Introduction
Genome-wide transcriptome analysis has revealed that
the majority (>98%) of human genes are non-protein coding genes
(1,2). Non protein-coding genes transcribe
non-coding RNAs (ncRNAs) that participate directly in developmental
and differential processes by regulating gene expression at
multiple levels, such as at post-transcriptional, translational and
epigenetic levels (3). Long ncRNAs
(lncRNAs) are a subgroup of ncRNAs that are longer than 200
nucleotides (4). Most of the
characterized lncRNAs are specifically expressed in certain types
of cells and tissues (5). However,
lncRNAs can also enter the circulating system to achieve systemic
regulation of gene expression. Thus, certain circulating lncRNAs
may be reflected in the RNA levels of lesions (6), indicating their potential role as
biomarkers for diseases. At present, the functions of most lncRNAs
remain unknown.
Colon cancer is one of the most commonly diagnosed
malignancies (7). Colon cancer
causes >60,000 deaths and >130,000 new cases are reported
every year in the United States (8).
The 5-year survival rate of patients with colon cancer at the early
stages following active treatment is >70% (9,10).
However, the treatment outcomes of patients at the advanced stages
remain poor due to the lack of radical treatment (9,10).
Therefore, novel therapeutic targets and prognostic markers are
required. The lncRNA mortal obligate RNA transcript (MORT) is
inhibited in numerous types of cancer in humans, such as ovarian
cancer and gastric cancer (11),
indicating its role as a tumor suppressor. The present study
investigated the involvement of lncRNA MORT in colon cancer and
observed its downregulation and its association with prognosis. A
primary aim of the present study was to investigate the interaction
between MORT and TGF-β signaling, which mediates diverse functions
in cancer biology by interacting with multiple downstream pathways
and regulating cancer cell behaviors (12,13).
Materials and methods
Patients and follow-up
The present study included 68 patients with colon
cancer, who were admitted to The Sixth Affiliated Hospital of Sun
Yat-sen University between July 2011 and July 2013. The inclusion
criteria were: i) Colon cancer diagnosed by pathological biopsies;
ii) understanding of the experimental principle and willingnes to
participate; and iii) informed consent. The exclusion criteria
were: Patients i) with other diseases; ii) who failed to complete
the 5-year follow-up; and iii) who died due to other causes during
follow-up. Follow-up was performed via telephone, every month for 5
years following admission. The patients included 12 individuals at
stage I, 18 at stage II, 20 at stage III and 18 at stage IV,
according to the staging guidelines of the American Joint Committee
on Cancer (14). There were 39 males
and 29 females with a mean age of 48.6±4.4 years (range, 32–66
years). Patients were treated with surgical resections and/or
chemotherapy according to their disease conditions. The Ethics
Committee of The Sixth Affiliated Hospital of Sun Yat-Sen
University approved this study.
Specimens and cell line
Tumor and adjacent (collected ≤3 cm from the tumor
border) tissues were obtained from all patients using fine needle
biopsies under the guidance of MRI. Blood (5 ml) was extracted from
each patient into EDTA-treated tubes one day after admission under
fasting conditions. The blood-collection tubes were centrifuged at
1,200 × g for 20 min at room temperature to isolate the plasma from
the blood. All samples were stored in a liquid nitrogen sink at
−80°C before use.
The colon cancer RKO cell line (American Type
Culture Collection) was used and cultured in Eagle's Minimum
Essential medium (MEM; Sigma-Aldrich; Merck KGaA) supplemented with
10% FBS (Sigma-Aldrich; Merck KGaA) at 5% CO2 and
37°C.
Cell transfection
PcDNA3.1 vectors expressing MORT (NCBI accession
no.; NR_036521.1) were designed and constructed by Sangon Biotech
Co., Ltd. Lipofectamine® 2000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) was used to transfect 10 nM vectors
into 1×106 cells. The non-transfected cells were the
control cells (C) and cells transfected with the empty vector were
the negative control (NC) cells. Cells were harvested 24 h after
transfection to perform subsequent experiments.
Total RNA extraction and reverse
transcription-quantitative (RT-q)PCR
Tissues were ground and mixed with RNAzol reagent
(Sigma-Aldrich; Merck KGaA) to extract total RNA. The RNAzol
reagent was also directly mixed with plasma from patients and cells
cultured in vitro to extract total RNA. Furthermore,
exogenous treatment with TGF-β1 (Sigma-Aldrich; Merck KGaA) at
doses of 5, 10 and 20 ng/ml at 37°C for 24 h was performed in
certain cases, prior to RNA extraction. SuperScript IV Reverse
Transcriptase (Thermo Fisher Scientific, Inc.) was used to perform
RT. The thermal protocol for the reverse transcription stage was
52°C for 30 min, followed by 80°C for 10 min. In order to detect
lncRNA MORT and transforming growth factor β1 (TGF-β1) mRNA,
qScript One-Step RT-qPCR kit (Quantabio) was used to prepare the
qPCR mixture. The ABI 7500 system was used to carry out all qPCR
reactions with GAPDH as the endogenous control. Thermocycling
conditions for the PCR reactions were: 95°C for 1 min, followed by
40 cycles of 95°C for 10 sec and 60°C for 35 sec. Primers of lncRNA
MORT, TGF-β1 and GAPDH were obtained from Sangon Biotech Co., Ltd.
MORT and TGF-β1 were normalized to GAPDH, according to the
2−ΔΔCq method (15). The
primer sequences were: MORT forward, 5′-GTGTCCGCCATAAAGTCGTT-3′;
MORT reverse, 5′-CTGCTATCATTCGCCATGAC-3′; TGF-β1 forward,
5′-AAGAAGTCACCCGCGTGCTA-3′; TGF-β1 reverse,
5′-TGTGTGATGTCTTTGGTTTTGTCA-3′; GAPDH forward,
5′-CTGCACCACCAACTGCTTAC-′3; and GAPDH reverse,
5′-CAGAGGTGCCATCCAGAGTT-3′.
Measurement of cell migration and
invasion rates
Transwell inserts (8 µl, Corning) were used to
analyze cell invasion and migration. Cells collected 24 h after
transfection were mixed with serum-free MEM (Sigma-Aldrich; Merck
KGaA) to prepare a single-cell suspension at a concentration of
5×104 cells/ml. The cell suspensions were added to upper
chamber of the 96-well plate (0.1 ml per well). MEM (20% FBS;
Sigma-Aldrich; Merck KGaA) was used to fill the lower chamber. In
order to mimic cell invasion in vivo, Matrigel (Merck KGaA)
was used to coat the membrane of the upper chamber at 37°C for 12 h
prior to performing the invasion assay. Uncoated membranes were
used for migration assay, but the same protocol was followed. The
plate was incubated at 37°C in 5% CO2 for 2 h.
Subsequently, the upper chamber membranes were stained at 25°C for
90 min with 0.5% crystal violet (Sigma-Aldrich; Merck KGaA). An
optical microscope was used to count the stained cells
(magnification, ×40).
Western blot assay
RIPA buffer (Invitrogen; Thermo Fisher Scientific,
Inc.) was used for total protein extraction, and the BCA assay
(Invitrogen; Thermo Fisher Scientific, Inc.) was used for protein
quantification. To denature proteins, protein samples were
incubated with boiling water for 10 min. After that, 10% SDS-PAGE
was used to separate proteins (30 µg per lane) and proteins were
transferred to PVDF membranes. PBS (Sigma-Aldrich; Merck KGaA)
containing 5% non-fat milk was used to coat membranes at room
temperature for 2 h. After that, GAPDH (1:1,000; cat. no. ab9845;
Abcam) and TGF-β1 (1:1,000; cat. no. ab92486; Abcam) primary
antibodies were used to incubate the membranes for 12 h at 4°C,
followed by incubation with secondary goat anti-rabbit (horseradish
peroxidase, 1:1,000; cat. no. ab6721; Abcam) for 2 h at room
temperature. Enhanced chemiluminescence system (ECL; GE Healthcare)
was used for signal production. All signals were analyzed using
Quantity One software v.4.6 (Bio-Rad Laboratories, Inc.).
Statistical analysis
Experiments were repeated three times to calculate
mean values ± standard deviation. Prism 6.01 software (GraphPad
Software, Inc.) was used to carry out all statistical analyses. The
association between the expression level of MORT in the biopsies
and plasma of patients with colon cancer was analyzed using linear
regression analysis. The expression of MORT was compared between
tumor and healthy tissues using the paired t-test. The associations
between clinicopathological factors of patients (age, gender and
clinical stages) and the plasma levels of MORT were analyzed using
the χ2 test. The expression of TGF-β1 and the cell
migration and invasion rates were compared among the different
groups of transfected cells using one-way ANOVA, followed by
Tukey's post hoc test (all data met the assumption of homogeneity
of variance). According to the expression levels of lncRNA MORT in
tumor tissues, the patients were divided into high- (n=31) and low-
(n=37) lncRNA MORT expression groups using the cut-off value of
2.27, identified by Youden's index. Survival curves were plotted
for both of these groups, based on the follow-up data, using the
Kaplan-Meier plotter (Prism 6; GraphPad Software, Inc.) and were
compared with the log rank test. P<0.05 was considered to
indicate a statistically significant difference.
Results
LncRNA MORT is downregulated in colon
tumor tissues
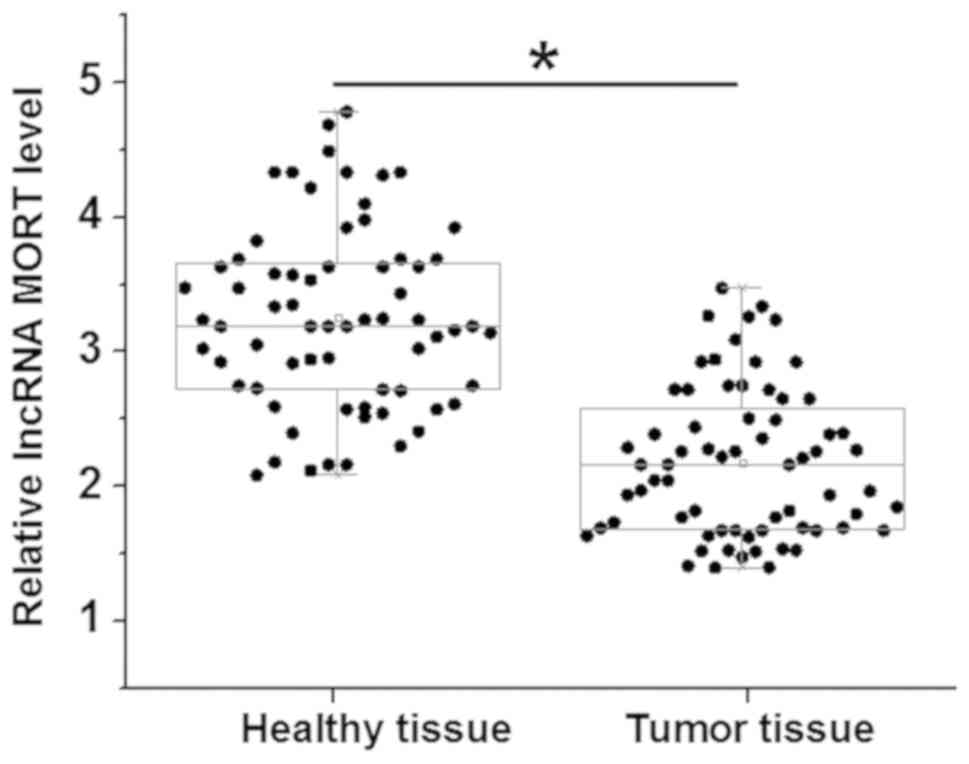
MORT was detected by RT-qPCR in 68 tumor and
adjacent healthy tissues from patients with colon cancer. The
differences in the levels of MORT expression between the two types
of tissue were analyzed using a paired t-test. Compared with that
in the healthy tissues, the expression of MORT was significantly
decreased in the tumor samples (P<0.05; Fig. 1). No significant differences were
found in the expression levels of MORT, in tumor and healthy
tissues, among patients at different clinical stages (data not
shown).
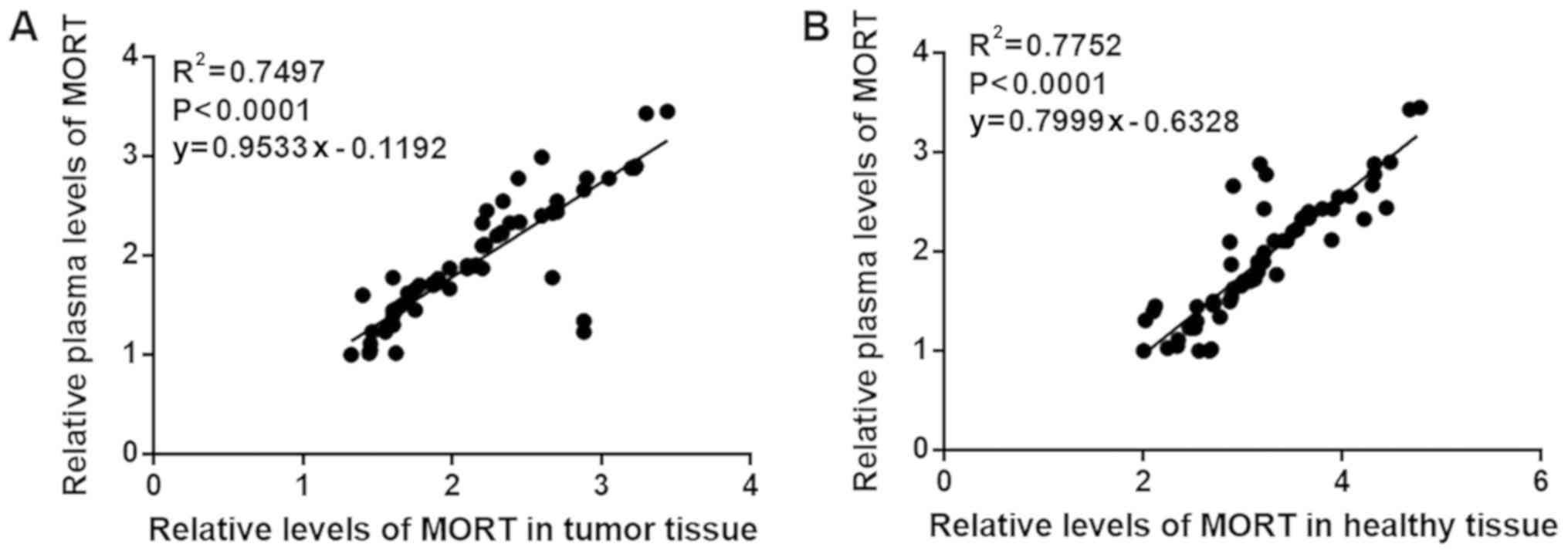
The expression level of lncRNA MORT in
tissue is linearly associated with plasma levels
The plasma lncRNA MORT levels in 68 patients with
colon cancer were measured using RT-qPCR. The association between
the expression levels of MORT in the biopsies and plasma of the
patients was analyzed by linear regression analysis. As shown in
Fig. 2A, MORT expression in tumor
tissues was significantly associated with the plasma levels
(P<0.0001). In addition, the expression of MORT in healthy
tissues was also significantly associated with the levels in plasma
(P<0.0001; Fig. 2B).
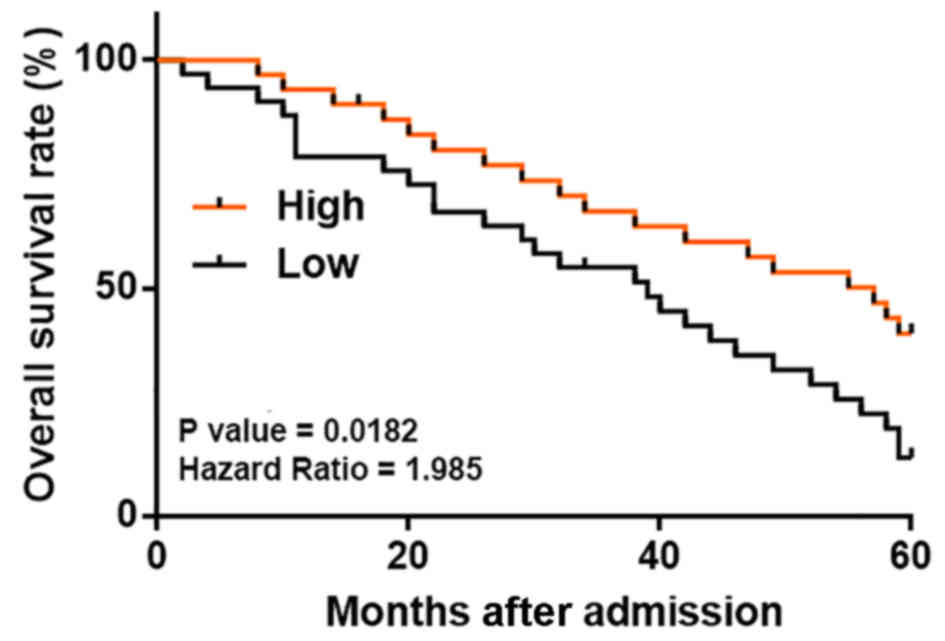
Low lncRNA MORT plasma levels are
associated with low overall survival (OS) rate in patients with
colon cancer
It was observed that the plasma levels of MORT were
not significantly associated with the patient age, gender and
clinical stage (all P>0.05; data not shown). The patients were
divided into high (n=31) and low (n=37) plasma MORT level groups.
Kaplan-Meier survival curves were plotted for both groups, based on
the follow-up data, and compared using the log rank test. As shown
in Fig. 3, the patients with low
plasma levels of lncRNA MORT had a significantly lower OS rate.
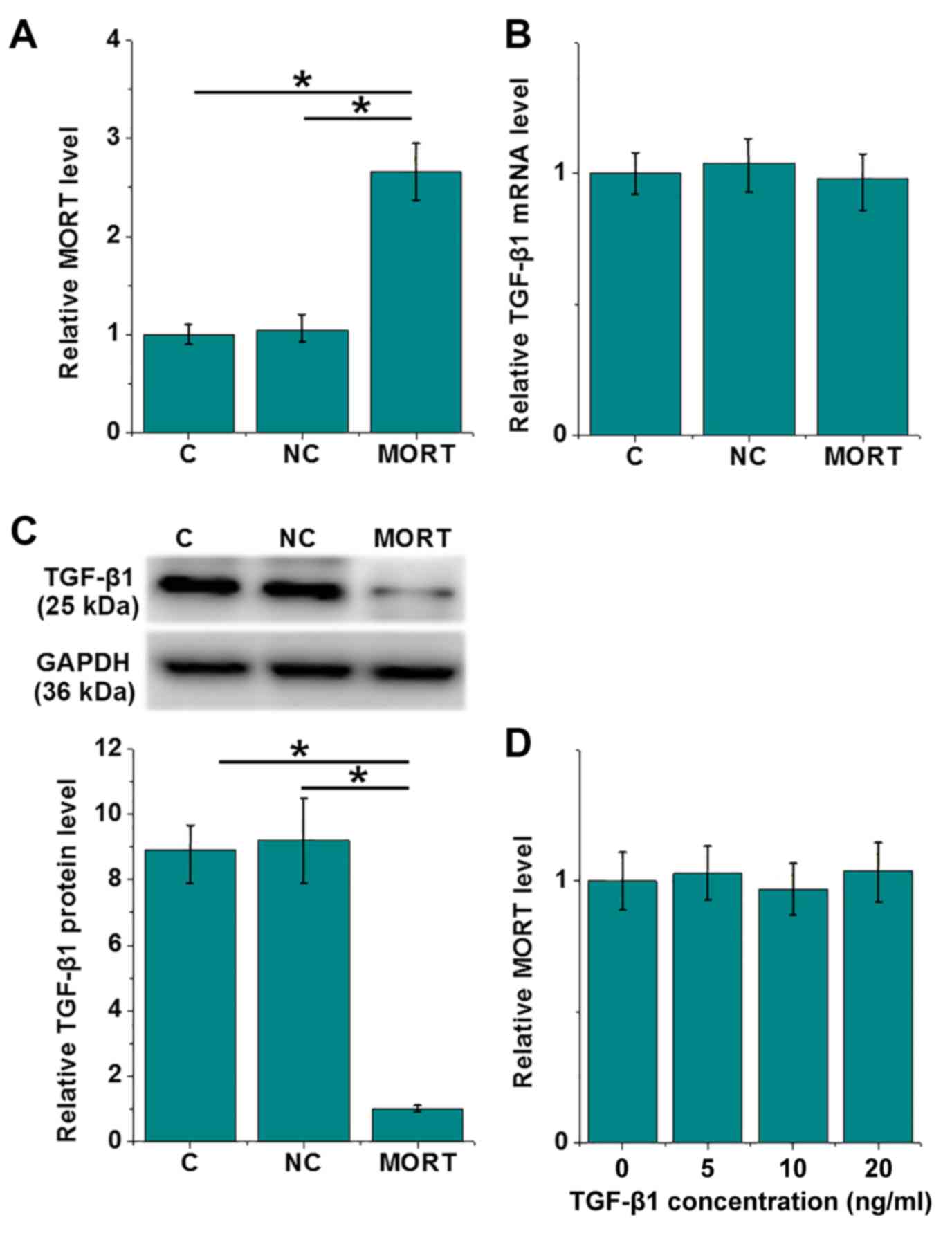
LncRNA MORT is an upstream inhibitor
of TGF-β1 in colon cancer RKO cells
The overexpression of MORT was achieved 24 h after
transfection of an overexpression plasmid in RKO cells (P<0.05;
Fig. 4A). Compared with the C and NC
cells, the overexpression of lncRNA MORT led to no significant
difference in the mRNA level of TGF-β1 (Fig. 4B), whereas its protein level was
significantly decreased (P<0.05; Fig.
4C). In contrast, exogenous treatment with TGF-β1 at doses of
5, 10 and 20 ng/ml for 24 h had no significant effect on the
expression of MORT (Fig. 4D).
Overexpression of LncRNA MORT
decreases migration and invasion of RKO cells by inhibiting
TGF-β1
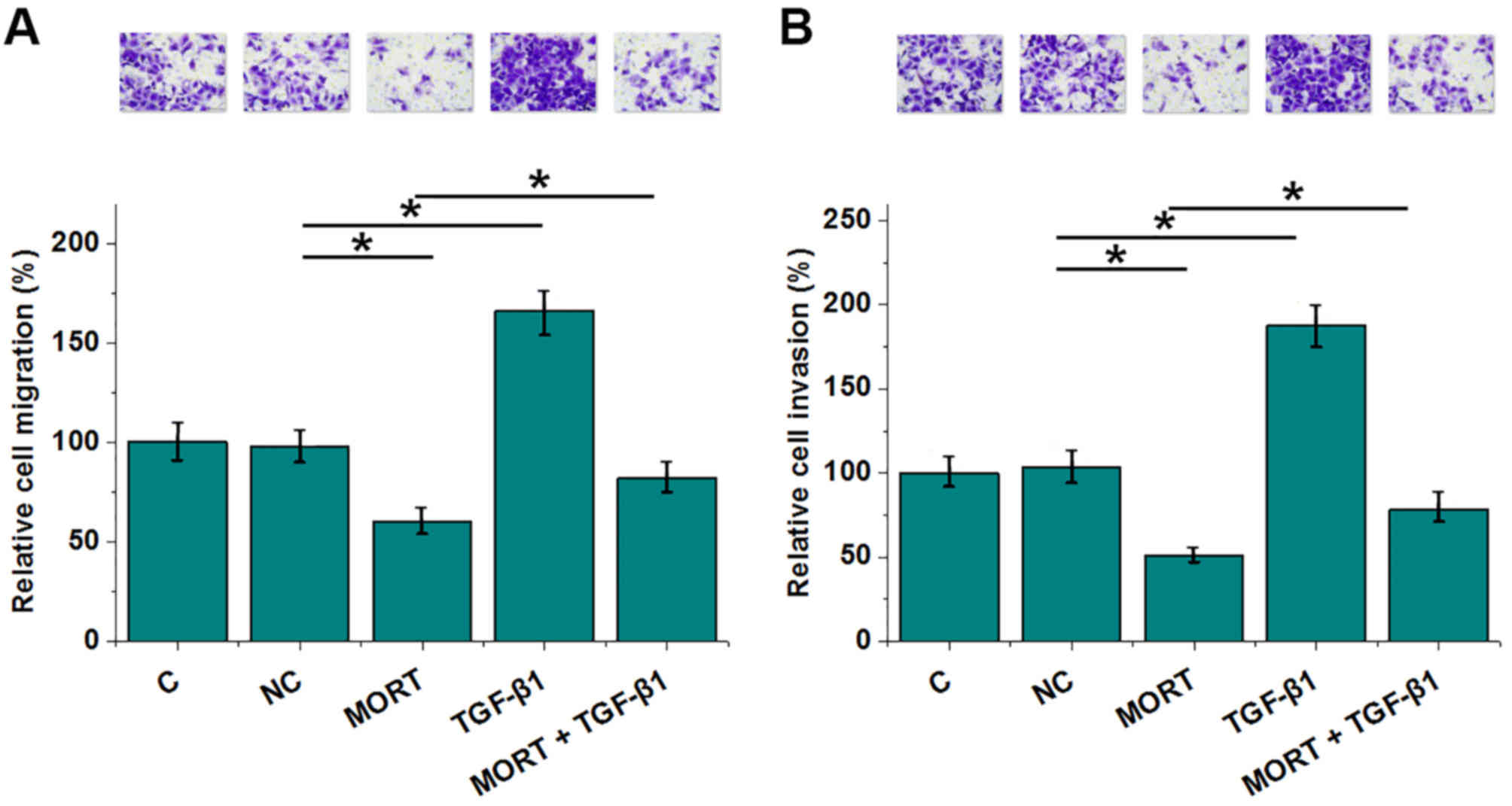
Compared with the negative control (NC) group,
lncRNA MORT overexpression resulted in decreased, whereas TGF-β1
treatment resulted in increased rates of migration (Fig. 5A) and invasion (Fig. 5B) of colon cancer cells (all
P<0.05). In addition, TGF-β1 significantly decreased the effects
of MORT overexpression (P<0.05).
Discussion
At present, the prognosis of patients with colon
cancer remains poor, especially for those at advanced stages
(7–10). The present study demonstrated
downregulation of lncRNA MORT in colon cancer and its association
with low OS rate in patients.
TGF-β signaling is a well-characterized signaling
transduction pathway in cancer biology (12,13). It
is generally considered that TGF-β signaling activation in most, if
not all, types of cancer inhibits tumor cell proliferation at the
early stages and promotes tumor metastasis at the later stages
(12,13). In clinical practices, TGF-β signaling
inhibition can also improve cancer treatment outcomes, such as
overall survival time (16).
However, the activation of TGF-β signaling can also promote the
development of colon cancer (17).
Consistent with previous studies, the present study reported
increased migration and invasion of colon cancer cells in response
to exogenous TGF-β1 treatment. The TGF-β pathway participates in
cancer biology by regulating downstream signaling molecules, such
as lncRNAs (18). The present study
suggests that the TGF-β pathway may be regulated by the lncRNA
MORT.
Several characterized lncRNAs are specifically
expressed in certain types of cells and tissues, indicating their
specific involvement in certain biological processes (5). However, lncRNAs can also enter the
circulating system to regulate gene expression globally (6). The present study detected the
expression levels of MORT in both tumor and healthy tissues, which
were linearly associated with the plasma levels. This indicates
that the lncRNA MORT expressed in tissues may be released into the
bloodstream. Therefore, MORT may serve as a regulator of gene
expression. The circulating levels of lncRNAs may be used as
markers to reflect diseases (6). The
findings from the present study suggest MORT as a potential
prognostic marker for colon cancer. Therefore, detecting the plasma
levels of MORT may be valuable for the design of follow-up care
after treatment. However, further clinical studies are required to
confirm this hypothesis. A limitation of the present study was that
it failed to elucidate the mechanism underlying the interaction
between MORT and TGF-β; consequently, further studies are required
to investigate this process.
In conclusion, lncRNA MORT is downregulated in colon
cancer and is associated with low OS rate. Moreover, overexpression
of lncRNA MORT inhibits migration and invasion of colon cancer
cells by inhibiting TGF-β protein expression.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
TZ, LW and ZZ conducted experiments, analyzed all
the data and were major contributors in writing the manuscript. NM,
YL and ZJ conducted experiments. QW and SC contributed to the study
design. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Approval was obtained from the Ethics Committee of
The Sixth Affiliated Hospital of Sun Yat-Sen University, Guangzhou,
China. Written informed consent was provided by all
participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mattick JS and Rinn JL: Discovery and
annotation of long noncoding RNAs. Nat Struct Mol Biol. 22:5–7.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mattick JS: Non-coding RNAs: The
architects of eukaryotic complexity. EMBO Rep. 2:986–991. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Qi P, Zhou XY and Du X: Circulating long
non-coding RNAs in cancer: Current status and future perspectives.
Mol Cancer. 15:392016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RGS, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liang B, Shahbaz M, Wang Y, Gao H, Fang R,
Niu Z, Liu S, Wang B, Sun Q, Niu W, et al: Integrinbeta6-targeted
immunoliposomes mediate tumor-specific drug delivery and enhance
therapeutic efficacy in colon carcinoma. Clin Cancer Res.
21:1183–1195. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mannucci S, Ghin L, Conti G, Tambalo S,
Lascialfari A, Orlando T, Benati D, Bernardi P, Betterle N, Bassi
R, et al: Magnetic nanoparticles from Magnetospirillum
gryphiswaldense increase the efficacy of thermotherapy in a model
of colon carcinoma. PLoS One. 9:e1089592014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vrba L and Futscher BW: Epigenetic
silencing of lncRNA MORT in 16 TCGA cancer types. F1000Res.
7:2112018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Derynck R, Akhurst RJ and Balmain A:
TGF-beta signaling in tumor suppression and cancer progression. Nat
Genet. 29:117–129. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Akhurst RJ and Derynck R: TGF-beta
signaling in cancer-a double-edged sword. Trends Cell Biol.
11:S44–S51. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hari DM, Leung AM, Lee JH, Sim MS, Vuong
B, Chiu CG and Bilchik AJ: AJCC Cancer Staging Manual 7th edition
criteria for colon cancer: Do the complex modifications improve
prognostic assessment? J Am Coll Surg. 217:181–190. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Colak S and Ten Dijke P: Targeting TGF-β
Signaling in Cancer. Trends Cancer. 3:56–71. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Peng X, Luo Z, Kang Q, Deng D, Wang Q,
Peng H, Wang S and Wei Z: FOXQ1 mediates the crosstalk between
TGF-β and Wnt signaling pathways in the progression of colorectal
cancer. Cancer Biol Ther. 16:1099–1109. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-β promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI
|