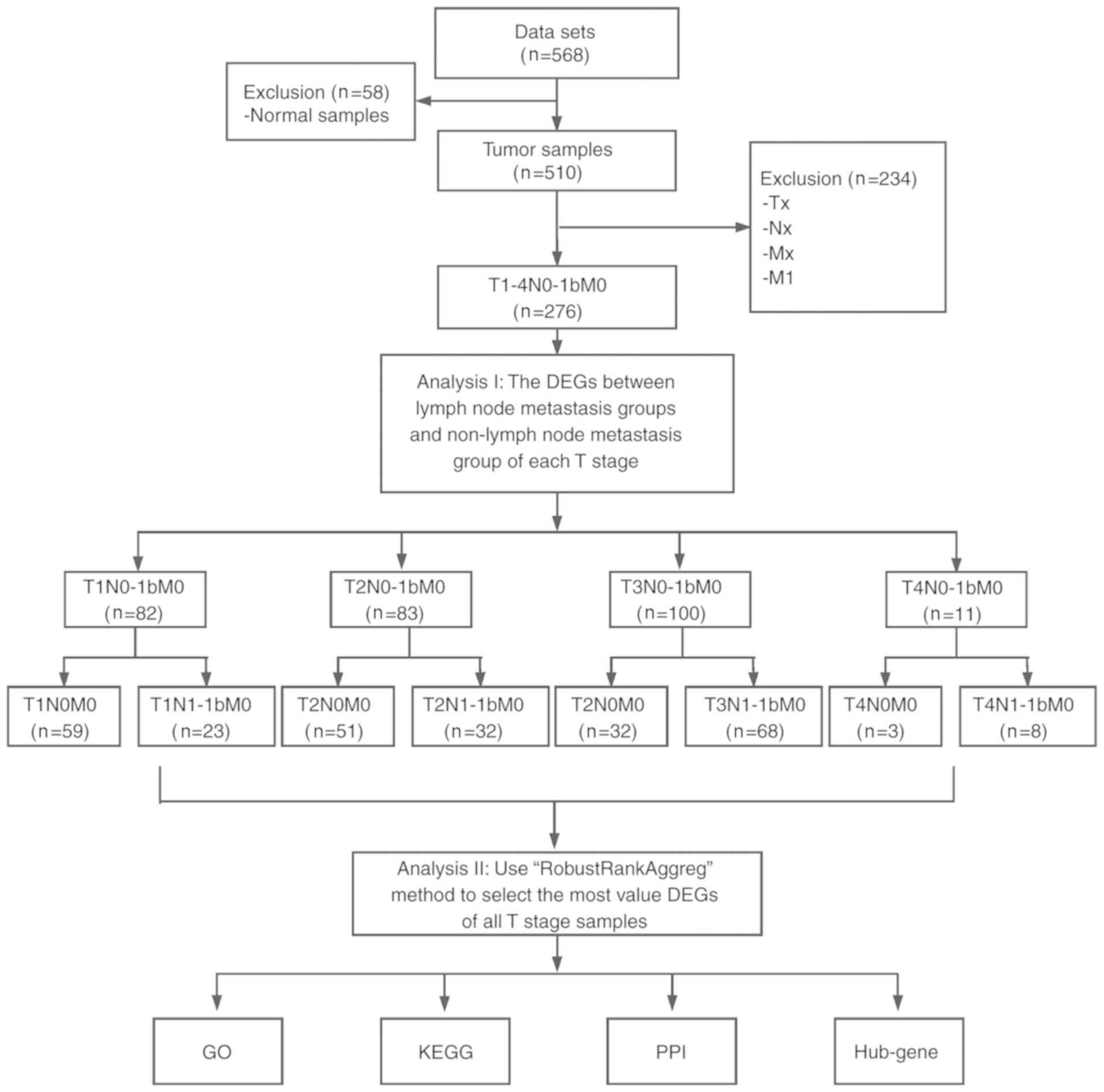
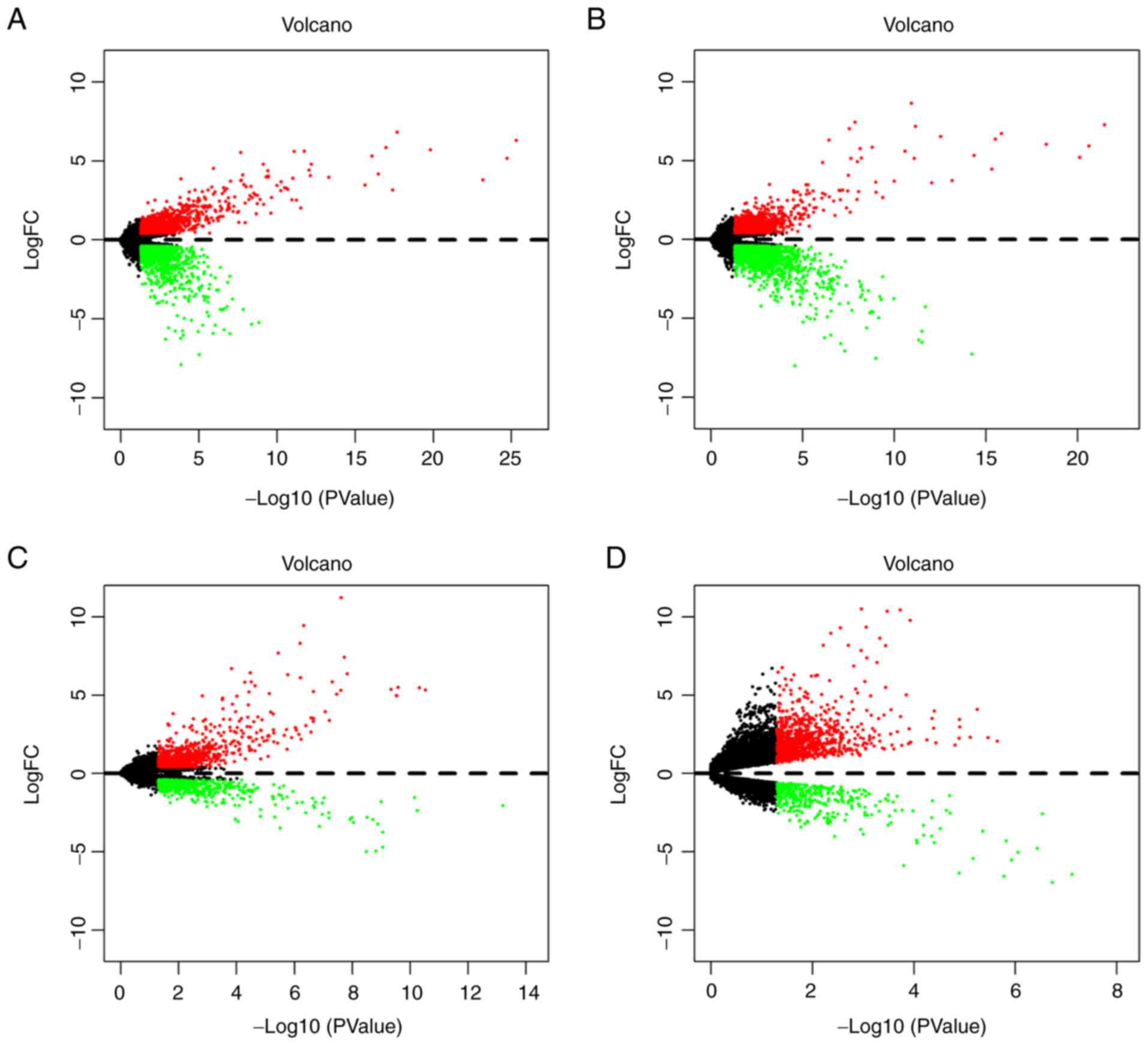
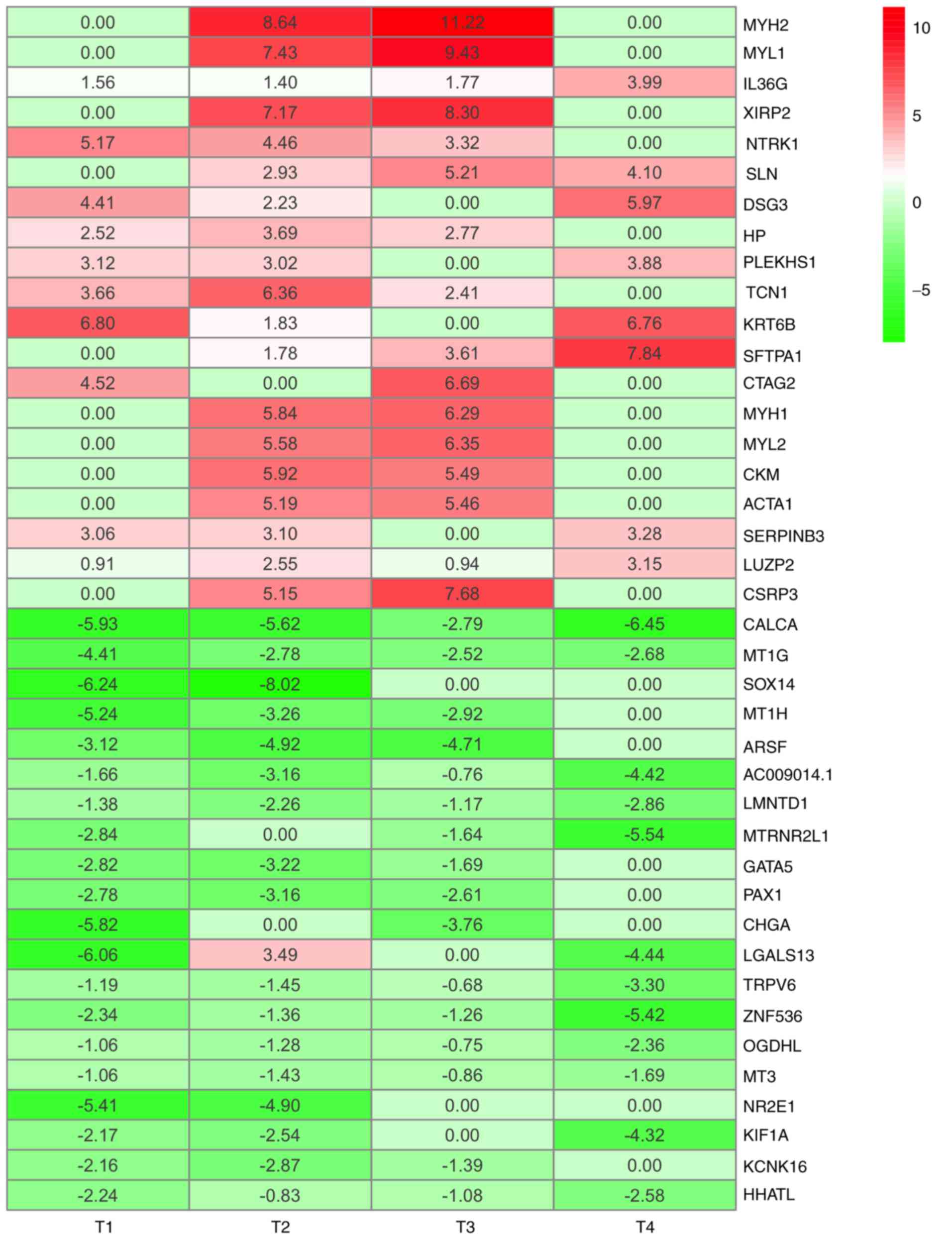
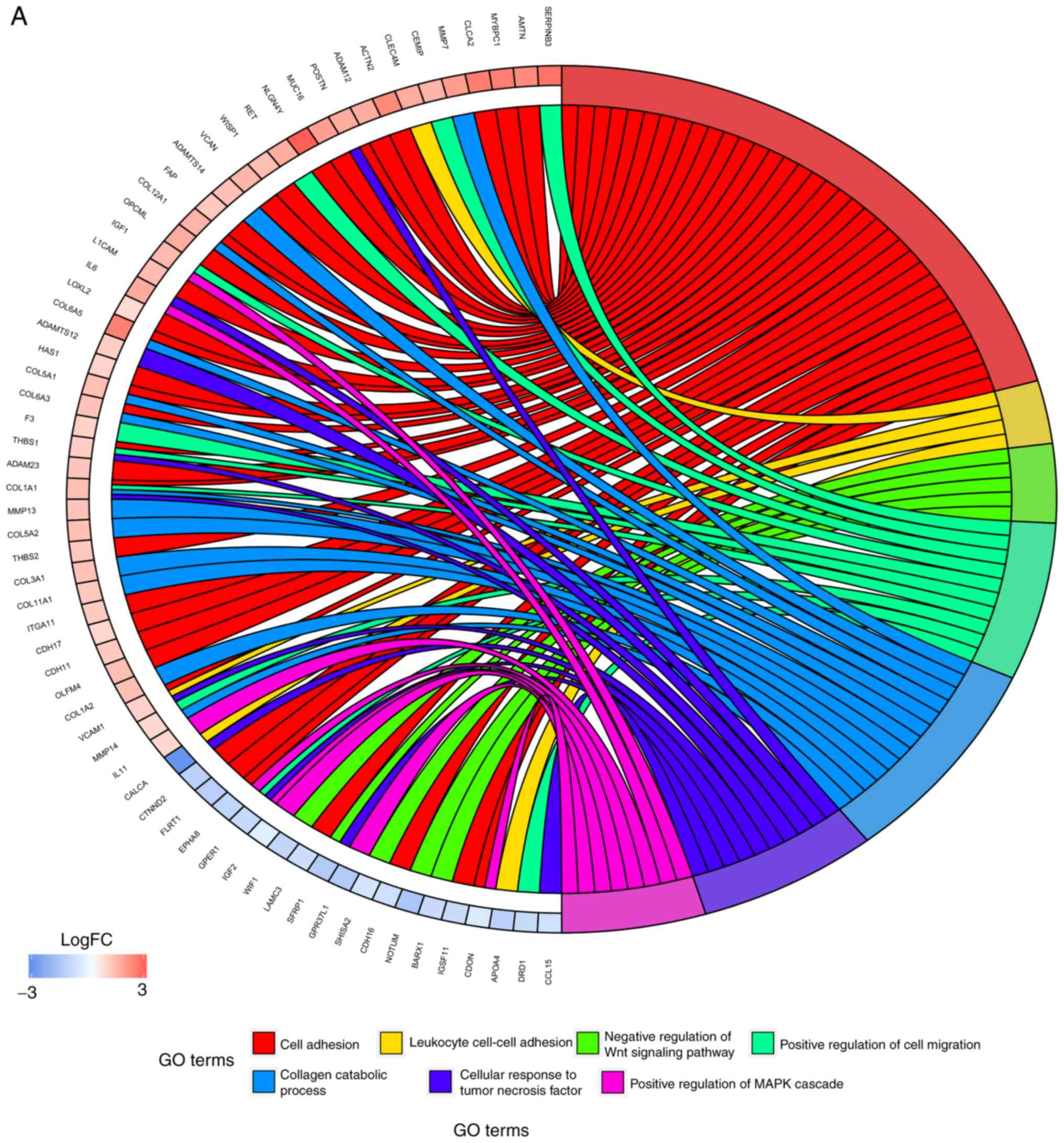
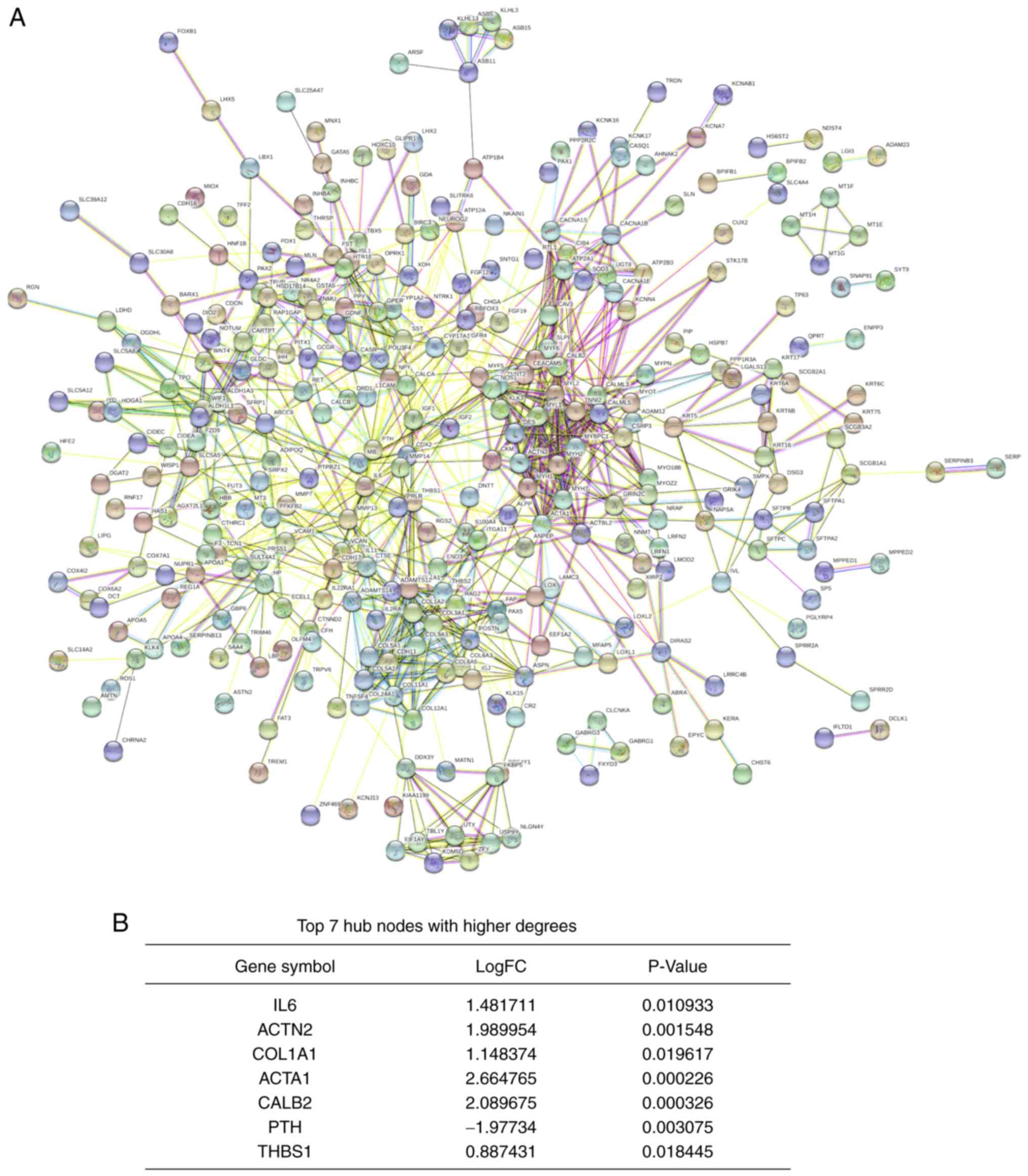
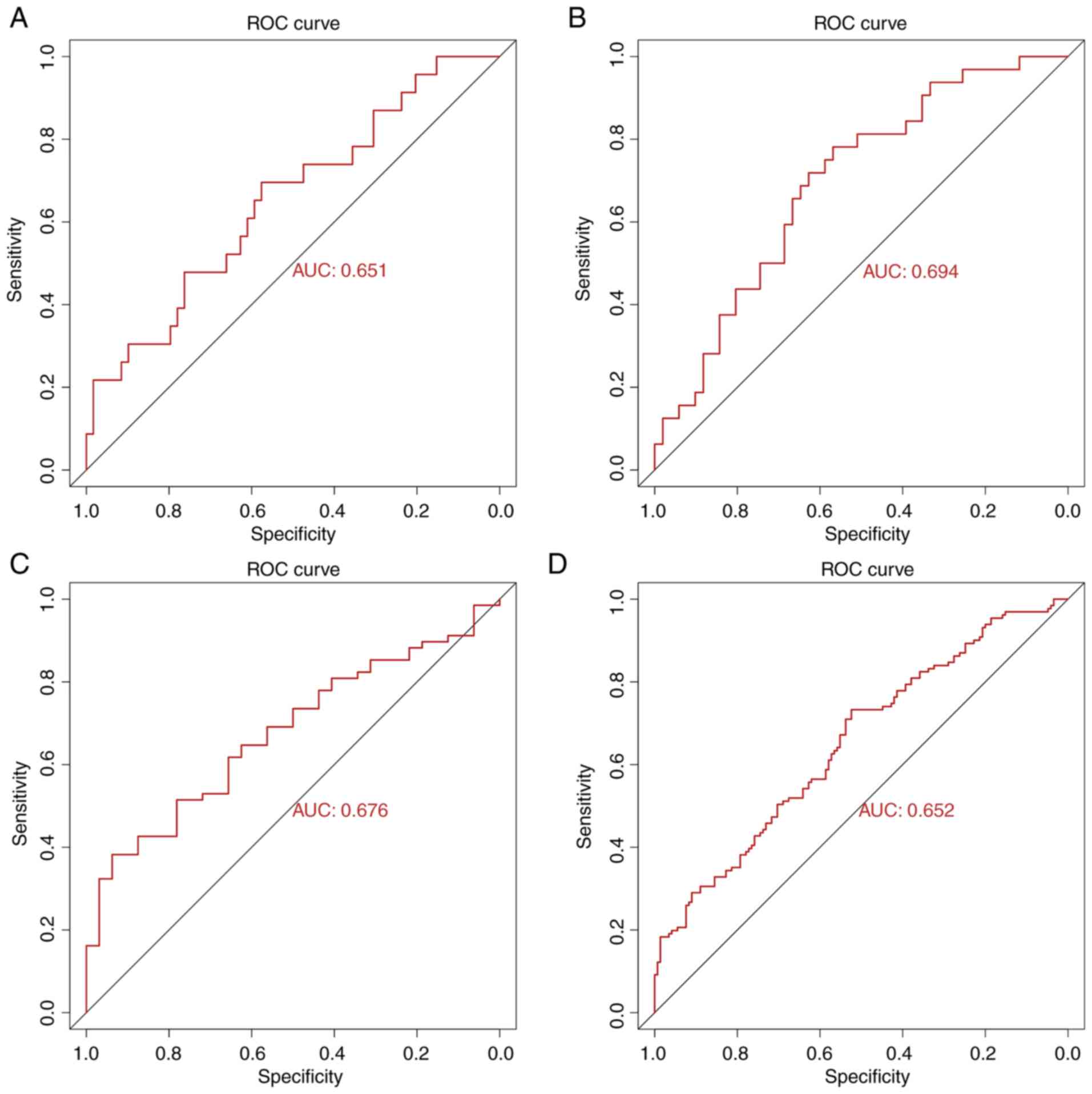
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lim H, Devesa SS, Sosa JA, Check D and
Kitahara CM: Trends in thyroid cancer incidence and mortality in
the United States, 1974–2013. JAMA. 317:1338–1348. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Davies L and Welch HG: Current thyroid
cancer trends in the United States. JAMA Otolaryngol Head Neck
Surg. 140:317–322. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Morris LG, Sikora AG, Tosteson TD and
Davies L: The increasing incidence of thyroid cancer: The influence
of access to care. Thyroid. 23:885–891. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang L, Wei WJ, Ji QH, Zhu YX, Wang ZY,
Wang Y, Huang CP, Shen Q, Li DS and Wu Y: Risk factors for neck
nodal metastasis in papillary thyroid microcarcinoma: A study of
1066 patients. J Clin Endocrinol Metab. 97:1250–1257. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Adam MA, Pura J, Goffredo P, Dinan MA,
Reed SD, Scheri RP, Hyslop T, Roman SA and Sosa JA: Presence and
number of lymph node metastases are associated with compromised
survival for patients younger than age 45 years with papillary
thyroid cancer. J Clin Oncol. 33:2370–2375. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang Q, Wang Z, Meng X, Duh QY and Chen
G: Predictors for central lymph node metastases in CN0 papillary
thyroid microcarcinoma (mPTC): A retrospect analysis of 1304 cases.
Asian J Surg. 42:571–576. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pisanu A, Saba A, Podda M, Reccia I and
Uccheddu A: Nodal metastasis and recurrence in papillary thyroid
microcarcinoma. Endocrine. 48:575–581. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jin WX, Ye DR, Sun YH, Zhou XF, Wang OC,
Zhang XH and Cai YF: Prediction of central lymph node metastasis in
papillary thyroid microcarcinoma according to clinicopathologic
factors and thyroid nodule sonographic features: A case-control
study. Cancer Manag Res. 10:3237–3243. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xia E, Wang Y, Bhandari A, Niu J, Yang F,
Yao Z and Wang O: CITED1 gene promotes proliferation, migration and
invasion in papillary thyroid cancer. Oncol Lett. 16:105–112.
2018.PubMed/NCBI
|
|
12
|
Jin Y, Jin W, Zheng Z, Chen E, Wang Q,
Wang Y, Wang O and Zhang X: GABRB2 plays an important role in the
lymph node metastasis of papillary thyroid cancer. Biochem Biophys
Res Commun. 492:323–330. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Collins A: The cancer genome atlas (TCGA)
pilot project. Cancer Res. 67:2007.
|
|
14
|
Perrier ND, Brierley JD and Tuttle RM:
Differentiated and anaplastic thyroid carcinoma: Major changes in
the American joint committee on cancer eighth edition cancer
staging manual. CA Cancer J Clin. 68:55–63. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
McCarthy DJ, Chen Y and Smyth GK:
Differential expression analysis of multifactor RNA-Seq experiments
with respect to biological variation. Nucleic Acids Res.
40:4288–4297. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kolde R, Laur S, Adler P and Vilo J:
Robust rank aggregation for gene list integration and
meta-analysis. Bioinformatics. 28:573–580. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Walter W, Sánchez-Cabo F and Ricote M:
GOplot: An R package for visually combining expression data with
functional analysis. Bioinformatics. 31:2912–2914. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47(D1): D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Robin X, Turck N, Hainard A, Tiberti N,
Lisacek F, Sanchez JC and Müller M: pROC: An open-source package
for R and S+ to analyze and compare ROC curves. BMC Bioinformatics.
12:772011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang N, Li Y, Wei J, Pu J, Liu R, Yang Q,
Guan H, Shi B, Hou P and Ji M: TBX1 functions as a tumor suppressor
in thyroid cancer through inhibiting the activities of the PI3K/AKT
and MAPK/ERK pathways. Thyroid. 29:378–394. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Choi C, Thi Thao Tran N, Van Ngu T, Park
SW, Song MS, Kim SH, Bae YU, Ayudthaya PDN, Munir J, Kim E, et al:
Promotion of tumor progression and cancer stemness by MUC15 in
thyroid cancer via the GPCR/ERK and integrin-FAK signaling
pathways. Oncogenesis. 7:852018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vitale G, Dicitore A, Pepe D, Gentilini D,
Grassi ES, Borghi MO, Gelmini G, Cantone MC, Gaudenzi G, Misso G,
et al: Synergistic activity of everolimus and
5-aza-2′-deoxycytidine in medullary thyroid carcinoma cell lines.
Mol Oncol. 11:1007–1022. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang H, Teng X and Liu Z, Zhang L and Liu
Z: Gene expression profile analyze the molecular mechanism of CXCR7
regulating papillary thyroid carcinoma growth and metastasis. J Exp
Clin Cancer Res. 34:162015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Duquette M, Sadow PM, Lawler J and Nucera
C: Thrombospondin-1 silencing down-regulates integrin expression
levels in human anaplastic thyroid cancer cells with BRAF(V600E):
New insights in the host tissue adaptation and homeostasis of tumor
microenvironment. Front Endocrinol (Lausanne). 4:1892013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yee KO, Connolly CM, Duquette M,
Kazerounian S, Washington R and Lawler J: The effect of
thrombospondin-1 on breast cancer metastasis. Breast Cancer Res
Treat. 114:85–96. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang T, Wang L, Liu D, Li P, Xiong H,
Zhuang L, Sun L, Yuan X and Qiu H: FGF7/FGFR2 signal promotes
invasion and migration in human gastric cancer through upregulation
of thrombospondin-1. Int J Oncol. 50:1501–1512. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Borsotti P, Ghilardi C, Ostano P, Silini
A, Dossi R, Pinessi D, Foglieni C, Scatolini M, Lacal PM, Ferrari
R, et al: Thrombospondin-1 is part of a Slug-independent motility
and metastatic program in cutaneous melanoma, in association with
VEGFR-1 and FGF-2. Pigment Cell Melanoma Res. 28:73–81. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jayachandran A, Anaka M, Prithviraj P,
Hudson C, McKeown SJ, Lo PH, Vella LJ, Goding CR, Cebon J and
Behren A: Thrombospondin 1 promotes an aggressive phenotype through
epithelial-to-mesenchymal transition in human melanoma. Oncotarget.
5:5782–5797. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhao J, Shi L, Zeng S, Ma C, Xu W, Zhang
Z, Liu Q, Zhang P, Sun Y and Xu C: Importin-11 overexpression
promotes the migration, invasion, and progression of bladder cancer
associated with the deregulation of CDKN1A and THBS1. Urol Oncol.
36:311.e1–311.e13. 2018. View Article : Google Scholar
|
|
34
|
Teraoku H, Morine Y, Ikemoto T, Saito Y,
Yamada S, Yoshikawa M, Takasu C, Higashijima J, Imura S and Shimada
M: Role of thrombospondin-1 expression in colorectal liver
metastasis and its molecular mechanism. J Hepatobiliary Pancreat
Sci. 23:565–573. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tzeng HT, Tsai CH, Yen YT, Cheng HC, Chen
YC, Pu SW, Wang YS, Shan YS, Tseng YL, Su WC, et al: Dysregulation
of Rab37-mediated cross-talk between cancer cells and endothelial
cells via thrombospondin-1 promotes tumor neovasculature and
metastasis. Clin Cancer Res. 23:2335–2345. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Han J, Meng Q, Xi Q, Zhang Y, Zhuang Q,
Han Y, Jiang Y, Ding Q and Wu G: Interleukin-6 stimulates aerobic
glycolysis by regulating PFKFB3 at early stage of colorectal
cancer. Int J Oncol. 48:215–224. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Luan S, An Z, Bi S, Chen L and Fan J:
Interleukin 6 receptor (IL-6R) was an independent prognostic factor
in cervical cancer. Histol Histopathol. 33:269–276. 2018.PubMed/NCBI
|
|
38
|
Shang GS, Liu L and Qin YW: IL-6 and TNF-α
promote metastasis of lung cancer by inducing
epithelial-mesenchymal transition. Oncol Lett. 13:4657–4660. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hansel DE, Rahman A, House M, Ashfaq R,
Berg K, Yeo CJ and Maitra A: Met proto-oncogene and insulin-like
growth factor binding protein 3 overexpression correlates with
metastatic ability in well-differentiated pancreatic endocrine
neoplasms. Clin Cancer Res. 10:6152–6158. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Guvakova MA, Adams JC and Boettiger D:
Functional role of alpha-actinin, PI 3-kinase and MEK1/2 in
insulin-like growth factor I receptor kinase regulated motility of
human breast carcinoma cells. J Cell Sci. 115:4149–4165. 2002.
View Article : Google Scholar : PubMed/NCBI
|