Introduction
It has been reported that >90% of head and neck
malignant tumor cases are squamous cell carcinoma (1–3). Head
and neck squamous cell carcinoma (HNSCC) mainly arises from the
epithelium of the oral cavity, oropharynx and larynx (4). Currently, treatment for HNSCC involves
surgery, radiotherapy and adjuvant chemotherapy. Despite continuous
improvements and updates in treatment, the five-year survival rate
of patients with HNSCC has not improved (5). This is partly due to the occurrence of
early tumor metastasis (6,7). Lymph node metastasis has been reported
in 50%, with distant metastasis observed in 20–30% of patients with
HNSCC in the two years following therapy (8,9).
However, almost two thirds of patients with HNSCC, who receive
combined treatment of chemo- and radiotherapy succumb within three
years; therefore, further improvements in the treatment and
management of HNSCC are required (10). Although a number of genes and
pathways are aberrantly expressed in HNSCC, including tumor protein
53, cyclin dependent kinase inhibitor 2A, Ras, nuclear factor κB
and epidermal growth factor receptor, an understanding of the
functional underlying mechanisms by which they regulate metastasis
and recurrence remains undetermined (11–15).
Therefore, it is crucial to identify proteins that regulate
metastasis and understand their functional mechanisms, which in
turn would promote the development of novel prognostic biomarkers
and therapeutic approaches for the disease.
Epithelial-to-mesenchymal transition (EMT) is a
crucial process in metastasis, as cells acquire mesenchymal and
fibroblast-like properties and indicate increased motility and
reduced adhesion (16–18). A number of pathways have been
reported to induce EMT, including transforming growth factor β1
(TGF-β1)-mediated signaling (19–21).
Although TGF-β1 activates multiple signaling pathways, the main one
for EMT regulation is as follows: TGF-β1 induces its own receptor,
leading to the activation of downstream signaling molecules, which
reduces SMAD family member 2 (Smad2)/Smad3 phosphorylation,
followed by Smad2/Smad3 forming heterodimers with Smad4 (22). This complex enters the nucleus,
upregulates nuclear transcription factors and inhibits the
epithelial cell marker E-cadherin, ultimately leading to EMT and
tumor cell invasion and metastasis (23). Therefore, the aim of the present
study was to identify a key regulator of TGF-β1-mediated cell
signaling to prevent EMT and metastasis.
Caveolin-1 (Cav-1), a 21–24 kDa integral membrane
protein and a principal structural component of caveolae, serves a
critical role in TGF-β-associated transactivation of epidermal
growth factor (EGF) receptor signaling in hepatocytes; in addition,
caveolin was reported to suppress TGF-β signaling, with TGF-β
receptors known to be endocytosed in a caveolin-dependent manner
(24–26). Previous studies have reported that
high Cav-1 expression levels are associated with tumor progression
and metastasis in prostate and pancreatic cancer, while inhibiting
metastasis and tumor progression (27–29).
Furthermore, Tu686 and 686LN cell lines, established from
pharyngeal squamous cell carcinoma and its metastatic lymph nodes,
respectively, were assessed, and it was reported that Cav-1
expression was reduced in 686LN cells compared with Tu686 cells,
with greater metastatic and invasive abilities exhibited in 686LN
cells (30). However, a
comprehensive understanding of the role of caveolin in cancer
metastasis is required.
In the present study, the role of Cav-1 in the
regulation of HNSCC metastasis was examined, and the mechanism by
which Cav-1 regulates TGF-β signaling in metastasis and EMT was
investigated by silencing Cav-1 using short hairpin (sh)RNA
technology.
Materials and methods
Cell lines
The HNSCC Tu686 cell line, provided by the Winship
Cancer Institute of Emory University (Atlanta, GA, USA) and
established from a primary base of tongue tumor (31), was used in the present study. The
cells were maintained as monolayer cultures in DMEM/F12 (1:1)
(Gibco/Brl; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (Gibco/Brl; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) at 37°C with 5%
CO2.
Construction of Cav-1 shRNA and the
cDNA plasmid vector
shRNA expressing vector pLKO.1-puro was purchased
from Addgene, Inc. (Cambridge, MA). Three putative candidate
sequences were designed using the Oligoengine software (version 2;
www.oligoengine.com), with their
specificities confirmed by nucleotide BLAST searches. The two
putative candidate sequences and a scramble sequence were as
follows: Sequence-1, sense
5′-CCGGGTACATCCATTATAAGCTGCTCGAGCAGCTTATAATGGATGTACTTTTTG-3′ and
antisense
5′-AATTCAAAAAGTACATCCATTATAAGCTGCTCGAGCAGCTTATAATGGATGTAC-3′;
Sequence-2, sense
5′-CCGGGCCGGCGACGACTTCTCCCCTCGAGGGGAGAAGTCGTCGCCGGCTTTTTG-3′ and
antisense
5′-AATTCAAAAAGCCGGCGACGACTTCTCCCCTCGAGGGGAGAAGTCGTCGCCGGC-3′;
Control-sequence, sense
5′-CCGGAGCGTTCACTCCCAACCTGCTCGAGCAGGTTGGGAGTGAACGCTTTTTTG-3′ and
antisense
5′-AATTCAAAAAAGCGTTCACTCCCAACCTGCTCGAGCAGGTTGGGAGTGAACGCT-3′.
A total of 4.0×104 cells were inoculated
per well in a 24-well plate for 24 h, and three uniformly
distributed wells were selected at 60% confluency. First, 500 µl
cell culture containing polybrene at a final concentration of 5
µg/ml was added to the three wells. Subsequently, the two
experimental groups (Cav-1 shRNA lentiviral particle suspension,
sequence-1 or sequence-2; 80 µl), the negative control group
(negative control lentiviral particle suspension; 80 µl) and the
green fluorescent protein (GFP) group (control group lentiviral
particle suspension containing the fluorescent GFP marker to
observe the transfection efficiency; 80 µl) were incubated at 37°C
with 5% CO2 overnight. The medium (DMEM; Gibco/Brl;
Thermo Fisher Scientific, Inc.) containing viral particles was
changed with fresh medium every 2 days. The selection medium
containing puromycin (5 µg/ml) was added to screen positive clones
from the third day. After four weeks, a positive clone cell line
(Tu686Cav-1RNAi+) was selected, cultured and
expanded.
Western blot analysis
Cells were lysed in radioimmunoprecipitation assay
buffer (Beijing Solarbio Science and Technology Co., Ltd., Beijing,
China), for protein extraction. Equal amounts of total protein (100
µg), measured by the bicinchoninic acid method, were separated by
12% SDS-PAGE electrophoresis and transferred onto polyvinylidene
fluoride membranes. The membrane was placed in a blocking buffer
(cat. no. P0023B; Beyotime Institute of Biotechnology) for 1 h. The
membrane was incubated with primary antibody (rabbit anti-human
Cav-1 polyclonal antibody; cat. no. PA1-064; 1:800; Santa Cruz
Biotechnology, Inc, Dallas, CA, USA) at room temperature for 2 h
followed by incubation with horseradish peroxidase-conjugated
rabbit anti-goat antibody (Beyotime Institute of Biotechnology,
Haiman, China; 1:1,000) at room temperature for 2 h. Other primary
antibodies included mouse anti-human E-cadherin polyclonal (Cell
Signaling Technology, Inc.; cat. no. sc-8426; 1:400), mouse
anti-human vimentin polyclonal (Cell Signaling Technology, Inc.;
cat. no. sc-32322; 1:200), mouse anti-human Smad2 monoclonal (Cell
Signaling Technology Inc.; cat. no. 3103s; 1:800), rabbit
anti-human p-Smad2 polyclonal (Cell Signaling Technology, Inc.;
cat. no. ser465/467; 1:800) and mouse anti-human β-actin monoclonal
(Beyotime Institute of Biotechnology; cat. no. AF0003; 1:1,000)
antibodies. Horseradish peroxidase-conjugated goat anti-mouse and
anti-rabbit secondary antibodies (Beyotime Institute of
Biotechnology; cat. nos. A0562 and A0568; 1:1,000) were incubated
for 2 h at 37°C. The membrane was subsequently washed and treated
with Pierce enhanced chemiluminescence (ECL) western blotting
substrate (Thermo Fisher Scientific, Inc.). Immunoreactive bands
were visualized by ECL on a LAS4000 imager and densitometric
analysis was performed using Image Quant LAS 500 (GE Healthcare,
Chicago, IL, USA).
CCK-8 assay
CCK-8 assay was used to evaluate proliferation.
Following transduction of control shRNA sequence and the
experimental sequence for 24 h, respectively, Tu686 cells were
seeded in 96-well plates at 5,000 cells/well. After 1–7 days of
culture at 37°C with 5% CO2, CCK-8 (Beyotime Institute of
Biotechnology) was added to the cells (10 µl/well), which were
further incubated for 1 h at 37°C. Absorbance was measured at a
wavelength of 450 nm on a microplate reader (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). The experiments were performed in
triplicate. Semi logarithmic curves for cell survival were drawn by
Microsoft Excel 2000 software (Microsoft Corporation, Redmond, WA,
USA).
Wound healing assay
Tu686 cells (1×106) were seeded in 6-cm
plates coated with 10 mg/ml type I collagen (Advanced Biomatrix).
Following incubation for 24 h, the monolayer was disrupted with a
cell scraper (1.2 mm width), and micrographs were acquired at 0 and
24 h on a phase contrast microscope (magnification, ×50; Olympus
Corporation, Tokyo, Japan). Experiments were performed in
triplicate, and four fields of view in each data point were
recorded in a blinded manner.
Transwell assay
For invasion assay, the upper chambers of Matrigel
pre-coated Transwell inserts (cat. no. ECM550; Chemicon
International, USA.) prior to addition of Tu686 cells
(2×104). The lower chambers were filled with 0.8 ml
conditioned DMEM supplemented with 10% fetal bovine serum
(Gibco/Brl; Thermo Fisher Scientific, Inc., Waltham, MA, USA) to
induce chemotaxis in Tu686 cells. After 24 h of incubation at 37°C,
the cells were fixed in 4% methanol at room temperature (25°C) for
10 min and stained with hematoxylin and eosin at room temperature
(25°C) for 30 min. Cells that invaded through the pores to the
lower side of the filter were counted under an inverted light
microscope (magnification, ×100; Olympus Corporation, Tokyo,
Japan). Three migration chambers were used for each condition and
the mean number of cells was calculated.
Activation of the TGF-β1 signaling
pathway in TU686 cells
Logarithmic growth was observed in Tu686 cells,
using serum for 12 h prior to withdrawal and culture for 24 h at
37°C, maintaining parameters, including cell confluence of 80–90%,
0.25% trypsin + 0.02% EDTA digestion and density adjustment to
2.5×104 cells/ml. The cells were maintained in 50 ml
disposable cell suspension culture bottles at 37°C and 5%
CO2 for 24 h with serum-free DMEM/F12 for 24 h.
Subsequently, five groups were set up with TGF-β1 (cat. no.
100–21/100-21C; PeproTech US, Rocky Hill, NJ, USA) at the final
concentrations of 5, 10, 15, 20 and 25 ng/ml, diluted with
serum-free DMEM/F12 to 4 ml, and incubated at 37°C with 5%
CO2 for 48 h. The cells were subsequently collected for
further experiments.
Immunofluorescence
For the immunofluorescence staining, Tu686 cells
(4×104) grown on coverslips were fixed with warm PHEMO
buffer (68 mM PIPES, 25 mM HEPES, pH 6.9, 15 mM EGTA, 3 mM
MgCl2), and 10% (v/v) DMSO containing 3.7% formaldehyde,
0.05% glutaraldehyde, and 0.5% Triton X-100 for 10 min at room
temperature (25°C). Cells were blocked with 5% BSA (cat. no.
sw3015; Beijing Solarbio Science and Technology Co., Ltd.) for 1 h
at room temperature (25°C) and incubated with primary at 37°C for 1
h or at 4°C overnight and subsequently with fluorophore-conjugated
secondary antibodies at 37°C for 1 h, including mouse anti-human
Cav-1 polyclonal antibody (cat. no. 610493, BD Biosciences, San
Jose, CA, USA; 1:800), mouse anti-human transforming growth factor
β1 (cat. no. sc-130348; 1:1,000; Santa Cruz Biotechnology, Inc.),
and fluorescein isothiocyanate- and rhodamine B
isothiocyanate-conjugated IgG antibodies (cat.no. zf-0311; 1:100;
OriGene Technologies, Inc., Beijing, China). Images were acquired
on a Zeiss LSM 510 META confocal microscope (magnification,
×630).
Statistical analysis
Statistical analysis was performed with the SPSS
17.0 software (SPSS, Inc., Chicago, IL, USA). Quantitative data are
expressed as the mean ± standard deviation. Statistical
significance was estimated by two-tailed Student's t-test or
one-way analysis of variance followed by the Student-Newman-Keuls
post hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
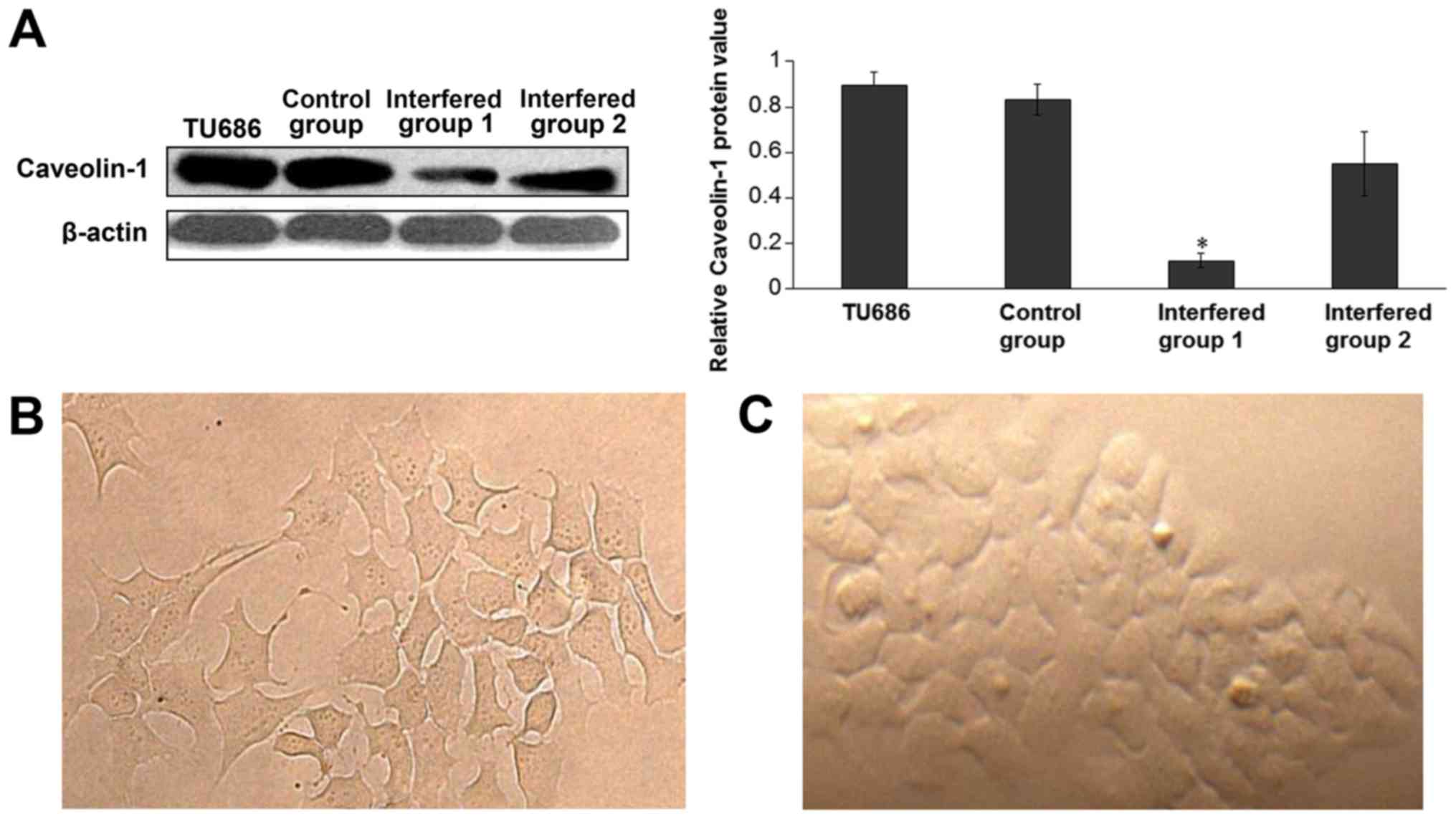
Silencing of Cav-1 expression by shRNA
in Tu686 cells
In the present study, shRNA was used to inhibit
Cav-1 expression. Candidate sequence 1 significantly inhibited the
protein expression of Cav-1 by >60% and candidate sequence 2
inhibited the expression by 30–40%. Control sequences caused no
detectable changes in Cav-1 expression (Fig. 1A). These results indicated that
candidate sequence 1 was more efficient compared with candidate
sequence 2. A change in cell morphology was also detected following
transfection under an inverted microscope (magnification, ×100;
Olympus Corporation, Tokyo, Japan), where cells with silenced Cav-1
displayed a spindle shape and pseudopodia, similar to fibroblasts
(Fig. 1B). Control cells are
presented in Fig. 1C.
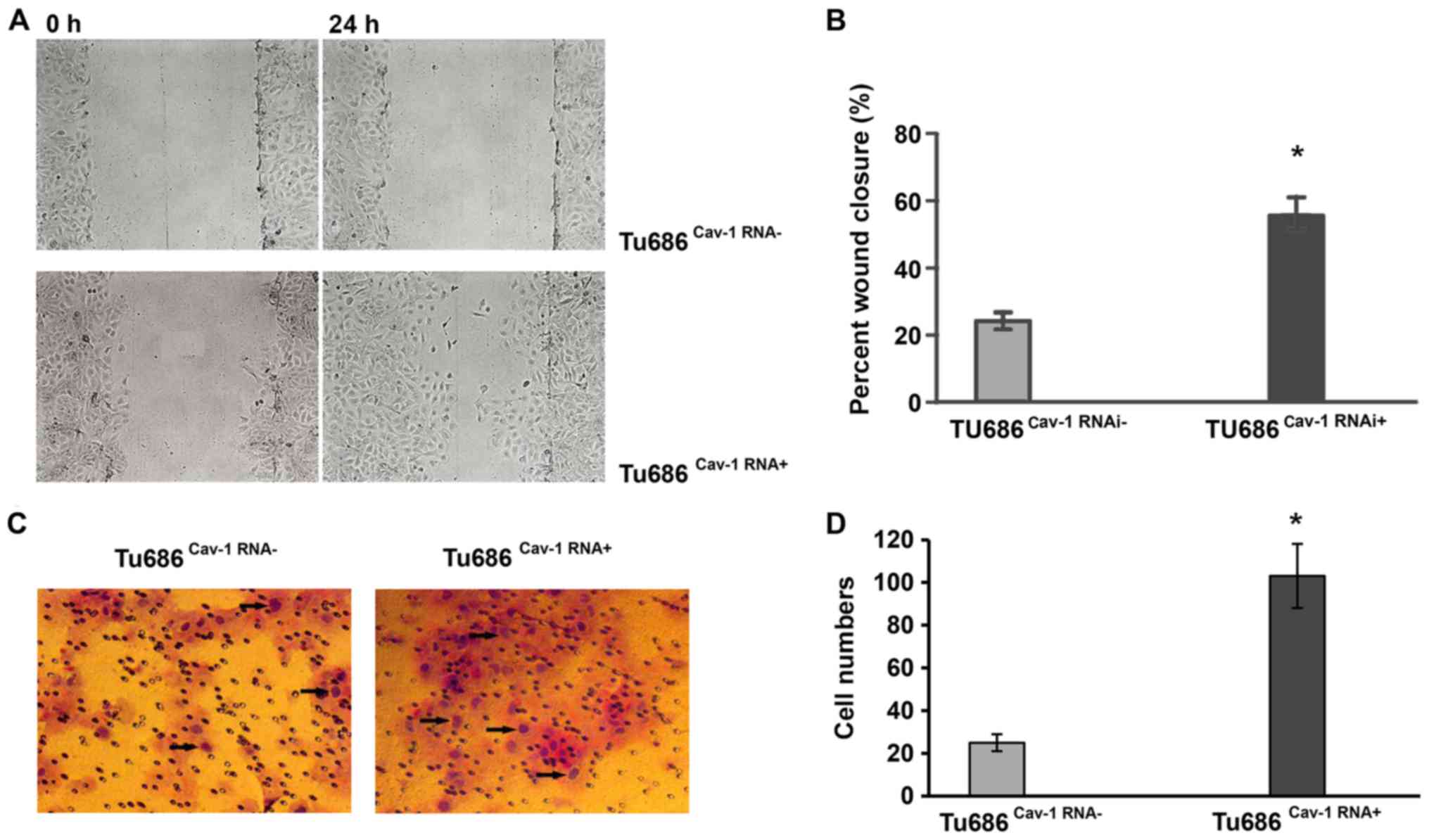
Cav-1 promotes migration but not
proliferation in Tu686 cells
Taking into consideration that upregulation of Cav-1
expression is associated with HNSCC metastasis (30), we hypothesized that Cav-1 may be
involved in the invasive phenotype of HNSCC by promoting cell
migration. To test this possibility, the effect of Cav-1 on the
metastasis of Tu686 cells was assessed. Wound healing and Transwell
assays were employed to determine the migration and invasion
abilities of Tu686 cells, respectively. Tu686Cav-1RNAi+
cells exhibited a 15% increase in wound closure compared with
Tu686Cav-1RNAi cells (P<0.05) in the wound healing
assay. In the Transwell assay, a significantly higher number of
Tu686Cav-1RNA+ cells passed through the Matrigel
compared with Tu686Cav-1RNA- cells, suggesting a role
for Cav-1 in the migration of Tu686 cells (P<0.05; Fig. 2).
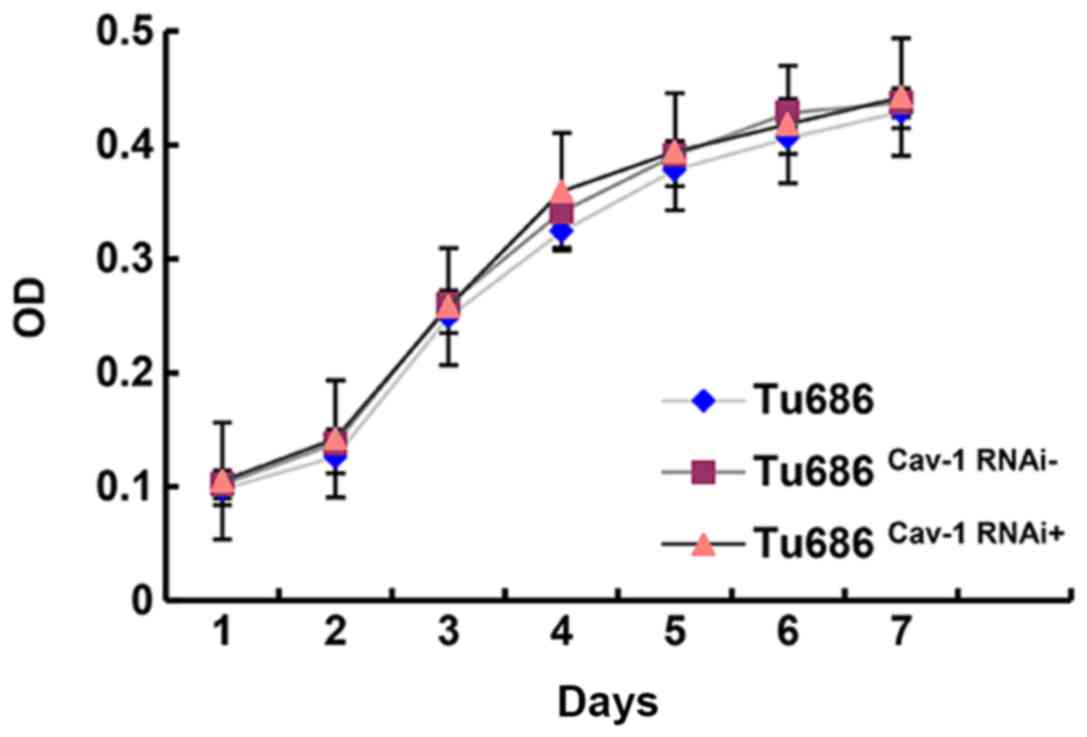
A CCK-8 assay was employed to assess the growth and
proliferation ability of Tu686, Tu686Cav-1RNAi− and
Tu686Cav-1RNAi+ cells. No statistical differences were
identified between the Tu686 and Tu686Cav-1RNAi− groups,
Tu686 and Tu686Cav-1RNAi+ groups, and
Tu686Cav-1RNAi− and Tu686Cav-1RNAi+ groups
(P>0.05; Fig. 3).
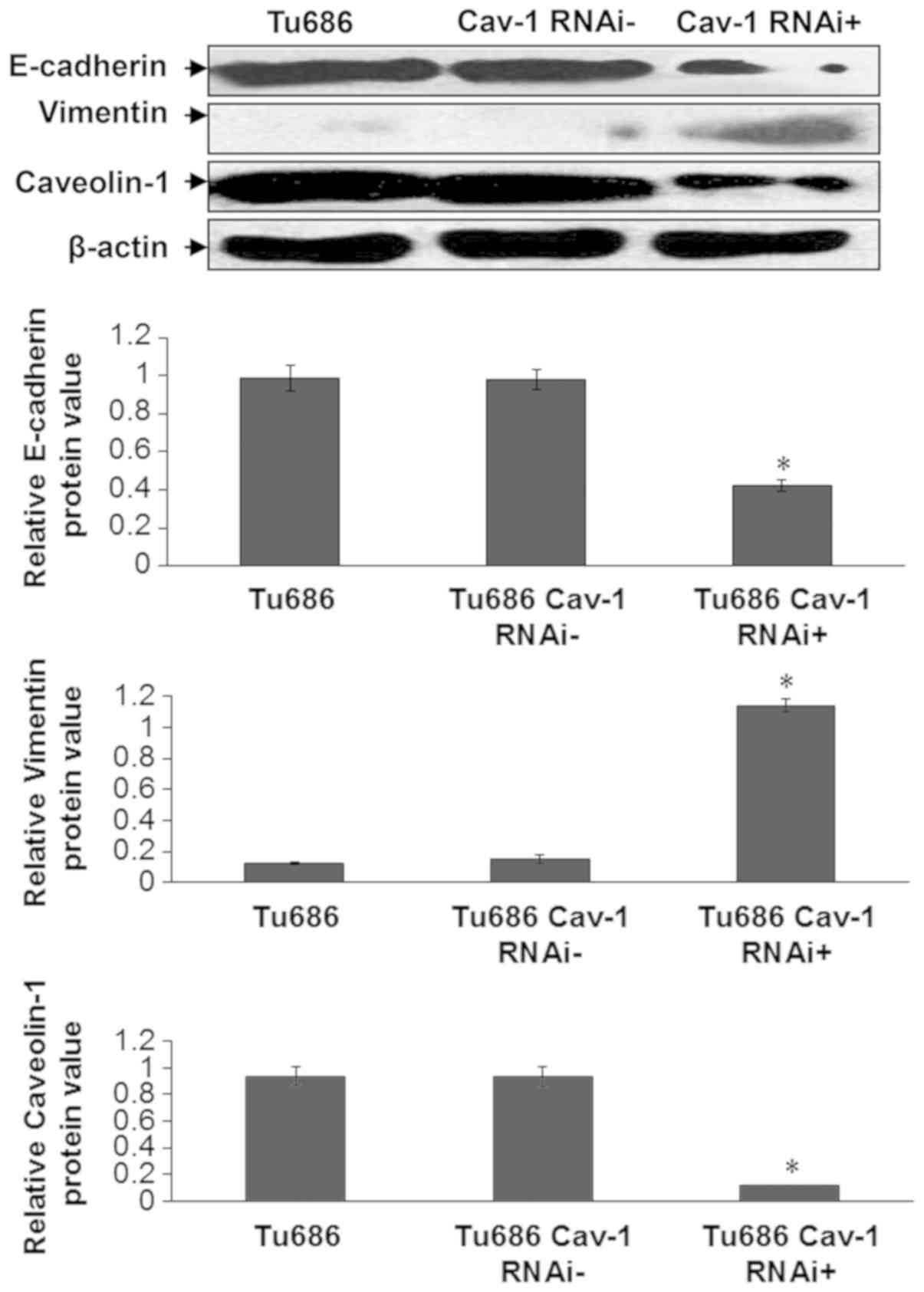
Cav-1 silencing upregulates vimentin
and reduces E-cadherin expression
EMT is marked by two crucial events: Loss of
E-cadherin, a transmembrane protein involved in cell-cell adhesion,
and an increase in the expression of vimentin, an intermediate
filament protein (32). Therefore,
the present study assessed the expression levels of these two
proteins following Cav-1 silencing. It was indicated that Cav-1
shRNA significantly reduced the expression of E-cadherin and
increased that of vimentin in Tu686 cells (Tu686Cav-1RNAi
+ vs. Tu686 cells and Tu686Cav-1RNAi−; P<0.05;
Fig. 4).
Cav-1 silencing induces TGF-β1
signaling
Confocal microscopy was used to determine the in
vivo localizations of Cav-1 and TGF-β1 receptors. As indicated
in Fig. 5A, TGF-β1R and Cav-1 were
co-localized inside the cells.
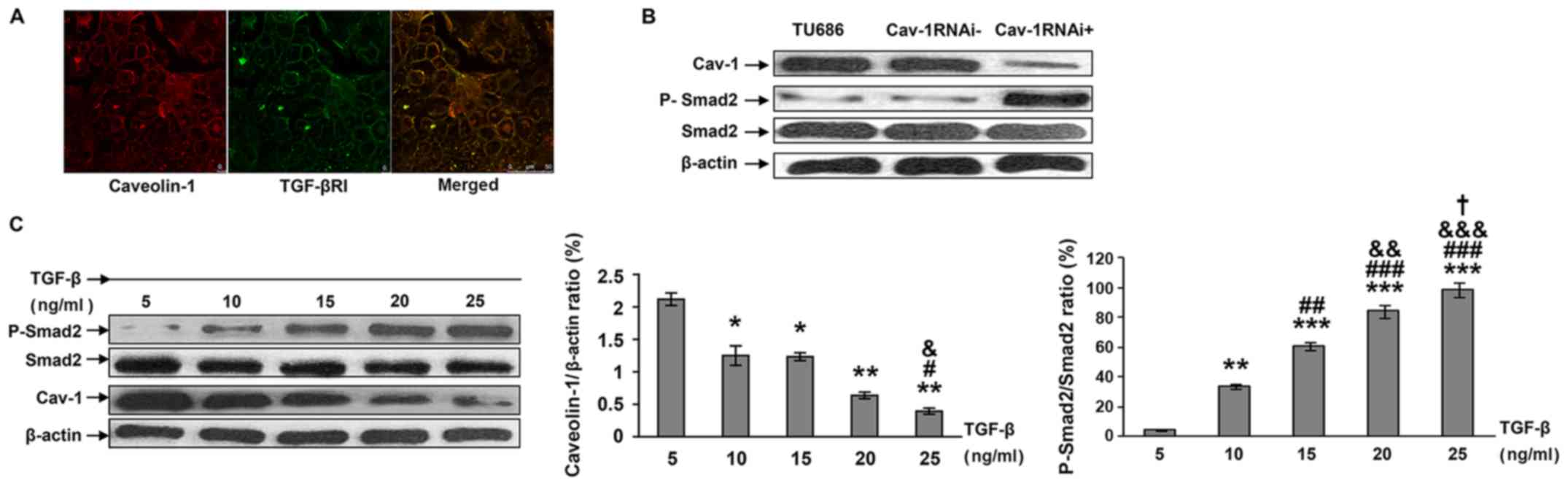 | Figure 5.Effects of Cav-1-knockdown on TGF-β1
signaling. (A) Double-labeled immunofluorescence was employed to
determine the localizations of Cav-1 and TGF-βRI in Tu686 cells.
Magnification, ×630. (B) Knockdown of Cav-1 resulted in increased
p-Smad2 levels. Tu686 cells were used as the blank control. (C)
Dose-dependent changes in Smad, p-Smad and Cav-1 protein levels
with increasing doses of TGF-β1 (5, 10, 15, 20 and 25 ng/ml)
following 48 h of culture. Quantification of results was performed
using one-way analysis of variance and the Student-Newman-Keuls
post hoc test. *P<0.04, **P<0.01 and ***P<0.001 vs. 5
ng/ml. #P<0.05, ##P<0.01 and
###P<0.001 vs. 10 ng/ml. &P<0.05,
&&P<0.01 and
&&&P<0.001 vs. 15 ng/ml.
†P<0.05 vs. 20 ng/ml. RNAi, RNA interference; Cav-1,
caveolin-1; Smad2, SMAD family member 2; TGF-β1, transforming
growth factor β1; p, phosphorylated; TGF-βRI, transforming growth
factor β1. |
It was also identified that stable silencing of
Cav-1 in Tu686 cells led to an increased expression of
phosphorylated Smad2, indicating an increased activation of the
TGF-β pathway (Fig. 5B). A
dose-dependent decrease in Cav-1 expression level was observed with
gradually increasing TGF-β1 levels. These findings suggest a mutual
regulation of TGF-β1 signaling and Cav-1 (Fig. 5C).
Discussion
The aim of the present study was to examine the
molecular regulation of HNSCC metastasis. The role of Cav-1 in
regulating TGF-β-induced EMT was assessed. Silencing of Cav-1
expression led to reduced E-cadherin expression, cell migration and
increased vimentin expression, all key markers of EMT (33). It was also indicated that Cav-1
downregulation resulted in increased TGF-β signaling, reflected by
increased phosphorylation of Smad2. Therefore, the present study
proposes that Cav-1 expression inhibits EMT by regulation of TGF-β
signaling.
In order to gain novel insights into the role of
Cav-1 in HNSCC, shRNA was employed to knockdown Cav-1 expression in
Tu686 cells. Cav-1 depletion resulted in increased cell migration,
with no significant effects on proliferation. In addition, changes
in cell morphology were observed, as cells were spindle-shaped and
a number of them presented with pseudopodia, similar to
fibroblasts. These findings of morphological changes are in
accordance with previous studies on EMT-induced shape changes
(34–36). Therefore, downregulation of Cav-1
expression resulted in phenotypic changes similar to EMT in Tu686
cells.
A number of studies have assessed the role of Cav-1
expression during EMT and cancer metastasis. Lu et al
(37) demonstrated that
EGF-stimulating factor in tumor cells significantly reduced Cav-1
expression, activated the β-catenin-T-cell factor/lymphoid
enhancer-binding factor transcription factor, decreased the levels
of the epithelial cell marker E-cadherin, blunted cell-cell
contact, and enhanced the cellular phenotypes of EMT and
metastasis. Bailey et al (38) indicated that Cav-1 regulation during
EMT is mediated by focal adhesion kinase.
Abnormalities in the TGF-β signaling pathway are
closely associated with tumor cell invasion and metastasis
(39). In breast cancer, activated
TGF-β signaling pathway can cause hypermethylation and subsequent
loss of expression of E-cadherin, cingulin, claudin 4 and
kallikrein related peptidase 10, resulting in EMT and breast cancer
metastasis (40). Araki et al
(41) reported that activated TGF-β1
signaling in breast cancer cells increases the expression of E3
ubiquitin ligase human murine double minute, leading to P53 gene
destabilization and the EMT phenotype in cells.
By using confocal microscopy, the present study
observed that Cav-1 and TGF-β1 receptors were co-localized on the
plasma membrane, indicating that the effects of Cav-1 on EMT may
involve TGF-β1 signaling. Schwartz et al (42) reported that TGF-β receptor (TGF-βR) I
and II co-localize with Cav-1 and nitric oxide synthase 3 (eNOS) in
human umbilical vein epithelial cells. They further indicated that
TGF-β1 interacts and regulates eNOS activity in the caveolae.
In the present study, Cav-1 could inhibit TGF-β
signaling by regulating the phosphorylation levels of its
downstream effector SMAD2. In addition, Cav-1 silencing
significantly increased the expression of phosphorylated Smad2,
indicating an induction of TGF-β1R II signaling. The present study
also indicated a dose-dependent decrease in Cav-1 levels with
increasing levels of TGF-β1, which further indicates the mutual
inhibitory regulation between the TGF-β1 signaling pathway and Cav
−1.
A limitation of the current study should be
mentioned. Although EMT is a complex event involving transcription
factors, cytoskeletal proteins and extracellular matrix components
among others (43), E-cadherin and
vimentin were only assessed in this experiment; therefore, further
comprehensive studies should examine the effects of Cav-1 on other
factors affecting EMT.
In conclusion, the present study indicated that
Cav-1 may regulate EMT by influencing cell invasion and migration
in HNSCC cells. It was further demonstrated that the aforementioned
regulation may involve the TGF-β1 signaling pathway. The current
study also provided a basis for a mutual association between Cav-1
and TGF-β signaling for metastasis regulation in HNSCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JS conceived and coordinated the study, designed and
performed the experiments, and wrote the manuscript. YL, CY, TX,
GN, BM and XZ performed data collection, data analysis and revised
the manuscript. All authors reviewed the results and approved the
final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
shRNA
|
short hairpin RNA
|
|
Cav-1
|
caveolin-1
|
|
EMT
|
epithelial-to-mesenchymal
transition
|
|
FACS
|
fluorescence activated cell sorting
analysis
|
|
HNSCC
|
head and neck squamous cell
carcinoma
|
|
TGF-β1
|
transforming growth factor β1
|
References
|
1
|
Sanderson RJ and Ironside JA: Squamous
cell carcinomas of the head and neck. BMJ. 325:822–827. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rezende TM, de Souza Freire M and Franco
OL: Head and neck cancer: Proteomic advances and biomarker
achievements. Cancer. 116:4914–4925. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chi AC, Day TA and Neville BW: Oral cavity
and oropharyngeal squamous cell carcinoma-an update. CA Cancer J
Clin. 65:401–421. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ho AS, Kraus DH, Ganly I, Lee NY, Shah JP
and Morris LG: Decision making in the management of recurrent head
and neck cancer. Head Neck. 36:144–151. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Garavello W, Ciardo A, Spreafico R and
Gaini RM: Risk factors for distant metastases in head and neck
squamous cell carcinoma. Arch Otolaryngol Head Neck Surg.
132:762–766. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Walsh JE, Lathers DM, Chi AC, Gillespie
MB, Day TA and Young MRl: mechanisms of tumor growth and metastasis
in head and neck squamous cell carcinoma. Curr Treat Options Oncol.
8:227–238. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jung AC, Ray AM, Ramolu L, Macabre C,
Simon F, Noulet F, Blandin AF, Renner G, Lehmann M, Choulier L, et
al: Caveolin-1-negative head and neck squamous cell carcinoma
primary tumors display increased epithelial to mesenchymal
transition and prometastatic properties. Oncotarget. 6:41884–41901.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ferlito A, Shaha AR, Silver CE, Rinaldo A
and Mondin V: Incidence and sites of distant metastases from head
and neck cancer. ORL J Otorhinolaryngol Relat Spec. 63:202–207.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Merlano M, Vitale V, Rosso R, Benasso M,
Corvò R, Cavallari M, Sanguineti G, Bacigalupo A, Badellino F,
Margarino G, et al: Treatment of advanced squamous-cell carcinoma
of the head and neck with alternating chemotherapy and
radiotherapy. N Eng J Med. 327:1115–1121. 1992. View Article : Google Scholar
|
|
11
|
Molinolo AA, Amornphimoltham P, Squarize
CH, Castilho RM, Patel V and Gutkind JS: Dysregulated molecular
networks in head and neck carcinogenesis. Oral Oncol. 45:324–334.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Choi P and Chen C: Genetic expression
profiles and biologic pathway alterations in head and neck squamous
cell carcinoma. Cancer. 104:1113–1128. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Koshizuka K, Hanazawa T, Fukumoto I,
Kikkawa N, Okamoto Y and Seki N: The microRNA signatures:
Aberrantly expressed microRNAs in head and neck squamous cell
carcinoma. J Hum Genet. 62:3–13. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hedberg ML, Goh G, Chiosea SI, Bauman JE,
Freilino ML, Zeng Y, Wang L, Diergaarde BB, Gooding WE, Lui VW, et
al: Genetic landscape of metastatic and recurrent head and neck
squamous cell carcinoma. J Clin Invest. 126:169–180. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gaykalova DA, Mambo E, Choudhary A,
Houghton J, Buddavarapu K, Sanford T, Darden W, Adai A, Hadd A,
Latham G, et al: Novel insight into mutational landscape of head
and neck squamous cell carcinoma. PLoS One. 9:e931022014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Heerboth S, Housman G, Leary M, Longacre
M, Byler S, Lapinska K, Willbanks A and Sarkar S: EMT and tumor
metastasis. Clin Transl Med. 4:62015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Larue L and Bellacosa A:
Epithelial-mesenchymal transition in development and cancer: Role
of phosphatidylinositol 3′ kinase/AKT pathways. Oncogene.
24:7443–7454. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Diepenbruck M and Christofori G:
Epithelial-mesenchymal transition (EMT) and metastasis: Yes, no,
maybe? Curr Opin Cell Biol. 43:7–13. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Heldin CH, Vanlandewijck M and Moustakas
A: Regulation of EMT by TGFβ in cancer. FEBS Lett. 586:1959–1970.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu J, Lamouille S and Derynck R:
TGF-beta-induced epithelial to mesenchymal transition. Cell Res.
19:156–172. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zavadil J and Böttinger EP: TGF-beta and
epithelial-to-mesenchymal transitions. Oncogene. 24:5764–5774.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zavadil J, Cermak L, Soto-Nieves N and
Böttinger EP: Integration of TGF-beta/Smad and Jagged1/Notch
signalling in epithelial-to-mesenchymal transition. EMBO J.
23:1155–1165. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Moreno-Caceres J, Caja L, Mainez J,
Mayoral R, Martín-Sanz P, Moreno-Vicente R, Del Pozo MÁ, Dooley S,
Egea G and Fabregat I: Caveolin-1 is required for TGF-β-induced
transactivation of the EGF receptor pathway in hepatocytes through
the activation of the metalloprotease TACE/ADAM17. Cell Death Dis.
5:e13262014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fridolfsson HN, Roth DM, Insel PA and
Patel HH: Regulation of intracellular signaling and function by
caveolin. FASEB J. 28:3823–3831. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Del Galdo F, Lisanti MP and Jimenez SA:
Caveolin-1, transforming growth factor-beta receptor
internalization, and the pathogenesis of systemic sclerosis. Curr
Opin Rheumatol. 20:713–719. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sloan EK, Stanley KL and Anderson RL:
Caveolin-1 inhibits breast cancer growth and metastasis. Oncogene.
23:7893–7897. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chatterjee M, Ben-Josef E, Thomas DG,
Morgan MA, Zalupski MM, Khan G, Andrew Robinson C, Griffith KA,
Chen CS, Ludwig T, et al: Caveolin-1 is associated with tumor
progression and confers a multi-modality resistance phenotype in
pancreatic cancer. Sci Rep. 5:108672015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sugie S, Mukai S, Yamasaki K, Kamibeppu T,
Tsukino H and Kamoto T: Significant association of caveolin-1 and
caveolin-2 with prostate cancer progression. Cancer Genomics
Proteomics. 12:391–396. 2015.PubMed/NCBI
|
|
30
|
Zhang H, Su L, Müller S, Tighiouart M, Xu
Z, Zhang X, Shin HJ, Hunt J, Sun SY, Shin DM and Chen ZG:
Restoration of caveolin-1 expression suppresses growth and
metastasis of head and neck squamous cell carcinoma. Br J Cancer.
99:1684–1694. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sacks PG: Cell, tissue and organ culture
as in vitro models to study the biology of squamous cell carcinomas
of the head and neck. Cancer Metastasis Rev. 15:27–51. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transition in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Moreno-Bueno G, Peinado H, Molina P,
Olmeda D, Cubillo E, Santos V, Palacios J, Portillo F and Cano A:
The morphological and molecular features of the
epithelial-to-mesenchymal transition. Nat Protoc. 4:1591–1613.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yu C, Liu Y, Huang D, Dai Y, Cai G, Sun J,
Xu T, Tian Y and Zhang X: TGF-β1 mediates epithelial to mesenchymal
transition via the TGF-β/Smad pathway in squamous cell carcinoma of
the head and neck. Oncol Rep. 25:1581–1587. 2011.PubMed/NCBI
|
|
36
|
Pi LM, Liu Y, Yu CY, Cai GM, Hunag DH, Qiu
YZ, Tian YQ and Zhang X: EGCG regulates TGF-β1-induced epithelial
mesenchymal transition in squamous cell carcinoma of head and neck.
Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 47:749–752. 2012.(In
Chinese). PubMed/NCBI
|
|
37
|
Lu Z, Ghosh S, Wang Z and Hunter T:
Downregulation of caveolin-1 function by EGF leads to the loss of
E-cadherin, increased transcriptional activity of beta-catenin, and
enhanced tumor cell invasion. Cancer Cell. 4:499–515. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bailey KM and Liu J: Caveolin-1
up-regulation during epithelial to mesenchymal transition is
mediated by focal adhesion kinase. J Biol Chem. 283:13714–13724.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Miyazono K: Transforming growth
factor-beta signaling in epithelial-mesenchymal transition and
progression of cancer. Proc Jpn Acad Ser B Phys Biol Sci.
85:314–323. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Papageorgis P, Lambert AW, Ozturk S, Gao
F, Pan H, Manne U, Alekseyev YO, Thiagalingam A, Abdolmaleky HM,
Lenburg M and Thiagalingam S: Smad signaling is required to
maintain epigenetic silencing during breast cancer progression.
Cancer Res. 70:968–978. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Araki S, Eitel JA, Batuello CN,
Bijangi-Vishehsaraei K, Xie XJ, Danielpour D, Pollok KE, Boothman
DA and Mayo LD: TGF-beta1-induced expression of human Mdm2
correlates with late-stage metastatic breast cancer. J Clin Invest.
120:290–302. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Schwartz EA, Reaven E, Topper JN and Tsao
PS: Transforming growth factor-beta receptors localize to caveolae
and regulate endothelial nitric oxide synthase in normal human
endothelial cells. Biochem J. 390:199–206. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
De Craene B and Berx G: Regulatory
networks defining EMT during cancer initiation and progression. Nat
Rev Cancer. 13:97–110. 2013. View Article : Google Scholar : PubMed/NCBI
|