesis’. The
DOWN genes were enriched in ‘central nervous system development’.
In the KEGG pathway analysis, UP genes were enriched in ‘bladder
cancer’ and DOWN genes were enriched in ‘endocytosis’. Based on
this, Cytohubba, a plug-in of Cytoscape, was used to analyze the
DEGs, and a total of 10 hub genes were identified, including PLP1,
MOBP, CNTN2, MOG, MBP, MAG, SOX10, CXCL8, VEGFA and PLLP. Only
CXCL8 and VEGFA were UP genes, and the other hub genes were DOWN
genes. In addition, GEPIA was used to assess the prognostic value
of the 10 hub genes. It was determined that the PLP1, MOBP, CNTN2,
MOG, MBP, MAG, SOX10 and PLLP genes did not reduce the overall
survival rate of patients with GBM. However, it was demonstrated
that CXCL8 and VEGFA were significantly associated with a reduction
in the overall survival rate of patients with GBM.
GO term analysis of the 10 hub genes demonstrated
that these genes were significantly enriched in terms such as ‘cell
maturation’, ‘substantia nigra development’ and ‘central nervous
system development’. In particular, three key genes, CNTN2, MOG and
MBP, were enriched in the ‘central nervous system development’
term. Rickman et al (17)
reported that CNTN2 gene expression is increased in middle and
high-grade gliomas. Abnormal CNTN2 gene expression is associated
with glioma (17). Inhibiting the
expression of CNTN2 can inhibit the migration of glioma (17,18).
Using the UniProt website (https://www.uniprot.org/), it has been demonstrated
that MOG gene expression was associated with cell adhesion
(19). In 1994, Nakagawa et
al (20) measured the expression
level of myelin basic protein (MBP) in the cerebrospinal fluid of
patients with various types of tumors, including glioblastoma,
using radioimmunoassay. It was reported that the expression level
of MBP was >4.0 ng/ml, which was associated with active
malignant tumors. In the pathways of ‘cell maturation’ and
‘substantia nigra development’, it was indicated that the SOX10,
PLP1, and MAG genes were associated with GBM. In 2011, Serão et
al (21) found that the SOX10
gene was a high-risk factor for GBM in different races, and Kong
et al (22) found that the
expression of the PLP1 gene was associated with the morphological
classification of GBM. Ljubimova et al (23) found that the MAG gene cannot be
expressed in normal brain tissue, but its expression level in GBM
was high. In this study by analyzing these genes in ‘cell
maturation’, ‘substantia nigra development’ and ‘central nervous
system development’ pathways, it was determined that these genes
are associated with GBM recurrence.
In the KEGG pathway analysis, the 10 hub genes were
enriched in ‘bladder cancer’ and ‘rheumatoid arthritis’. Both
pathways involved the two genes VEGFA and CXCL8. The two genes are
involved in the angiogenesis of the pathway and promote the
progression of these two diseases (24,25). A
previous study demonstrated that the use of an anti-rheumatoid
arthritis drug (auranofin) inhibits the invasiveness of GBM cells
(26). Based on the consideration of
these pathways, Lanzardo et al (27) revealed that αVβ3 is a target marker
of rheumatoid arthritis. Additionally, it has been demonstrated
that αVβ3 could be used as a marker of GBM molecular imaging
(27). The enrichment of these two
pathways suggested that the PBZ is more susceptible to the
expression of these two genes, which leads to the formation of
blood vessels and the possibility of GBM metastasis.
The VEGFA gene can encode angiogenic factors, and
its products can alter the microenvironment of endothelial cells
and promote angiogenesis (28). In
numerous tumors, gene expression products of VEGFA not only
stimulate tumor growth, metastasis and survival, but also stimulate
hemopoietic cells, endothelial cells and neuronal cells (29–31).
Desbaillets et al (32)
reported that hypoxia stimulates VEGFA expression in tumor cells in
the pseudopalisades region. Wong et al (33) demonstrated that GBM is a type of
tumor that is rich in blood vessels and that VEGFA is upregulated
in cancer, providing insight to develop anti-VEGF and other
anti-angiogenic medicines. Gong et al (34) found that the growth and invasiveness
of GBM are associated with the regulation of matrix
metalloproteinase 2 (MMP2) by VEGFA. It has also been suggested
that blocking MMP2 expression could inhibit angiogenesis when the
anti-VGEFA gene is expressed (34).
Egorova et al (35) have
demonstrated that small interfering RNA can be used to target and
knockout VEGFA gene expression in tumor and endothelial cells.
Bevacizumab (an antibody that specifically binds to VEGFA) has been
found to reduce vascular density and heterogeneity of tumors, but
also increases the invasiveness of tumors and causes cognitive
impairment in patients (36,37).
The CXCL8 gene has a number of aliases, one of which
is interleukin (IL)-8, and is a chemokine with 77 amino acids.
Previous studies have demonstrated that it can promote the
formation of blood vessels (38–41).
Furthermore, Desbaillets et al (32) identified IL-8 gene expression in
necrotic cells surrounding GBM, and demonstrated that hypoxia and
anoxia could promote IL-8 gene expression. Additionally, the
deficiency of glucose and amino acids, and the increase of reactive
oxygen species could promote IL-8 expression (42). Ahn et al (43) reported that necrotic cells induce
NF-κB/activator protein-1, which could promote IL-8 expression in
tumor cells. Previous studies have demonstrated that IL-8 promotes
the growth and metastasis of GBM by stimulating its receptors C-X-C
chemokine receptor type (CXCR)1 and CXCR2 (44–46). A
recent study has demonstrated that bradykinin can induce IL-8
expression through the B1 bradykinin receptor (B1R), whereas B1R
promotes the transfer of STAT3 and transcription factor SP-1 to the
nucleus, and promotes the metastasis of GBM cells (47). Hasan et al (48) analyzed 17 matched pairs of primary
and rGBM samples; IL-8 expression was increased in 11 pairs of rGBM
samples. Furthermore, blocking IL-8 gene expression may reduce the
expression of key glioma-initiating cells genes, temozolomide
(TMZ)-induced increases in transcription factors (Sox2 and C-myc),
and enhance the efficacy of TMZ therapy to prolong survival time
(48). In accordance with
Desbaillets et al (32), this
study also found that the IL-8 gene is highly expressed in the PBZ,
which may be a signal of migration and proliferation for tumor
cells. By targeting CXCR2 using the inhibitor SB225002 (49), knockout of cyclophilin A can reduce
the activity of NF-κB, inhibit the expression of IL-8, and prevent
the metastasis and growth of tumors (50).
CXCL8 and VEGFA genes serve an important role in the
growth, proliferation and metastasis of tumors in GBM (29,38–41).
However, the principle of using anti-VEGF agents to reduce tumor
perfusion, and cause ischemia and hypoxia (51), may additionally promote the
expression of vascular endothelial growth factor and IL-8 following
hypoxia, and in turn stimulate the invasiveness of tumors. At
present, bevacizumab is mainly used in the treatment of rGBM;
however, it is not able to prolong overall survival (36), and the recurrence of GBM mainly
depends on the surgery (52).
Combined with the results of the present study, it may be suggested
that both anti-VEGF and anti-IL-8 therapy may be used to improve
the prognosis of patients. At present, it was determined that both
the VEGFA and CXCL8 genes are highly expressed in the PBZ of GBM,
which can cause peripheral angiogenesis, migration and
proliferation of tumor cells (29,38–41).
Additionally, it was determined that through data analysis, the
high expression levels of both VEGFA and CXCL8 can reduce the
overall survival rate of patients. Taking into consideration the
results of Desbaillets et al (32) and this study, it is suggested that
high expression of CXCL8 and VEGFA genes were associated with the
recurrence of GBM. However, the association of GBM with recurrence
requires further investigation. The gene-related pathways have
become increasingly clear. However, further studies are required to
understand these pathways and to develop appropriate drugs to
improve the prognosis of patients.
In conclusion, through a comprehensive
bioinformatics analysis, 10 hub DEGs between PBZ and TC were
identified, including PLP1, MOBP, CNTN2, MOG, MBP, MAG, SOX10,
VEGFA, CXCL8 and PLLP. The GO and KEGG enrichment analyses revealed
that the hub genes were mainly enriched in ‘central nervous system
development’ and ‘rheumatoid arthritis’. According to literature,
the genes in these pathways have a specific association with tumor
invasion, metastasis and angiogenesis (24,25), and
pathway and term analyses can provide further insights for the
development of future drugs and treatments. Through survival
analysis of the 10 hub genes, it was demonstrated that high
expression levels of the VEGFA and CXCL8 genes are associated with
a reduced survival time of patients with GBM, and they were
expressed differently in the tumor and normal samples. The present
study could provide targets for future therapies and novel insights
to block gene expression, which may reduce the recurrence rate of
tumors.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13276)
and (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE116520).
Authors' contributions
XLu, SX and FY designed the study. XLu, SX, YZ, TT,
YX, XLi and BW performed the bioinformatics analysis and
interpretation of the data. XLu, SX and TT wrote the manuscript. FY
revised the manuscript and gave final approval of the version to be
published. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
GBM
|
glioblastoma
|
|
PBZ
|
peritumoral brain zone
|
|
TC
|
tumor core
|
|
rGBM
|
recurrent glioblastoma
|
|
GEO
|
Gene Expression Omnibus
|
|
DEGs
|
differentially expressed genes
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
PPI
|
protein-protein interaction
|
|
UP
|
upregulated
|
|
DOWN
|
downregulated
|
|
GEPIA
|
Gene Expression Profiling Interactive
Analysis
|
|
TCGA
|
The Cancer Genome Atlas
|
|
GTEx
|
Genotype-Tissue Expression
|
|
HR
|
hazard ratio
|
|
PLP1
|
proteolipid protein 1
|
|
MOBP
|
myelin-associated oligodendrocyte
basic protein
|
|
CNTN2
|
contactin 2
|
|
CXCL8
|
C-X-C motif chemokine ligand 8
|
|
MOG
|
myelin oligodendrocyte
glycoprotein
|
|
VEGFA
|
vascular endothelial growth factor
A
|
|
MBP
|
myelin basic protein
|
|
MAG
|
myelin associated glycoprotein
|
|
SOX10
|
SRY-box 10
|
|
PLLP
|
plasmolipin
|
|
IL-8
|
interleukin 8
|
|
MMP2
|
matrix metalloproteinase 2
|
|
CXCR
|
C-X-C chemokine receptor
|
|
B1R
|
B1 bradykinin receptor
|
|
TMZ
|
temozolomide
|
References
|
1
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 World Health Organization
Classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mangiola A, Saulnier N, De Bonis P,
Orteschi D, Sica G, Lama G, Pettorini BL, Sabatino G, Zollino M,
Lauriola L, et al: Gene expression profile of glioblastoma
peritumoral tissue: An ex vivo study. PLoS One. 8:e571452013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jayamanne D, Wheeler H, Cook R, Teo C,
Brazier D, Schembri G, Kastelan M, Guo L and Back MF: Survival
improvements with adjuvant therapy in patients with glioblastoma.
ANZ J Surg. 88:196–201. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lassman AB, van den Bent MJ, Gan HK,
Reardon DA, Kumthekar P, Butowski N, Lwin Z, Mikkelsen T, Nabors
LB, Papadopoulos KP, et al: Safety and efficacy of depatuxizumab
mafodotin + temozolomide in patients with EGFR-amplified, recurrent
glioblastoma: Results from an international phase I multicenter
trial. Neuro Oncol. 21:106–114. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Franceschi E, Minichillo S and Brandes AA:
Pharmacotherapy of glioblastoma: Established treatments and
emerging concepts. CNS Drugs. 31:675–684. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mangiola A, de Bonis P, Maira G, Balducci
M, Sica G, Lama G, Lauriola L and Anile C: Invasive tumor cells and
prognosis in a selected population of patients with glioblastoma
multiforme. Cancer. 113:841–846. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Long H, Liang C, Zhang X, Fang L, Wang G,
Qi S, Huo H and Song Y: Prediction and analysis of key genes in
glioblastoma based on bioinformatics. Biomed Res Int.
2017:76531012017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ruiz-Ontañon P, Orgaz JL, Aldaz B,
Elosegui-Artola A, Martino J, Berciano MT, Montero JA, Grande L,
Nogueira L, Diaz-Moralli S, et al: Cellular plasticity confers
migratory and invasive advantages to a population of
glioblastoma-initiating cells that infiltrate peritumoral tissue.
Stem Cells. 31:1075–1085. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Van Meter T, Dumur C, Hafez N, Garrett C,
Fillmore H and Broaddus WC: Microarray analysis of MRI-defined
tissue samples in glioblastoma reveals differences in regional
expression of therapeutic targets. Diagn Mol Pathol. 15:195–205.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lama G, Mangiola A, Anile C, Sabatino G,
De Bonis P, Lauriola L, Giannitelli C, La Torre G, Jhanwar-Uniyal
M, Sica G and Maira G: Activated ERK1/2 expression in glioblastoma
multiforme and in peritumor tissue. Int J Oncol. 30:1333–1342.
2007.PubMed/NCBI
|
|
11
|
Kruthika BS, Jain R, Arivazhagan A,
Bharath RD, Yasha TC, Kondaiah P and Santosh V: Transcriptome
profiling reveals PDZ binding kinase as a novel biomarker in
peritumoral brain zone of glioblastoma. J Neurooncol. 141:315–325.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang DW, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang DW, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rickman DS, Tyagi R, Zhu XX, Bobek MP,
Song S, Blaivas M, Misek DE, Israel MA, Kurnit DM, Ross DA, et al:
The gene for the axonal cell adhesion molecule TAX-1 is amplified
and aberrantly expressed in malignant gliomas. Cancer Res.
61:2162–2168. 2001.PubMed/NCBI
|
|
18
|
Eckerich C, Zapf S, Ulbricht U, Müller S,
Fillbrandt R, Westphal M and Lamszus K: Contactin is expressed in
human astrocytic gliomas and mediates repulsive effects. Glia.
53:1–12. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Solly SK, Daubas P, Monge M, Dautigny A
and Zalc B: Functional analysis of the mouse myelin/oligodendrocyte
glycoprotein gene promoter in the oligodendroglial CG4 cell line. J
Neurochem. 68:1705–1711. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nakagawa H, Yamada M, Kanayama T,
Tsuruzono K, Miyawaki Y, Tokiyoshi K, Hagiwara Y and Hayakawa T:
Myelin basic protein in the cerebrospinal fluid of patients with
brain tumors. Neurosurgery. 34:825–833. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Serão NV, Delfino KR, Southey BR, Beever
JE and Rodriguez-Zas SL: Cell cycle and aging, morphogenesis, and
response to stimuli genes are individualized biomarkers of
glioblastoma progression and survival. BMC Med Genomics. 4:492011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kong J, Cooper LA, Wang F, Gao J, Teodoro
G, Scarpace L, Mikkelsen T, Schniederjan MJ, Moreno CS, Saltz JH
and Brat DJ: Machine-based morphologic analysis of glioblastoma
using whole-slide pathology images uncovers clinically relevant
molecular correlates. PLoS One. 8:e810492013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ljubimova JY, Wilson SE, Petrovic LM,
Ehrenman K, Ljubimov AV, Demetriou AA, Geller SA and Black KL:
Novel human malignancy-associated gene (MAG) expressed in various
tumors and in some tumor preexisting conditions. Cancer Res.
58:4475–4479. 1998.PubMed/NCBI
|
|
24
|
Reis ST, Leite KR, Piovesan LF,
Pontes-Junior J, Viana NI, Abe DK, Crippa A, Moura CM, Adonias SP,
Srougi M and Dall'Oglio MF: Increased expression of MMP-9 and IL-8
are correlated with poor prognosis of Bladder Cancer. BMC Urol.
12:182012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pathak JL, Bakker AD, Verschueren P, Lems
WF, Luyten FP, Klein-Nulend J and Bravenboer N: CXCL8 and CCL20
enhance osteoclastogenesis via modulation of cytokine production by
human primary osteoblasts. PLoS One. 10:e01310412015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kast RE: Glioblastoma invasion, cathepsin
B, and the potential for both to be inhibited by auranofin, an old
anti-rheumatoid arthritis drug. Cent Eur Neurosurg. 71:139–142.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lanzardo S, Conti L, Brioschi C,
Bartolomeo MP, Arosio D, Belvisi L, Manzoni L, Maiocchi A, Maisano
F and Forni G: A new optical imaging probe targeting αVβ3 integrin
in glioblastoma xenografts. Contrast Media Mol Imaging. 6:449–458.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bergers G and Benjamin LE: Tumorigenesis
and the angiogenic switch. Nat Rev Cancer. 3:401–410. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu Y, Hooper AT, Zhong Z, Witte L, Bohlen
P, Rafii S and Hicklin DJ: The vascular endothelial growth factor
receptor (VEGFR-1) supports growth and survival of human breast
carcinoma. Int J Cancer. 119:1519–1529. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kane NM, Xiao Q, Baker AH, Luo Z, Xu Q and
Emanueli C: Pluripotent stem cell differentiation into vascular
cells: A novel technology with promises for vascular
re(generation). Pharmacol Ther. 129:29–49. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Calvo CF, Fontaine RH, Soueid J, Tammela
T, Makinen T, Alfaro-Cervello C, Bonnaud F, Miguez A, Benhaim L, Xu
Y, et al: Vascular endothelial growth factor receptor 3 directly
regulates murine neurogenesis. Genes Dev. 25:831–844. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Desbaillets I, Diserens AC, Tribolet N,
Hamou MF and Van Meir EG: Upregulation of interleukin 8 by
oxygen-deprived cells in glioblastoma suggests a role in leukocyte
activation, chemotaxis, and angiogenesis. J Exp Med. 186:1201–1212.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wong ML, Prawira A, Kaye AH and Hovens CM:
Tumour angiogenesis: Its mechanism and therapeutic implications in
malignant gliomas. J Clin Neurosci. 16:1119–1130. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gong J, Zhu S, Zhang Y and Wang J:
Interplay of VEGFa and MMP2 regulates invasion of glioblastoma.
Tumour Biol. 35:11879–11885. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Egorova AA, Maretina MA and Kiselev AV:
VEGFA gene silencing in CXCR4-expressing cells via siRNA delivery
by means of targeted peptide carrier. Methods Mol Biol. 1974:57–68.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Keunen O, Johansson M, Oudin A, Sanzey M,
Rahim SA, Fack F, Thorsen F, Taxt T, Bartos M, Jirik R, et al:
Anti-VEGF treatment reduces blood supply and increases tumor cell
invasion in glioblastoma. Proc Natl Acad Sci USA. 108:3749–3754.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Latzer P, Shchyglo O, Hartl T, Matschke V,
Schlegel U, Manahan-Vaughan D and Theiss C: Blocking VEGF by
bevacizumab compromises electrophysiological and morphological
properties of hippocampal neurons. Front Cell Neurosci. 13:1132019.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Koch A, Polverini P, Kunkel S, Harlow LA,
DiPietro LA, Elner VM, Elner SG and Strieter RM: Interleukin-8 as a
macrophage-derived mediator of angiogenesis. Science.
258:1798–1801. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Strieter RM, Kunkel SL, Elner VM, Martonyi
CL, Koch AE, Polverini PJ and Elner SG: Interleukin-8. A corneal
factor that induces neovascularization. Am J Pathol. 141:1279–1284.
1992.PubMed/NCBI
|
|
40
|
Keane MP and Strieter RM: The role of CXC
chemokines in the regulation of angiogenesis. Chem Immunol.
72:86–101. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tada M, Suzuki K, Yamakawa Y, Sawamura Y,
Sakuma S, Abe H, van Meir E and de Tribolet N: Human glioblastoma
cells produce 77 amino acid interleukin-8 (IL-8(77)). J Neurooncol.
16:25–34. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Brat DJ, Bellail AC and Van Meir EG: The
role of interleukin-8 and its receptors in gliomagenesis and
tumoral angiogenesis. Neuro Oncol. 7:122–133. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ahn SH, Park H, Ahn YH, Kim S, Cho MS,
Kang JL and Choi YH: Necrotic cells influence migration and
invasion of glioblastoma via NF-κB/AP-1-mediated IL-8 regulation.
Sci Rep. 6:245522016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sharma I, Singh A, Siraj F and Saxena S:
IL-8/CXCR1/2 signalling promotes tumor cell proliferation, invasion
and vascular mimicry in glioblastoma. J Biomed Sci. 25:622018.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Infanger DW, Cho Y, Lopez BS, Mohanan S,
Liu SC, Gursel D, Boockvar JA and Fischbach C: Glioblastoma stem
cells are regulated by interleukin-8 signaling in a tumoral
perivascular niche. Cancer Res. 73:7079–7089. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Dwyer J, Hebda JK, Le Guelte A, Galan-Moya
EM, Smith SS, Azzi S, Bidere N and Gavard J: Glioblastoma
cell-secreted interleukin-8 induces brain endothelial cell
permeability via CXCR2. PLoS One. 7:e455622012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu YS, Hsu JW, Lin HY, Lai SW, Huang BR,
Tsai CF and Lu DY: Bradykinin B1 receptor contributes to
interleukin-8 production and glioblastoma migration through
interaction of STAT3 and SP-1. Neuropharmacology. 144:143–154.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hasan T, Caragher SP, Shireman JM, Park
CH, Atashi F, Baisiwala S, Lee G, Guo D, Wang JY, Dey M, et al:
Interleukin-8/CXCR2 signaling regulates therapy-induced plasticity
and enhances tumorigenicity in glioblastoma. Cell Death Dis.
10:2922019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Angara K, Borin TF, Rashid MH, Lebedyeva
I, Ara R, Lin PC, Iskander A, Bollag RJ, Achyut BR and Arbab AS:
CXCR2-expressing tumor cells drive vascular mimicry in
antiangiogenic therapy-resistant glioblastoma. Neoplasia.
20:1070–1082. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sun S, Wang Q, Giang A, Cheng C, Soo C,
Wang CY, Liau LM and Chiu R: Knockdown of CypA inhibits
interleukin-8 (IL-8) and IL-8-mediated proliferation and tumor
growth of glioblastoma cells through down-regulated NF-κB. J
Neurooncol. 101:1–14. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Folkman J: Tumor angiogenesis: Therapeutic
implications. N Engl J Med. 285:1182–1186. 1971. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Davis ME: Glioblastoma: Overview of
disease and treatment. Clin J Oncol Nurs. 20 (Suppl 5):S2–S8. 2016.
View Article : Google Scholar : PubMed/NCBI
|