Introduction
Lung cancer is a leading contributor to
cancer-related death worldwide. Non-small cell lung cancer (NSCLC)
accounts for ~80% of all lung cancer cases, including squamous cell
carcinoma and adenocarcinoma (1,2).
Although great progress has been made in chemotherapy and surgery,
NSCLC patients still have poor prognosis and a 5-year survival rate
of <15% due to latent symptoms at the early stage and the high
malignant potential of NSCLC (3).
Therefore, to improve clinical efficacy of NSCLC therapies, it is
necessary to seek biomarkers involved in the occurrence and
development of NSCLC and clarify the pathogenesis of NSCLC.
MicroRNA (miRNA) is an endogenous non-coding RNA
(4) that regulates gene expression
at the post-transcriptional level by binding to the 3′-
untranslated region (3′-UTR) of target mRNA (5). miRNA participates in a variety of
biological processes, including cell proliferation,
differentiation, invasion, angiogenesis and apoptosis (6). Abnormal expression of miRNA has been
reported to play a critical role in the occurrence and development
of tumors. Wan et al (7)
suggested that miR-27b expression is notably downregulated in NSCLC
tissues and cells, and expression of LIMK1 is upregulated to
inhibit the proliferation and invasion of tumors. miR-205 can be
used as a new therapeutic target due to its downregulation in
glioma and its inhibition of the migration and invasion of tumor
cells through targeting YAP1. The latest evidence links many miRNAs
to the regulation of the occurrence and progression of NSCLC
(8), and the abnormally expressed
miRNA is involved in tumor progression in NSCLC as an oncogene or
tumor inhibiting factor (9). Despite
the headway in research on miRNA in NSCLC, the relationship between
them has not been well-established and requires further efforts.
miR-15a-3p is found downregulated in cervical cancer while it
inhibits tumor cell proliferation, induces cell apoptosis, and
raises the sensitivity of tumor cells to radiotherapy by regulating
TPD (10). Jin et al
(11) found that miR-15a may be a
molecular therapeutic target for thyroid cancer, which inhibits
RET/AKT signaling pathways to inhibit metastasis and invasion of
thyroid cancer However, the effect of miR-15a on the biological
function of NSCLC and its mechanism of action in NSCLC are still
unclear.
The current study focused on the role of miR-15a in
NSCLC metastasis and in the proliferation, metastasis and invasion
of NSCLC by targeted-regulating of mothers against decapitaplegic
homolog3 (Smad3) expression, providing fundamental theoretical
basis for further understanding the occurrence and development
mechanism of NSCLC and prognosis evaluation of NSCLC patients.
Materials and methods
Main reagents, instruments and cell
lines
Annexin V-FITC, MTT kits, and HRP-labeled Goat
Anti-Rabbit IgG (A0208) were from Beyotime Biotechnology; RPMI-1640
medium, fetal bovine serum, penicillin-streptomycin and trypsin
from Gibco; Thermo Fisher Scientific, Inc.; TRIzol reagent and
Transwell cell culture plates from Corning Inc.; Promega M-MLV
reverse transcription kits from Promega Corporation; YBR Premix Ex
Taq from Takara Biotechnology Co., Ltd.; miR-15a overexpression
plasmid was synthesized by Guangzhou RiboBio Co., Ltd.;
Lipofectamine® 3000 Transfection kit was from
Invitrogen; Thermo Fisher Scientific, Inc.; Smad3 protein (rabbit
anti-human Smad3 monoclonal antibody, ab40854) from Abcam; GAPDH
antibody (mouse anti-human GAPDH monoclonal antibody, SC-32233)
from Santa Cruz Biotechnology, Inc.; Immobilon Western HRP from
Thermo Fisher Scientific, Inc.
Human NSCLC cell lines (A549, H1299, and H1975) and
the normal lung cells (BEAS-2B) were all from Shanghai Institute of
Biochemistry and Cell Biology, CAS. The cells were cultured in DMEM
(Corning Inc.) medium containing 10% fetal bovine serum (FBS)
(Thermo Fisher Scientific, Inc.) and 1% streptomycin (Corning Inc.)
at 37°C with the concentration of 5% CO2.
Clinical specimens
Fifty patients with NSCLC who underwent surgical
treatment in the thoracic surgery department of Shandong Provincial
Chest Hospital (Jinan, China) between January 2016 and December
2018 were enrolled. Inclusion criteria: The patients received
surgical treatment in the above hospital and had primary lesions,
and all specimens were pathologically confirmed as NSCLC. Exclusion
criteria: those who received radiotherapy, chemotherapy or
interventional therapies before treatment, and those who had other
metastases before treatment. Tumor tissues and para-cancerous
tissue of NSCLC patients were collected (2 cm away from tumor). The
specimens were stored in a liquid nitrogen container 10 min after
surgery in vitro for subsequent steps. The study was
examined and approved by the Ethics Committee of Shandong
Provincial Chest Hospital. Signed informed consents were obtained
from the patients and/or the guardians.
Cell culture and transfection
FBS, penicillin-streptomycin, and RPMI-1640 basal
medium were prepared into RMPI-1640 complete medium with 10% FBS
and 1% penicillin-streptomycin. The cells were cultured at 37°C
with the concentration of 5% CO2. The cells were
inoculated in a 6-well plate with an inoculation density of
~2.5×106 cells/well, and incubated in a constant
temperature incubator. Logarithmically growing cells were chosen
and inoculated in a culture plate, the cell confluence was ~80%
before transfection. The cells were divided into the empty plasmid
group (miR-NC group), and the transfection simulation sequence
group (5′-CTCAACTGGTGTCGTGGAGTC-3′) (miR-15a mimic group). After 36
h of transfection, the cells were trypsinized and then collected
for subsequent steps.
Detection of the expression level of
miR-15a mRNA before and after transfection via RT-PCR
Total RNA was extracted from tissues and cells using
TRIzol reagent, quantitatively detected in terms of content with an
ultraviolet spectrophotometer, and then reverse transcribed to
obtain cDNA and the transcribed cDNA was amplified by RT-PCR. The
primer sequences are shown in Table
I. RT-PCR reactions were performed with 10 µl SYBR Premix Ex
Taq, 0.4 µl forward primer, 0.4 µl reverse primer, 2 µl cDNA, and
7.2 µl sterilized distilled water. Pre-denaturation lasted for 10
min at 95°C, denaturation for 30 sec at 95°C, annealing for 30 sec
at 60°C and extension for 30 sec at 74°C. The circle was repeated
40 times.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Primer | Primer sequence |
|---|
| GAPDH | F,
5′-GTGGACCTCATGCTACAT-3′ |
|
| R,
5′-TGTGAGGGAGATGCTCAGTG-3′ |
| miR-15a | F,
5′-TCCAGCTGGCAGCATG-3′ |
|
| R,
5′-GTCGTGGAGTCACTCG-3′ |
Detection of the proliferation of
tumor cells via the MTT assay
Trypsinization was carried out 36 h post
transfection to collect cells of each group, and then the cells
were inoculated in 96-well plates with 5×103 cells/well,
respectively. OD value was measured at 490 nm 4 h after 20 µl of
MTT solution was added to each well at days 2, 3, 4 and 5 of
inoculation, and a cell growth curve was plotted. The trial was
repeated 3 times taking the average OD value. The cell growth
inhibitory concentration (IC) = (1 - average OD value of miR-15a
mimic group / average OD value of miR-NC group) × 100%.
Detection of the migration of tumor
cells via wound healing assay
The cells in the logarithmic growth phase were
cultured until confluence of 80%, and gently pushed to generate
wounds on the surface. Then PBS was used to wash the cells 3 times.
Complete medium was replaced, recording the wounds and the cell
culture was continued. After 24 h of culture, the wounds were
photographed and recorded to compare their width, and to
statistically analyze the cell migration of each group.
Detection of the invasion of tumor
cells via Transwell invasion assay
Trypsinization was carried out 36 h post
transfection to collect cells of each group, and then the cells
were inoculated in 24-well plates with 5×103 cells/well,
respectively. Serum-free medium (200 µl) containing
penicillin-streptomycin was added to the upper layer of Transwell
cell culture insert, and 400 µl of complete culture medium
containing 10% FBS and 1% penicillin-streptomycin to the lower
layer, to culture the cells for 12 h at 37°C with the concentration
of 5% CO2. The cell culture insert was then washed 3
times with PBS to remove non-migrated cells. The cells were fixed
with 4% paraformaldehyde solution for 10 min, and then washed with
PBS 3 times. Subsequently, the cells were dyed with 0.5% crystal
violet solution for 10–15 min and rinsed with PBS 3 times. The
final step was to count migrated cells. The trial was repeated 3
times to average the values.
Prediction of the miR-15a target
gene
Prediction of the human miR-15a target genes on
TargetScan (http://www.Targetscan.org) showed
that the higher the score of the binding of mRNA to miR-15a seed
region, the greater the possibility of the binding.
Detection of the expression level of
Smad3 protein via western blotting
Precooled 1X PBS was used to collect and wash the
cells twice; the cells were centrifuged at 1,200 × g at 4°C for 5
min, precipitated, and lysed with 100 RIPA lysate. After
centrifugation, the cells were isolated by adding 10 µl of protein
to 10% polyacrylamide gel. The isolated protein was transferred
onto the PVDF membrane by a wet transfer method (current 300 mA,
1.5 h) and then sealed at room temperature for 1 h before western
blotting was carried out. Primary Smad3 protein and GAPDH antibody
were diluted at 1:2,000 with 5% fat-free milk, and hybridized
overnight at 4°C, and the PVDF membrane was washed 3 times with 1X
PBST for 5 min each time. The second antibody was diluted at
1:5,000 with 5% fat-free milk powder, and incubated for 2 h at room
temperature. The PVDF membrane was washed 3 times with 1X PBST for
5 min each time. ECL luminescent solution was prepared, developed
and exposed. Then, the strip quantitative analysis was performed
(Gelpro Analyzer, Media Cybernetics, Inc.).
Statistical analysis
IBM SPSS Statistics 20.0 was used to make
statistical analysis of the collected data, with GraphPad Prism 8
to draw statistical charts. All data were obtained by 3 independent
trials. The measurement data were expressed as the mean ± standard
deviation (mean ± SD), whereas the count data were represented as a
percentage (%). The t-test was used to analyze the differences
between the two groups, variance analysis for differences among
groups, and Pearson's correlation coefficient for the correlation
between variables. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression level of miR-15a mRNA in
NSCLC tissues and cells
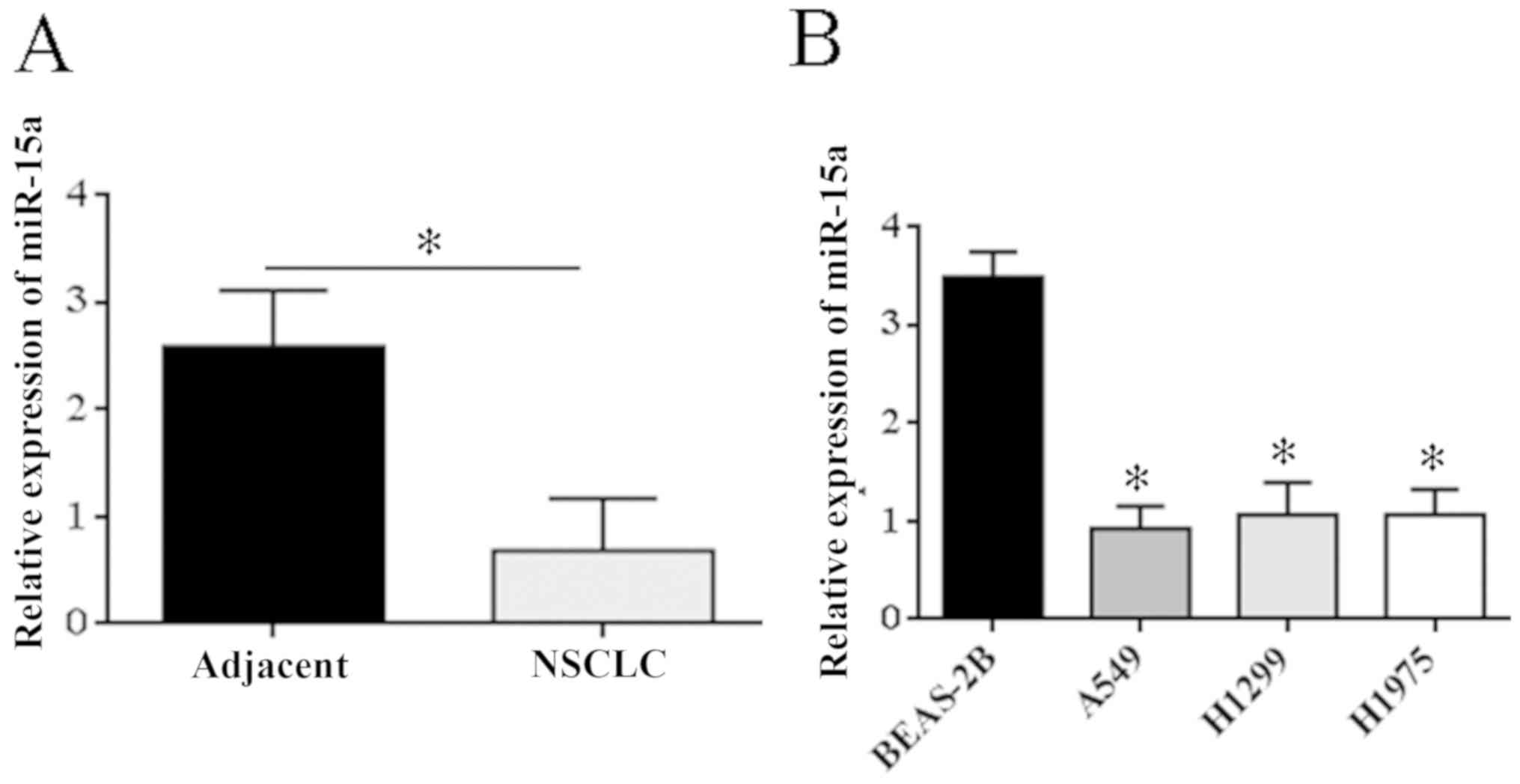
RT-PCR results showed that expression of miR-15a
mRNA was significantly lower in NSCLC tissue (P=0.023) compared
with para-carcinoma tissues (Fig.
1A). Expression level of miR-15a mRNA was detected in three
NSCL cell lines (H1975, A549, and H1299) and in normal lung cells
(BEAS-2B), and it was found that expression of miR-15a mRNA was
significantly lower in NSCLC cell lines than that in the normal
lung cells (P<0.05) (Fig.
1B).
Expression level of miR-15a mRNA in
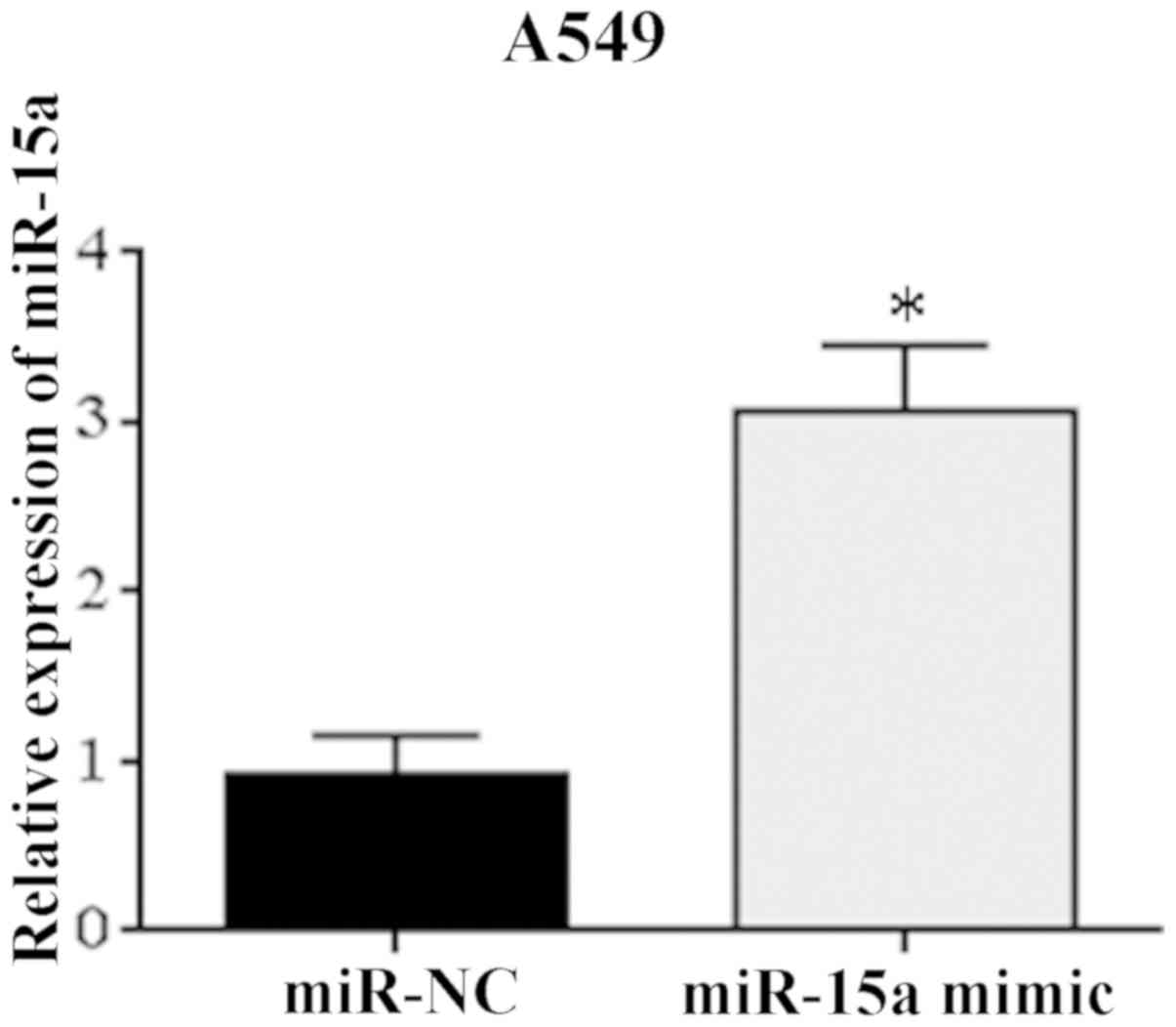
A549 cells after transfection
A549 cells with the lowest relative expression of
miR-15a were transfected with miR-15a mimic. RT-PCR indicated
significantly higher expression level of miR-15a mRNA in A549 cells
relative to miR-NC group after transfection (Fig. 2). The difference was statistically
significant (P=0.043). The results showed that the tumor cell
models were successfully transfected with miR-15a and could be used
in subsequent trials.
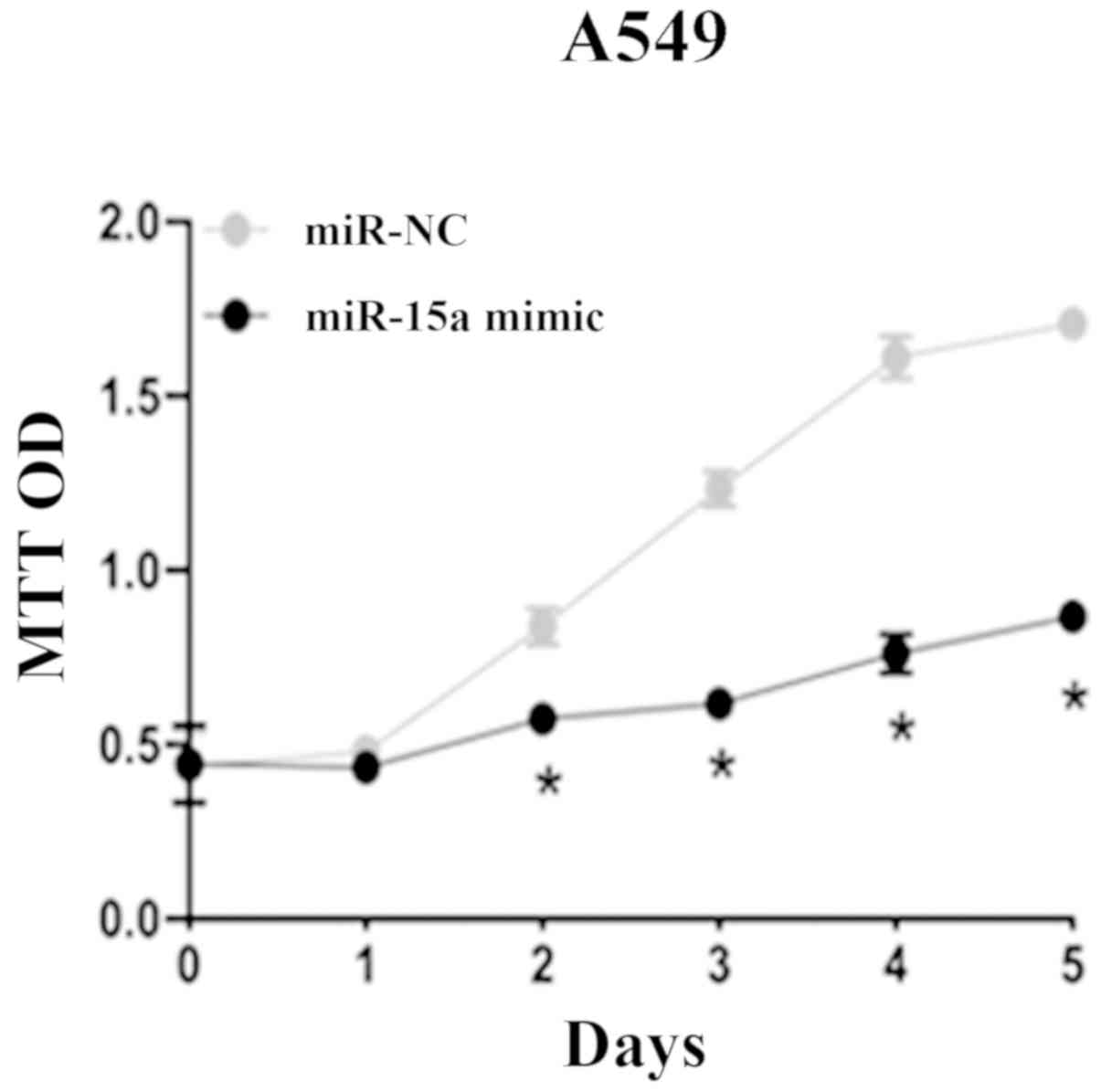
Overexpression of miR-15a
significantly inhibits the proliferation of NSCLC cells
Via the MTT assay, it was found that compared with
miR-NC group, A549 cells which were transfected with miR-15a mimic
had significantly reduced cell viability and proliferation
significantly slowed down on the 2nd, 3rd, 4th and 5th days after
adding MTT solution (P=0.038). This indicated that miR-15a could
significantly inhibit the proliferation of NSCLC cell lines
(Fig. 3).
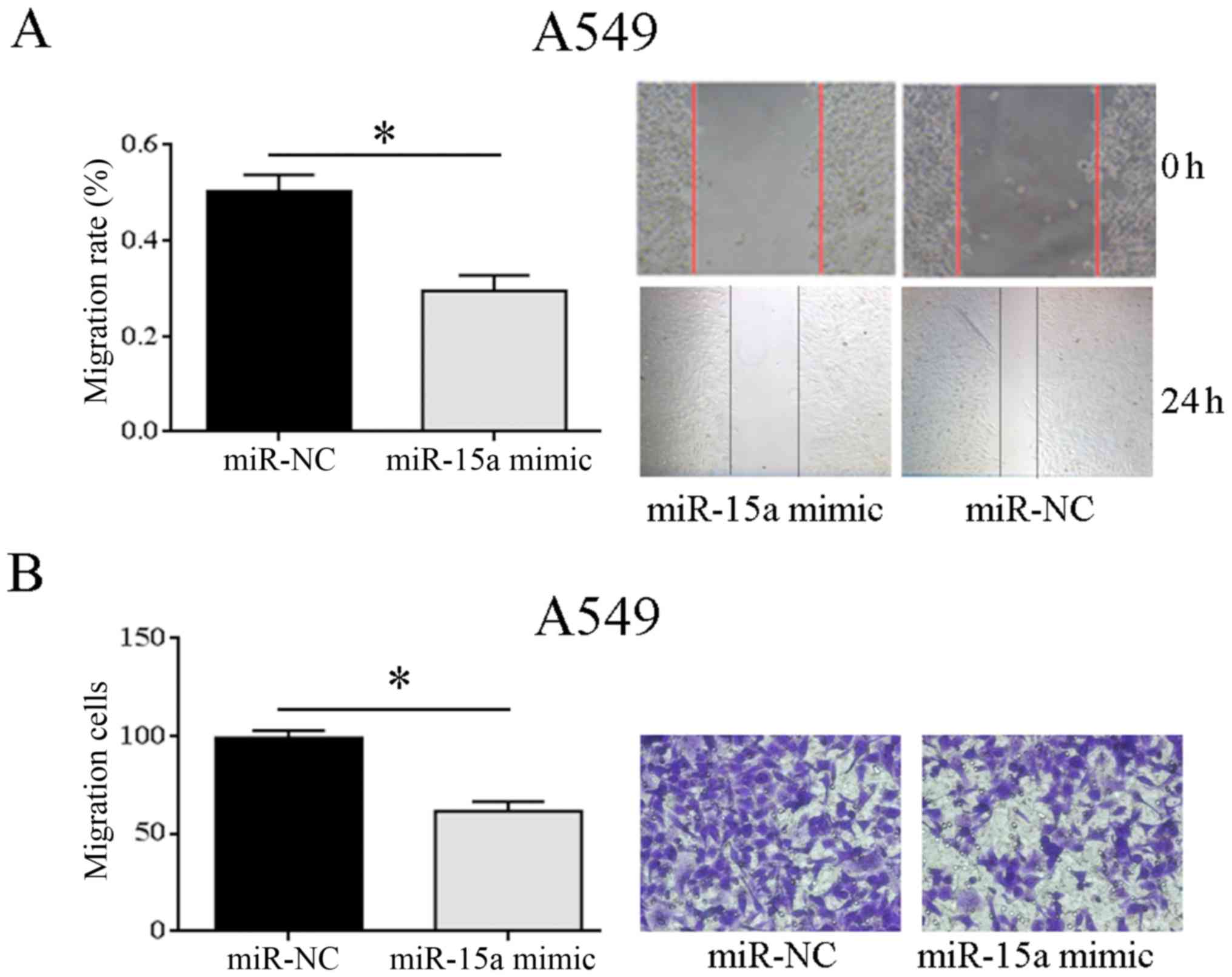
Effect of miR-15a overexpression on
migration and invasion of NSCLC
The wound healing assay showed that A549 cells had
significantly reduced migration than miR-NC group after
transfection of miR-15a mimic, and the difference was statistically
significant (P=0.033). It suggested that overexpression of miR-15a
can significantly inhibit the migration of NSCLC cell lines
(Fig. 4A). According to the
Transwell invasion assay, the cell invasion of miR-15a mimic group
was significantly reduced in comparison with that of miR-NC group
(P=0.025), indicating significant inhibition of the invasion
ability of NSCLC cell line by miR-15a overexpression (Fig. 4B). The above showed that
overexpression of miR-15a may weaken the cell migration and
invasion of A549 cells, and inhibit tumor cell metastasis.
miR-15a regulates Smad3 protein
expression
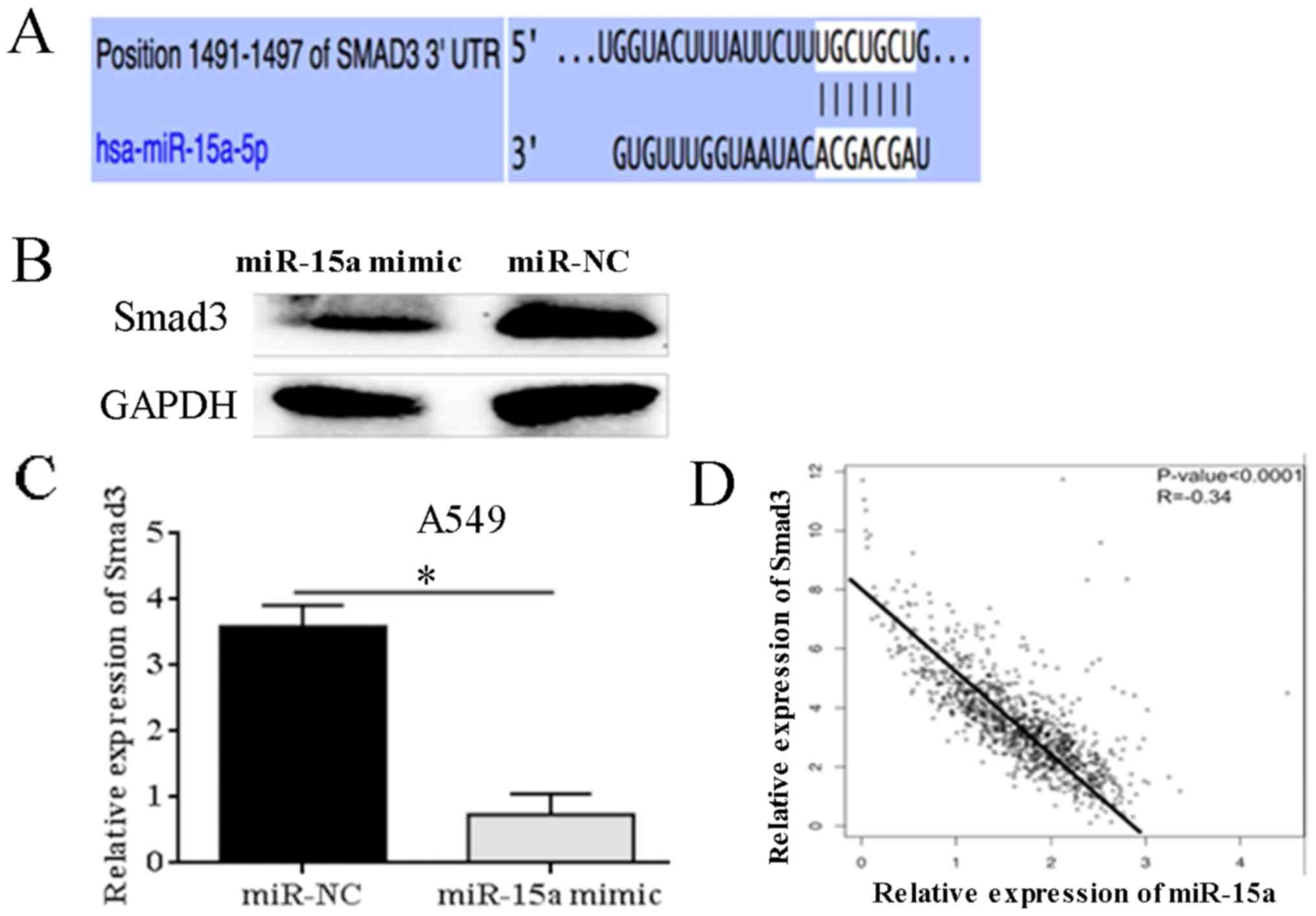
Bioinformatics analysis predicted the binding sites
of miR-15a on Smad3 (Fig. 5A).
Western blotting results showed that miR-15a was overexpressed in
A549 cells, significantly reducing the expression level of Smad3
protein (P=0.031) (Fig. 5B and C).
Pearson correlation analysis showed that miR-15a mRNA level was
significantly negatively correlated with Smad3 expression level,
suggesting that miR-15a and Smad3 may have negative regulatory
relationships (r=−0.34, P<0.0001) (Fig. 5D).
Discussion
In this study, NSCLC cell lines were transfected
with miR-15a to investigate the function of miR-15a in the
occurrence and development of NSCLC. It was found that miR-15a
served as a tumor inhibiting factor in NSCLC. Overexpression of
miR-15a inhibited cell proliferation, migration and invasion of
NSCLC. The findings showed that Smad3 is a target gene of miR-15a
in NSCLC.
miRNA is an endogenous non-coding small RNA, which
can bind to 3′-UTR of target mRNA to inhibit transcription of
target genes or degrade target mRNA fragments, and regulate its
expression at a post-transcription level (9,12).
Increasing evidence links miRNA to occurrence and development of
cancers (13). Due to organ
specificity, miRNA differs in different organs in terms of types
and proportions. miRNAs are related to the functional regulation of
organs, so miRNAs can be used as specific biological markers for
many different diseases. Therefore, research on the role of
NSCLC-specific miRNAs in its occurrence and development process can
provide new insights into the study of the occurrence and
development of NSCLC as well as new schemes for clinical treatment
of NSCLC.
The miR-15a gene is located at human chromosome
13q14 and was first reported to be abnormally expressed in cancer
in 2002. The deletion of miR-15a is associated with poor prognosis
of patients with chronic lymphocytic leukemia (14,15).
miR-15a is the first miRNA reported to be involved in tumor
development, which is of great significance. Subsequent studies
have reported the expression and mechanism of miR-15a in tumors.
miR-15a, as a tumor inhibiting factor, is downregulated in
melanoma, colorectal cancer, bladder cancer, prostate cancer and
other solid tumors (16–19). MicroRNA-15 (miR-15) family, as
upstream regulatory molecules, regulates different target mRNAs and
plays a crucial role in the occurrence and development of tumors.
Janaki Ramaiah et al (20)
found that miR-15 inhibits the proliferation of breast cancer cells
and induces apoptosis by targeting p70S6 kinase. Pouliot et
al (21) screened miRNA using
high-throughput screening to restore the sensitivity of
cisplatin-resistant cells. It was found that targeted regulating of
the expression of Wee1 and CHK1 by miR-15a could restore the
sensitivity of cisplatin-resistant cells. Bozok et al
(22) and Çalışkan et al
(23) confirmed that miR-15a
enhanced the anti-tumor effect of platinum chemotherapeutic drugs
in drug-resistant NSCLC. In other studies, miRNA-15 family
suppressed cell metastasis by regulating EMT process in malignant
cancer cells (24–26). He (27) found that knockout of miR-15a may
promote proliferation and invasion of lung cancer cells, inhibit
cell apoptosis, and induce EMT. In this study, miR-15a was found
expressed at significantly lower level in NSCLC tissues by
comparison with that in para-carcinoma tissues. miR-15a expression
was also relatively low in NSCLC cell lines in in vitro cell
trials, indicating that the low expression of miR-15a may promote
the occurrence of NSCLC. In addition, based on the expression of
miR-15a in the three NSCLC cell lines, A549 cell line with the
lowest miR-15a expression was transfected with miR-15a mimic to
construct a NSCLC cell line model with overexpression of miR-15a.
Metastasis is one of the most important malignant biological
characteristics of tumor cells. It was found herein through wound
healing assay and Transwell invasion assay that overexpression of
miR-15 effectively inhibited the migration and invasion ability of
NSCLC cells. Combined with previous studies, it was shown that
miR-15a has universal anti-tumor effect on a variety of tumors. Its
anti-tumor mechanism is not related to the origin of tumor tissue
and has potential clinical development value. This drove the
research team to find how miR-15a regulates the migration and
invasion of NSCLC cells.
It is predicted on online bioinformatics databases
(Pictar, Targetscan, miRanda) that miR-15a may be one of the genes
regulating transforming growth factor-β (TGF-β) signal pathways,
and may be bound to 3′UTR of Smad3. As a main transcription factor
of TGF-β signal transduction, Smad3 acts as a tumor inhibiting
factor and oncogene in the process of tumor occurrence and
development. TGF-β signaling pathways participate in normal
physiological processes such as growth and development,
inflammatory responses, and immune regulation, as well as in tumor
development. Jin et al (18)
found that miR-15a/16 inhibits prostate cancer metastasis and
invasion by inhibiting TGF-β signaling pathways. Underexpression or
mutation of Smad3 will lead to interruption of TGF-β signaling,
making cells beyond the growth inhibition of TGF-β signal pathways
and eventually develop into tumor cells (28–30).
Previous studies have shown that Smad3, as a negative growth signal
regulator, regulates the expression of TGF-β superfamily during the
occurrence and development of varying tumors, accommodates the
abnormal growth cycle of cells and protects the body (31,32). In
this study, western blotting and Pearson's correlation coefficient
showed that overexpression of miR-15a significantly reduced the
expression of Smad3, and the two had negative regulatory
correlation. Combined with in vitro cell trial, it was shown
that miR-15a can target downregulation of Smad3 to inhibit the
proliferation and metastasis of NSCLC cells.
In conclusion, it was found that miR-15a is
differentially expressed in NSCLC tissues and cells. miR-15a may
inhibit the proliferation, migration and invasion of NSCLC cells
through targeted regulation of Smad3 expression. This provides
theoretical basis for the pathogenesis of NSCLC. miR-15a is
expected to become a potential new target for NSCLC
biotherapies.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SG detected the migration of tumor cells via wound
healing assay and wrote the manuscript, ML interpreted and analyzed
the data. JL designed the study and performed the experiment. YL
was responsible for the analysis and discussion of the data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Shandong Provincial Chest Hospital (Jinan, China). Patients who
participated in this study had complete clinical data. Signed
informed consents were obtained from the patients and/or the
guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
van Klaveren RJ: Lung cancer screening.
Eur J Cancer. 47 (Suppl 3):S147–S155. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bouyssou JM, Manier S, Huynh D, Issa S,
Roccaro AM and Ghobrial IM: Regulation of microRNAs in cancer
metastasis. Biochim Biophys Acta. 1845:255–265. 2014.PubMed/NCBI
|
|
6
|
Zagryazhskaya A and Zhivotovsky B: miRNAs
in lung cancer: A link to aging. Ageing Res Rev. 17:54–67. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wan L, Zhang L, Fan K and Wang J: miR-27b
targets LIMK1 to inhibit growth and invasion of NSCLC cells. Mol
Cell Biochem. 390:85–91. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhan M, Qu Q, Wang G and Zhou H: Let-7c
sensitizes acquired cisplatin-resistant A549 cells by targeting
ABCC2 and Bcl-XL. Pharmazie. 68:955–961. 2013.PubMed/NCBI
|
|
9
|
Zhang N, Wei X and Xu L: miR-150 promotes
the proliferation of lung cancer cells by targeting P53. FEBS Lett.
587:2346–2351. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu Y, Huang J, Xu H and Gong Z:
Over-expression of miR-15a-3p enhances the radiosensitivity of
cervical cancer by targeting tumor protein D52. Biomed
Pharmacother. 105:1325–1334. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jin J, Zhang J, Xue Y, Luo L, Wang S and
Tian H: miRNA-15a regulates the proliferation and apoptosis of
papillary thyroid carcinoma via regulating AKT pathway. OncoTargets
Ther. 12:6217–6226. 2019. View Article : Google Scholar
|
|
12
|
Molina-Pinelo S, Gutiérrez G, Pastor MD,
Hergueta M, Moreno-Bueno G, García-Carbonero R, Nogal A, Suárez R,
Salinas A, Pozo-Rodríguez F, et al: MicroRNA-dependent regulation
of transcription in non-small cell lung cancer. PLoS One.
9:e905242014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
O'Connell RM, Rao DS, Chaudhuri AA and
Baltimore D: Physiological and pathological roles for microRNAs in
the immune system. Nat Rev Immunol. 10:111–122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Codony C, Crespo M, Abrisqueta P,
Montserrat E and Bosch F: Gene expression profiling in chronic
lymphocytic leukaemia. Best Pract Res Clin Haematol. 22:211–222.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pekarsky Y and Croce CM: Role of miR-15/16
in CLL. Cell Death Differ. 22:6–11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Aqeilan RI, Calin GA and Croce CM: miR-15a
and miR-16-1 in cancer: Discovery, function and future
perspectives. Cell Death Differ. 17:215–220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Musumeci M, Coppola V, Addario A, Patrizii
M, Maugeri-Saccà M, Memeo L, Colarossi C, Francescangeli F, Biffoni
M, Collura D, et al: Control of tumor and microenvironment
cross-talk by miR-15a and miR-16 in prostate cancer. Oncogene.
30:4231–4242. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jin W, Chen F, Wang K, Song Y, Fei X and
Wu B: miR-15a/miR-16 cluster inhibits invasion of prostate cancer
cells by suppressing TGF-β signaling pathway. Biomed Pharmacother.
104:637–644. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bonci D, Coppola V, Musumeci M, Addario A,
Giuffrida R, Memeo L, D'Urso L, Pagliuca A, Biffoni M, Labbaye C,
et al: The miR-15a-miR-16-1 cluster controls prostate cancer by
targeting multiple oncogenic activities. Nat Med. 14:1271–1277.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Janaki Ramaiah M, Lavanya A, Honarpisheh
M, Zarea M, Bhadra U and Bhadra MP: miR-15/16 complex targets p70S6
kinase 1 and controls cell proliferation in MDA-MB-231 breast
cancer cells. Gene. 552:255–264. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pouliot LM, Chen YC, Bai J, Guha R, Martin
SE, Gottesman MM and Hall MD: Cisplatin sensitivity mediated by
WEE1 and CHK1 is mediated by miR-155 and the miR-15 family. Cancer
Res. 72:5945–5955. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bozok Çetintaş V, Tetik Vardarlı A, Düzgün
Z, Tezcanlı Kaymaz B, Açıkgöz E, Aktuğ H, Kosova Can B, Gündüz C
and Eroğlu Z: miR-15a enhances the anticancer effects of cisplatin
in the resistant non-small cell lung cancer cells. Tumour Biol.
37:1739–1751. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Çalışkan M, Güler H and Bozok Çetintaş V:
Current updates on microRNAs as regulators of chemoresistance.
Biomed Pharmacother. 95:1000–1012. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Renjie W and Haiqian L: miR-132, miR-15a
and miR-16 synergistically inhibit pituitary tumor cell
proliferation, invasion and migration by targeting Sox5. Cancer
Lett. 356B:568–578. 2015. View Article : Google Scholar
|
|
25
|
Shi L, Jackstadt R, Siemens H, Li H,
Kirchner T and Hermeking H: p53-induced miR-15a/16-1 and AP4 form a
double-negative feedback loop to regulate epithelial-mesenchymal
transition and metastasis in colorectal cancer. Cancer Res.
74:532–542. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gao W, Wang Y, Wang W and Shi L: The first
multiplication atom-bond connectivity index of molecular structures
in drugs. Saudi Pharm J. 25:548–555. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
He J: Knocking down miR-15a expression
promotes the occurrence and development and induces the EMT of
NSCLC cells in vitro. Saudi J Biol Sci. 24:1859–1865. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li H, Xu D, Toh BH and Liu JP: TGF-beta
and cancer: Is Smad3 a repressor of hTERT gene? Cell Res.
16:169–173. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu W, Zeng F, Li S, Li G, Lai X, Wang QJ
and Deng F: Crosstalk of protein kinase C ε with Smad2/3 promotes
tumor cell proliferation in prostate cancer cells by enhancing
aerobic glycolysis. Cell Mol Life Sci. 75:4583–4598. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Paul D, Dixit A, Srivastava A, Tripathi M,
Prakash D, Sarkar C, Ramanujam B, Banerjee J and Chandra PS:
Altered transforming growth factor beta/SMAD3 signalling in
patients with hippocampal sclerosis. Epilepsy Res. 146:144–150.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ooshima A, Park J and Kim SJ:
Phosphorylation status at Smad3 linker region modulates
transforming growth factor-β-induced epithelial-mesenchymal
transition and cancer progression. Cancer Sci. 110:481–488. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang Y, Xiang J, Wang J and Ji Y:
Downregulation of TGF-β1 suppressed proliferation and increased
chemosensitivity of ovarian cancer cells by promoting BRCA1/Smad3
signaling. Biol Res. 51:582018. View Article : Google Scholar : PubMed/NCBI
|