Introduction
Breast cancer is one of the most common types of
cancer in women that is responsible for high number of
cancer-associated mortality cases worldwide (1). Each year, ~1.7 million new patients are
diagnosed with breast cancer, and >0.5 million women die from
breast cancer worldwide (2). Early
diagnosis and treatment could reduce the mortality rate of patients
with breast cancer. However, numerous patients are diagnosed at an
advanced stage, which is the major reason for frequent mortality
(3). The main treatments for breast
cancer include chemotherapy, surgery, targeted therapy, hormone
replacement therapy and radiation therapy (4). There are numerous causes of breast
cancer, including lifestyle (obesity, alcohol, Western-style diet)
(5), depression and anxiety disorder
(6) and gene mutations, including
breast cancer (BRCA) gene mutation (7); however, the mechanisms underlying
breast cancer pathogenesis remain unclear.
MicroRNAs (miRs) represent a class of 21–23
nucleotides long small non-coding RNAs that can act as oncogenes
(8) or tumor suppressors (9) in various types of cancer, including
gastrointestinal (10), liver
(11), pancreatic (12), ovarian (13), lung (14) and breast cancer (15). miRs can bind directly to the
3′-untranslated region (UTR) of target genes to negatively regulate
protein expression (16). It has
been reported that miRs can regulate the proliferation,
differentiation, migration, metastasis and apoptosis of cancer
cells. In addition, numerous miRs are associated with breast cancer
pathogenesis. For example, miR-454-3p can promote breast cancer
metastasis by regulating Wnt/β-catenin signaling pathway (15). Furthermore, miR-29a contributes to
breast cancer cell epithelial-mesenchymal transition, migration and
invasion by targeting suppressor of variegation 4–20 homolog 2
(17). In addition, miR-449b-5p
suppresses the proliferation and invasion of breast cancer cells by
regulating the cell cycle-related and expression elevated protein
in tumor-mediated Wnt/β-catenin signaling (18).
The miR-519 family is located on human chromosome 9
and includes miR-519a-3p, miR-519b-3p, miR-519c-3p, miR-519a-5p and
miR-519b-5p. miR-519a-3p, miR-519b-3p and miR-519c-3p have similar
sequences and share the same seed sequence (19). It has been reported that the miR-519
family is associated with cancer development. For example, miR-519
suppresses nasopharyngeal carcinoma cell proliferation by targeting
the oncogene upregulated gene 4/upregulator of cell proliferation
(20). However, the role of miR-519
in the development of breast cancer remains unknown. The present
study demonstrated that miR-519 was downregulated in primary breast
cancer tissues. The findings from this study suggested that miR-519
may regulate breast cancer cell proliferation by targeting human
antigen R (HUR).
Materials and methods
Clinical breast cancer tissues
A total of 20 pairs of breast cancer tissues and
adjacent normal breast tissues were collected from patients with
breast cancer at the Department of General Surgery of The Fourth
Hospital of Hebei Medical University between January 2017 and
October 2018. All patients (age range, 36–84 years; average age, 58
years) underwent primary surgical treatment without any
preoperative chemotherapy or radiotherapy. Adjacent normal breast
tissues were obtained from the margins of tumors (≥5 cm from the
tumor). The tissues were flash-frozen in liquid nitrogen and stored
at −80°C. All clinicopathological information was recorded. Each
patient had read and signed informed consent for tissue donation.
This study was approved by the Ethical Review Committee of Hebei
Medical Hospital.
Breast cancer cell culture
The human breast cancer cell line MCF-7 was
purchased from the American Type Culture Collection. MCF-7 cells
were cultured in RPMI-1640 supplemented with 10% fetal bovine serum
(FBS; both HyClone; GE Healthcare Life Sciences), 80 U/ml
penicillin (Gibco; Thermo Fisher Scientific, Inc.) and 100 µg/ml
streptomycin (Gibco; Thermo Fisher Scientific, Inc.) and placed at
37°C in a 95% humidified atmosphere containing 5%
CO2.
Cell transfection
The miR-519 family contains miR-519b-3p, miR-519c-3p
and miR-519a. The miR oligos for miR-519 mimic, miR mimic negative
control, miR-519 inhibitor and miR inhibitor negative control were
provided by Shanghai GenePharma Co., Ltd. The sequences of the miRs
were as follows: miRNA mimics negative control forward,
5′-UUCUCCGAACGUGUCACGUUU-3′ and reverse,
5′-ACGUGACACGUUCGGAGAAUU-3′; miR-519a mimic forward
5′-AAAGUGCAUCCUUUUAGAGUGU-3′ and reverse,
5′-ACACUCUAAAAGGAUGCACUU-3′; miR-519b mimic forward
5′-AAAGUGCAUCCUUUUAGAGGUU-3′ and reverse,
5′-AACCUCUAAAAGGAUGCACUU-3′; miR-519c mimic forward,
5′-AAAGUGCAUCCUUUUAGAAGGAU-3′ and reverse,
5′-AUCCUUCUAAAAGGAUGCACUU-3′; miR inhibitor negative control
forward 5′-UUCUCCGAACGUGUCACGUUU-3′ and reverse
5′-ACGUGACACGUUCGGAGAAUU-3′; miR-519a inhibitor,
5′-UCACACUCUAAAAGGAUGCACUU-3′; miR-519b inhibitor,
5′-UCCUCUAAAAGGAUGCACUU-3′; and miR-519c inhibitor,
5′-UCAUCCUUCUAAAAGGAUGCACUU-3′. One day prior to transfection,
MCF-7 cells were seeded in 6-well plates at the density of
3×105 cells/well in 1.6 ml RPMI-1640 medium containing
10% FBS. miR mimics or inhibitors for miR-519a, miR-519b and
miR-519c were individually diluted into diethyl pyrocarbonate
(DEPC)-treated H2O to the concentration of 20 nM and
mixed together in a 1:1:1 ratio (v/v). A volume of 4 µl miR-519
mimics or inhibitors (containing mimics or inhibitors for miR-519a,
miR-519b and miR-519c) were diluted in serum-free RPMI-1640 medium
(100 µl). Then, HiperFect transfection reagent (2 µl) was diluted
in serum-free RPMI-1640 medium (100 µl). Then, the two reagents
were mixed together and added to each well. The cells were
harvested 48 h following transfection for subsequent
experiments.
Small interfering (si)RNA targeting HUR (cat. no.
sc-35619) was obtained from Santa Cruz Biotechnology, Inc. and is a
pool of three HUR-specific siRNAs. The sequences of siRNAs were as
follows: Si-HUR-1 forward, 5′-GCGAGGUUGAAUCUGCAAAUU-3′ and reverse,
5′-UUUGCAGAUUCAACCUCGCUU-3′; si-HUR-2 forward,
5′-GACCAUGACAAACUAUGAAUU-3′ and reverse,
5′-UUCAUAGUUUGUCAUGGUCUU-3′; si-HUR-3 forward,
5′-GCUGGUGCAUCUUCAUCUAUU-3′ and reverse,
5′-UAGAUGAAGAUGCACCAGCUU-3′; siRNA negative control forward,
5′-UUCUCCGAACGUGUCACGUUU-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′. One day prior to transfection, MCF-7
cells were seeded in 6-well plates at the density of
3×105 cells/well in 1.6 ml RPMI-1640 medium containing
10% FBS. siRNA oligos were individually diluted into diethyl
pyrocarbonate (DEPC)-treated H2O to the concentration of
20 nM and mixed together in a 1:1:1 ratio (v/v). A volume of 4 µl
siRNAs (containing three siRNAs) were diluted in serum-free
RPMI-1640 medium (100 µl). Then, HiperFect transfection reagent (2
µl) was diluted in serum-free RPMI-1640 medium (100 µl). The two
reagents were mixed together and added to each well. The cells were
harvested 48 h following transfection for subsequent
experiments.
Adenovirus vector transfection
Recombinant adenoviruses expressing HUR (AD-HUR) and
control adenovirus vector containing GFP (AD-CON) were purchased
from Shanghai GeneChem Co., Ltd. One day prior to transfection,
MCF-7 cells were seeded in 6-well plates at the density of
3.5×105 cells/well in 2 ml RPMI-1640 medium containing
10% FBS. The adenovirus transfection was performed at a
multiplicity of infection) of 30. At 48 h following infection, the
cells were harvested.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
After 48 h of miR oligo transfection, total RNA was
extracted from MCF-7 cells using TRIzol® (Invitrogen;
Thermo Fisher Scientific, Inc.). RNA (1 µg) was reverse transcribed
into cDNA. RNA (1 µg), miR-specific stem-and-loop RT-primers (1
µl), dNTP (2 µl; 10 mM) and DEPC-treated H2O (6 µl) were
mixed together and incubated at 70°C for 10 min, then cooled on
ice. Recombinant RNase inhibitor (0.5 µl; 40 U/µl), M-MuLV reverse
transcriptase (0.5 µl; 200 U/µl; New England BioLabs, Inc.), 2 µl
10X reverse transcriptase buffers and 6 µl DEPC-treated
H2O were added. The RT reactions were performed as
follows: 42°C for 60 min, 95°C for 5 min, and 4°C forever. The
sequences of the RT primers were as follows: miR-519a RT,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACACC-3′; miR-519b
RT, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAACCTC-3′;
miR-519c RT,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACATCCTC-3′; and U6
RT, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAAATATG-3′.
The expression levels of miR-519a, miR-519b and miR-519c were
measured by qPCR using a SYBR Green II kit (Takara Bio, Inc.).
Then, 2 µl cDNA, 0.8 µl forward primer, 0.8 µl reverse primer, 10
µl SYBR Green mix and 6.4 µl ddH2O were mixed together.
The qPCR conditions were as follows: 95°C for 30 sec, followed by
40 cycles at 95°C for 30 sec and 60°C for 20 sec. U6 was used as a
housekeeping gene to calculate the relative level of miR-519 by
using the 2−∆∆Cq method (21). The sequences of primers used for qPCR
were as follows: miR-519a/b, forward 5′-GCGCAAAGTGCATCCTTTTA-3′;
miR-519c, forward 5′-GCGCAAAGTGCATCTTTTTA-3′; U6, forward,
5′-GCGCGTCGTGAAGCGTTC-3′; and universe reverse,
5′-GTGCAGGGTCCGAGGT-3′
Western blotting
After 48 h of transfection with miRNA oligos, total
protein from MCF-6 cells were harvested. Cells were washed three
times with 2 ml ice cold PBS and scraped in 0.5 ml ice cold PBS.
Cells were centrifuged at 2,000 × g for 10 min at 4°C. Total
protein was extracted by using 100 µl of RIPA Buffer per well (Cell
Signaling Technology, Inc.). The cell lysate was centrifuged at
10,000 × g for 10 min at 4°C. Supernatant fraction was collected,
and protein concentration was determined using a bicinchoninic acid
protein assay kit (Thermo Fisher Scientific, Inc.). Proteins (15
µg) were separated by 10% SDS-PAGE and transferred onto 0.2 µm pore
diameter polyvinylidene fluoride membranes (EMD Millipore).
Membranes were blocked with 5% non-fat milk diluted in TBS for 2 h
at room temperature and incubated with primary antibodies (1:1,000)
overnight at 4°C. The primary antibodies against BCL-2 (cat. no.
3498), BAX (cat. no. 5023), HUR (cat. no. 12582) and GAPDH (cat.
no. 5174) were purchased from CST (Cell Signaling Technology,
Inc.). Membranes were washed five times with TBS containing 0.1%
Tween-20 (TBST) and incubated with horseradish
peroxidase-conjugated anti-rabbit IgG secondary antibody (cat. no.
7074; Cell Signaling Technology, Inc.; 1:5,000) for 2 h at room
temperature. Membranes were washed five times with TBST and bands
were detected using enhanced chemiluminescence substrate (EMD
Millipore). Relative expression levels of proteins were normalized
to endogenous control GAPDH using. ImageJ version 1.42 software
(National Institutes of Health). Each experiment was performed in
triplicate and repeated three times.
Luciferase reporter activity
assay
It was reported that HUR mRNA contains two miR-519
binding sites: 886–907 nucleotides (nt) in the coding region (CR)
and 4332- 4357 nt in the 3′-untranslated region (UTR) (19). Two DNA fragments containing six
repeats of binding sites in the CR or UTR were synthesized by
Shanghai Shenggong Biology Engineering Technology Service, Ltd..
The DNA fragments were inserted into the pmirGLO vector (Promega
Corporation) to construct recombinant vectors (pmirGLO-CR and
pmirGLO-UTR). For the luciferase assay, MCF-6 cells were seeded in
96-well plates at the density of 5,000 cells per well in 100 µl
RPMI-1640 medium containing 10% FBS one day before transfection.
Then, recombinant luciferase reporter vectors and control vectors
were transfected into MCF-7 cells using Effectene reagent (Qiagen
China Co., Ltd.) for 48 h. The luciferase activity was measured
using a dual-luciferase reporter assay system (Promega
Corporation). Luciferase activity was normalized to Renilla
luciferase activity. A total of six samples were measured for each
group. The experiment was repeated three times.
MTT assay
MCF-7 cell proliferation was determined using MTT
assay (Sigma-Aldrich; Merck KGaA). MCF-7 cells were cultured in
96-well plate at the density of 5,000 cells per well in 100 µl
RPMI-1640 medium containing 10% FBS. After 24, 48 and 72 h, 20 µl
MTT (10 mg/ml) was added into each well for 4 h. Supernatant was
discarder and 150 µl DMSO was added to dissolve purple formazan
crystals. Absorbance was read at 490 nm on a microplate reader
(Bio-Rad Laboratories, Inc.).
Colony formation assay
One day following transfection of miRNA mimics or
inhibitors, ~300 MCF-7 cells were seeded in each well of 6-well
plate and cultured at 37°C for 2 weeks. The medium was removed once
a week and replaced with fresh medium containing the miRNAs mimics
or inhibitors transfection mixture. After 14 days, cells were
washed three times with PBS and fixed with 4% polymerized
formaldehyde (Beijing Solarbio Science & Technology Co., Ltd.)
for 20 min at room temperature. Cells were stained with 2.5%
crystal violet staining solution (Beijing Solarbio Science &
Technology Co., Ltd.) for 30 min at room temperature. Cells were
washed with PBS three times and air-dried. The colonies that
contained >50 cells were counted with the naked eye in each
well. The relative colony number was calculated as the ratio of
cells transfected with miR-519 mimics (519 m) to cells transfected
with negative control mimics (NCm), or cells transfected with
miR-519 inhibitors (519i) to cells transfected with negative
control inhibitors (NCi). All experiments were carried out in
triplicate each time and repeat three times.
Statistical analysis
SPSS 13.0 statistical software package (SPSS, Inc.)
was used to perform statistical analysis. All data are presented as
the means ± standard error of the mean. The non-parametric
Mann-Whitney U test was used to compare two groups. One-way
analysis of variance followed by Turkey's post hoc test was used to
compare three or more groups. P<0.05 was considered to indicate
a statistically significant difference.
Results
miR-519 is downregulated in breast
cancer tissues
The sequences of miR-519a-3p, miR-519b-3p and
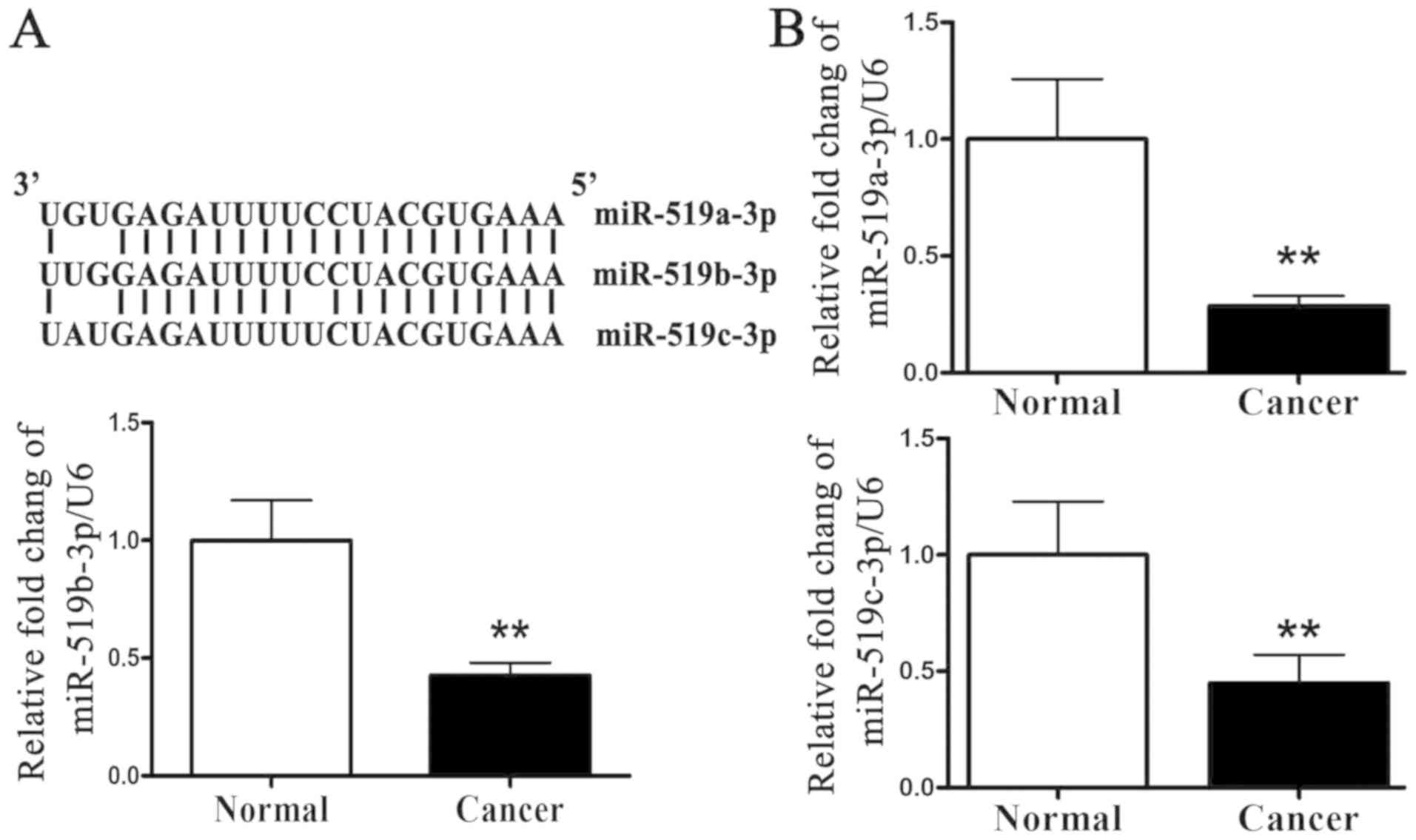
miR-519c-3p were highly conserved (Fig.
1A). miR-519 expression level was analyzed in 20 breast cancer
tissues and adjacent normal tissues by RT-qPCR. The results
presented in Fig. 1B demonstrated
that miR-519a-3p, miR-519b-3p and miR-519c-3p expression level was
significantly lower in breast cancer tissues compared with adjacent
normal tissues. miR-519 may therefore serve a role in breast cancer
development.
miR-519 regulates breast cancer cell
proliferation
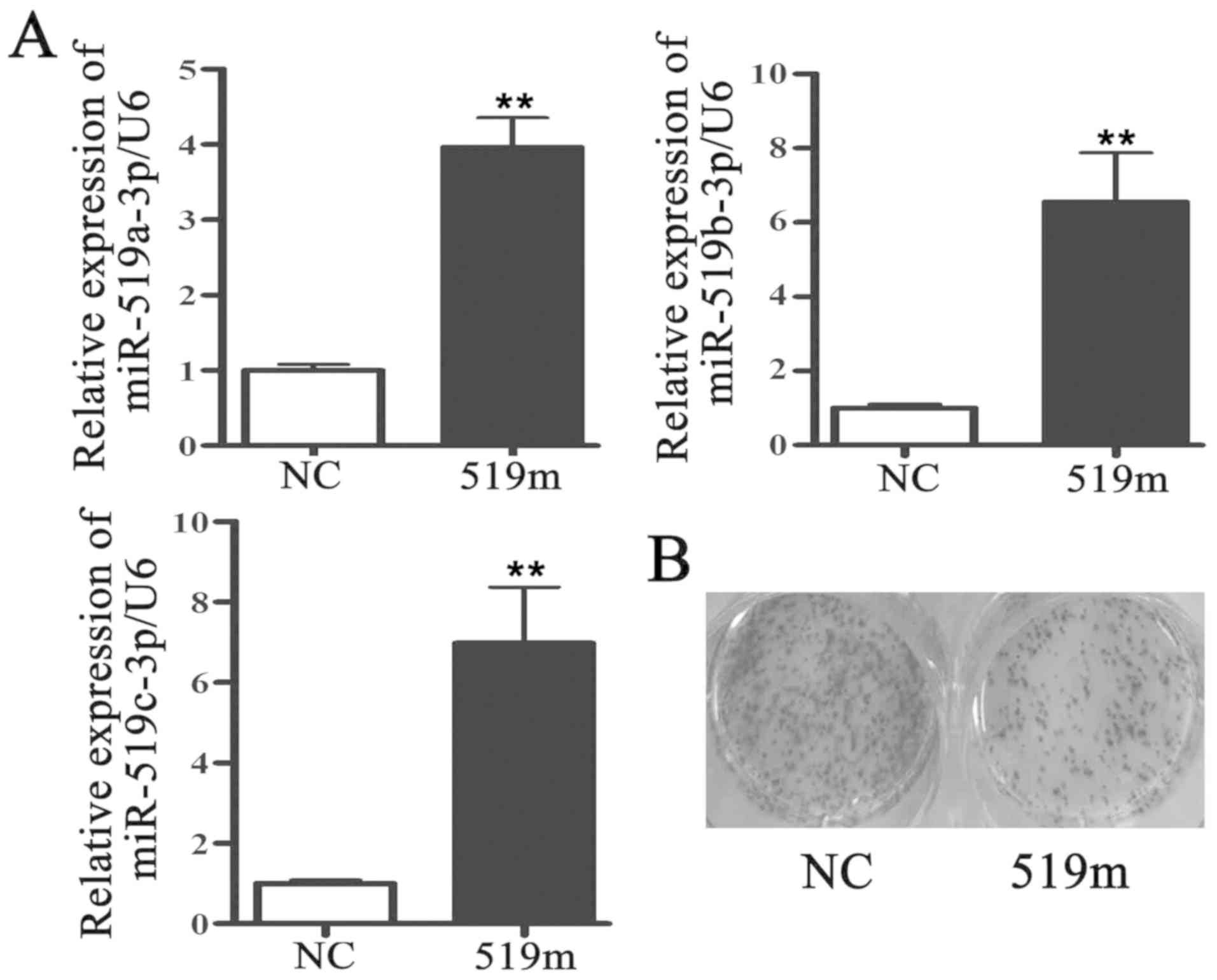
miR-519a-3p, miR-519b-3p and miR-519c-3p shared
similar sequences, and their expression levels were decreased in
breast cancer tissues. The mimics of miR-519a-3p, miR-519b-3p and
miR-519c-3p were then mixed equally and transfected into MCF-7
cells for 48 h. The results from RT-qPCR demonstrated that
miR-519a-3p, miR-519b-3p and miR-519c-3p expression levels were
significantly increased in MCF-7 cells following transfection with
miR-519 mimics compared with MCF-7 cells transfected with negative
control mimics (Fig. 2A). The
results from MTT and colony formation assays demonstrated that
MCF-7 cell proliferation was inhibited following transfection with
miR-519 mimic (Fig. 2B and C). To
verify the effect of miR-519 on MCF-7 cell proliferation, the
inhibitors of miR-519a-3p, miR-519b-3p and miR-519c-3p were mixed
equally and transfected into MCF-7 cells for 48 h. The results from
RT-qPCR demonstrated that miR-519a-3p, miR-519b-3p and miR-519c-3p
expression levels were significantly lower in MCF-7 cells
transfected with miR-519 inhibitors compared with MCF-7 cells
transfected with negative control inhibitors (Fig. 2D). The results from MTT and colony
formation assays demonstrated that MCF-7 cell proliferation was
increased following transfection with miR-519 inhibitors (Fig. 2E and F). These results suggested that
miR-519 could regulate MCF-7 cell proliferation.
miR-519 directly targets HUR
It has been reported that miRs execute their
biological functions by directly regulating downstream target
genes. Furthermore, it was predicted that HUR mRNA possesses two
binding sites (19). One was at
886–980 nt in the coding region of HUR and the other was at
4332–4337 nt in the 3′UTR of HUR (Fig.
3A). A luciferase activity assay was used to determine whether
miR-519 could bind directly to HUR mRNA. The results demonstrated
that miR-519 mimic transfection significantly decreased the
luciferase activity of cells transfected with a pmiRGLO vector
containing the binding sites (pmiRGLO-3′utr) (Fig. 3B, left panel) compared with cells
transfected with pmiRGLO-3′UTR only. Furthermore, luciferase
activity was significantly inhibited in MCF-7 cells co-transfected
with pmiRGLO-3′utr and miR-519 mimics (Fig. 3B, middle panel) compared with MCF-7
cells transfected with pmiRGLO control vector and miR-519 mimic.
Transfection with miR-519 inhibitors had no effect on luciferase
activity (Fig. 3B, right panel). The
results from western blotting demonstrated that miR-519
overexpression significantly decreased HUR and BCL-2 protein levels
and significantly increased BAX protein level (Fig. 3C). Conversely, HUR and BCL-2 protein
levels were significantly increased and BAX protein level was
significantly decreased in MCF-7 cells transfected with miR-519
inhibitors (Fig. 3D). HUR may
therefore be a direct target gene of miR-519.
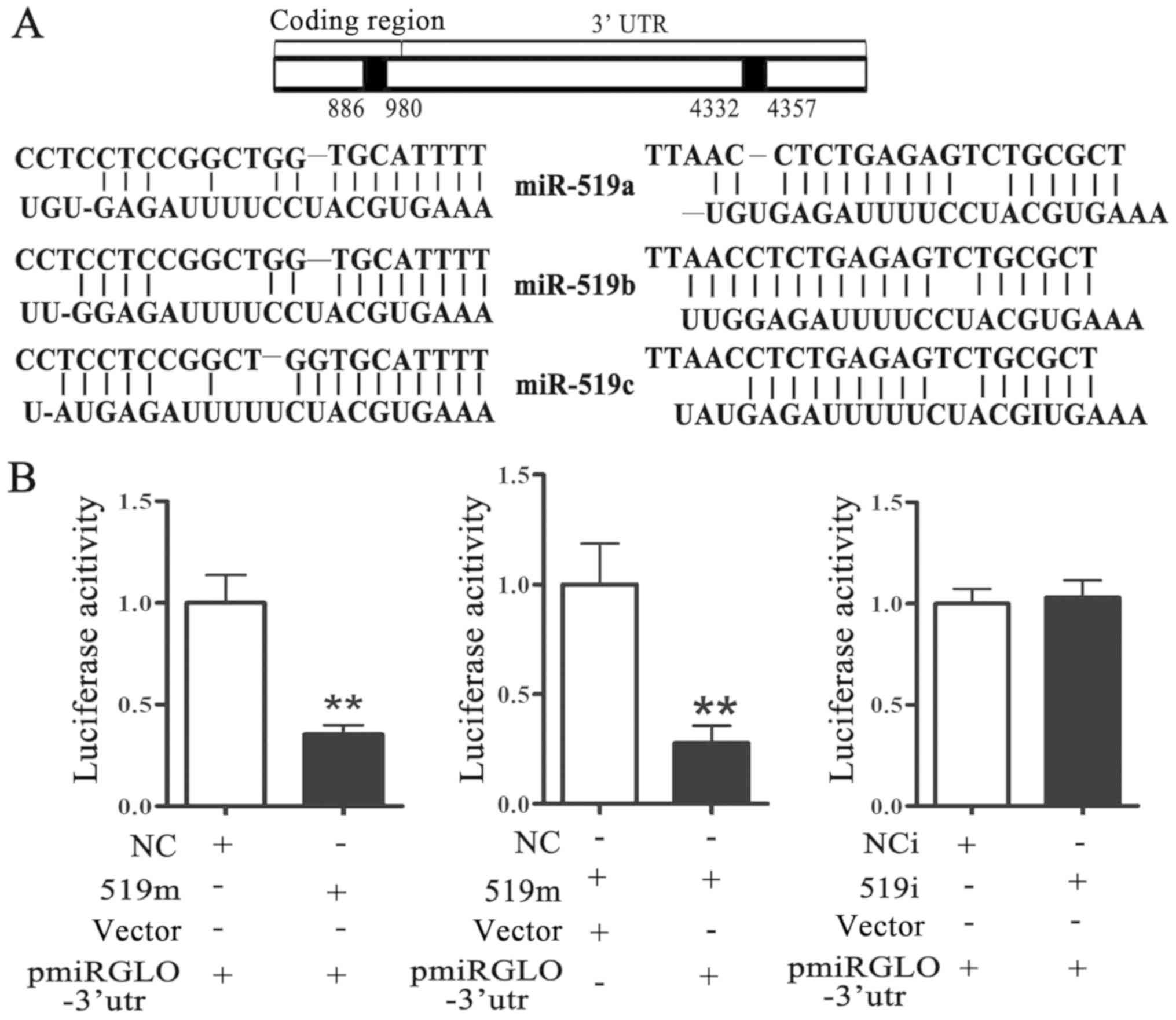 | Figure 3.miR-519 directly targets HUR. (A) A
bioinformatics database was used to predict the binding sites of
miR-519 on HUR mRNA. One was at 886–980 nt in the coding region and
the other was at 4332–4357 nt in the 3′-UTR. (B) Luciferase
activity assay was performed in MCF-7 transfected with negative
control mimics (NC group), miR-519 mimics (miR-519m group),
negative control inhibitors (NCi group) or miR-519 inhibitors
(miR-519i group). Western blotting was used to measure HUR, BAX and
BCL-2 protein levels in MCF-7 cells transfected with (C) miR-519
mimics or (D) miR-519 inhibitors. *P<0.05, **P<0.01 vs.
control group. n=5. 519i, miR-519 inhibitors; 519m, miR-519 mimics;
HUR, human antigen R; NC, negative control; NCi, negative control
inhibitors; nt, nucleotide. |
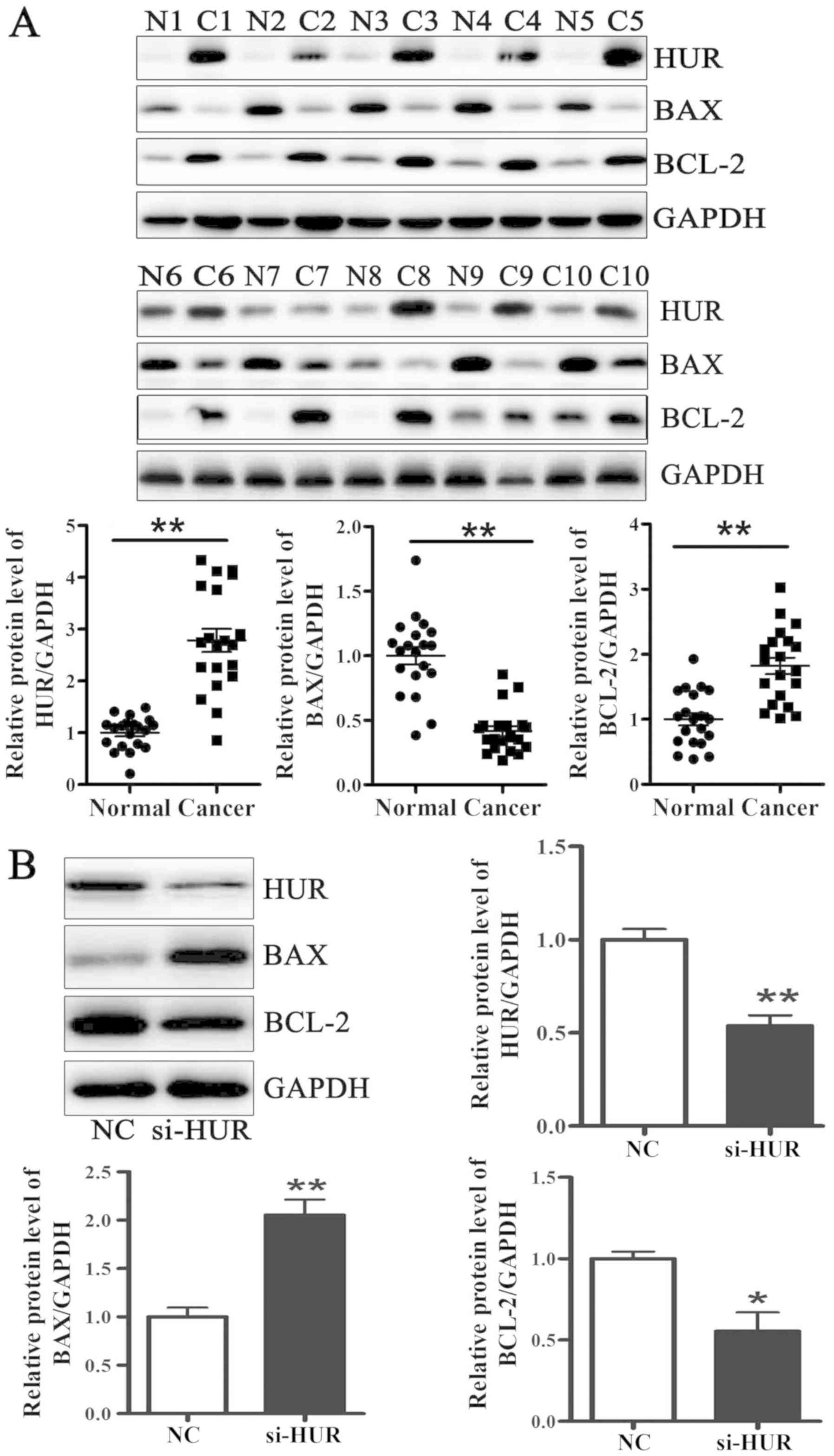
HUR protein level is upregulated in
breast cancer tissues
HUR protein level was analyzed in 20 pairs of breast
cancer and adjacent normal tissues. The results demonstrated that
HUR and BCL-2 protein levels were significantly higher, whereas BAX
protein level was significantly lower in breast cancer tissues
compared with adjacent normal tissues. (Fig. 4A). Next, HUR-specific siRNA (si-HUR)
was transfected into MCF-7 cells. As presented in Fig. 4B, HUR protein level was decreased to
~50% following si-HUR transfection. Furthermore, BCL-2 protein
level was significantly lower, and the BAX protein level was
significantly higher in MCF-7 cells transfected with si-HUR
compared with cells transfected with negative control siRNA (NC).
HUR may therefore serve a role in breast cancer development.
miR-519 regulates MCF-7 cell
proliferation via targeting HUR
To verify whether miR-519 could regulate MCF-7 cell
proliferation by negatively regulating HUR, HUR protein expression
was up- or downregulated in MCF-7 cells. An adenovirus vector
expressing HUR was constructed and transfected into MCF-7 cells.
The results demonstrated that HUR overexpression could reverse the
effects of miR-519 mimic on BAX and BCL-2 protein levels (Fig. 5A) and MCF-7 cell proliferation
(Fig. 5B and C). In addition,
HUR-specific siRNA was designed and transfected into MCF-7 cells to
silence HUR protein expression. The results demonstrated that HUR
downregulation could rescue the effects of miR-519 inhibitors on
BAX and BCL-2 protein levels (Fig.
5D). The results from colony formation and MTT assays indicated
that HUR silencing could reverse the effects of miR-519 on MCF-7
cell proliferation. Taken together, these results suggested that
miR-519 may regulate MCF-7 cell proliferation by targeting HUR.
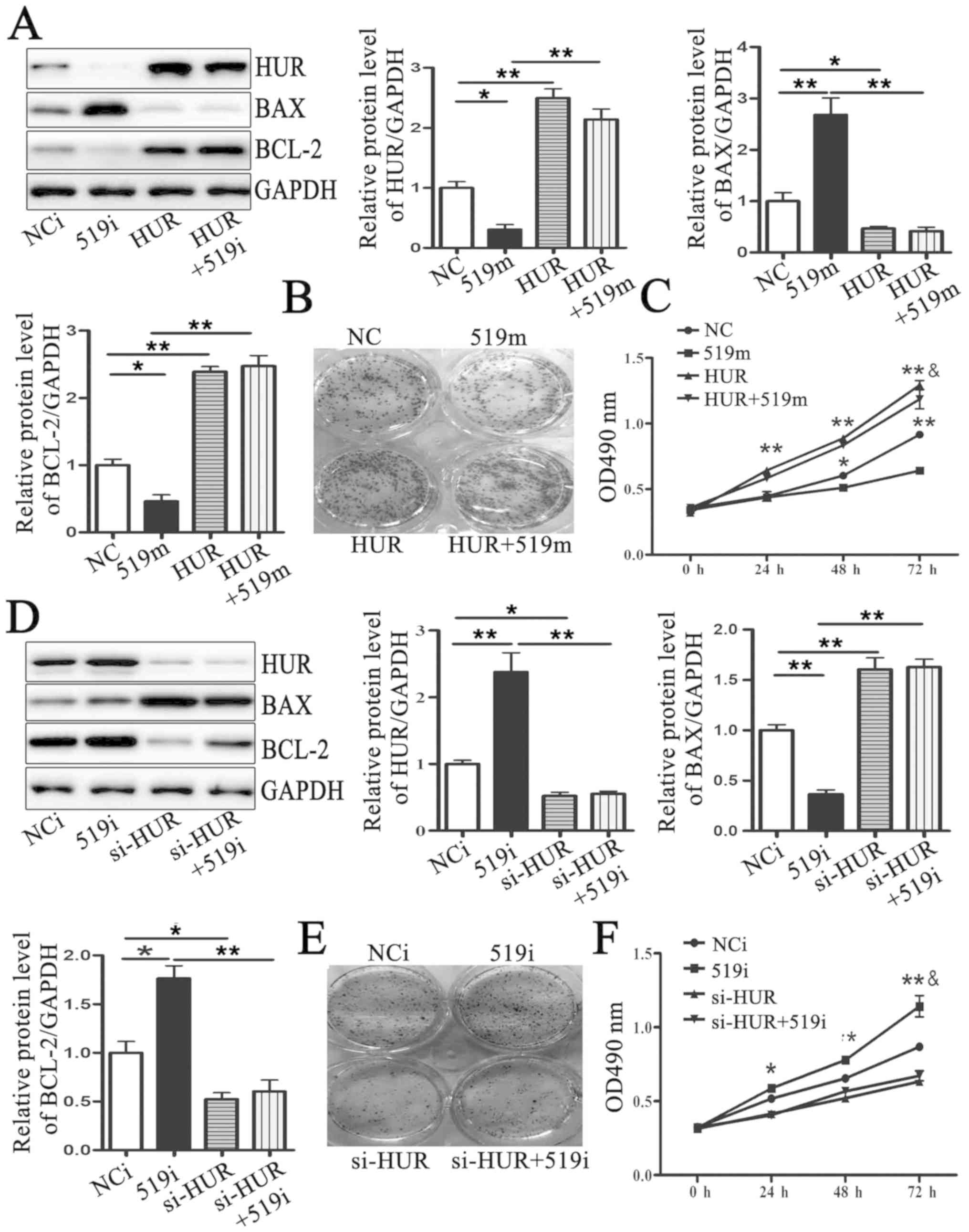 | Figure 5.miR-519 regulates MCF-7 cell
proliferation by targeting HUR. (A) HUR, BAX and BCL-2 protein
levels were measured by western blotting in MCF-7 cells
co-transfected with an adenovirus vector expressing HUR (HUR) and
miR-519 mimics (519m). (B) Colony formation in MCF-7 cells
transfected with negative control mimics (NC group), miR-519 mimics
(519m group), an adenovirus vector expressing HUR (HUR group) or an
adenovirus vector expressing HUR and miR-519 mimics (HUR+519m
group). (C) MCF-7 cell proliferation determined by MTT in MCF-7
cells transfected with negative control mimics (NC group), miR-519
mimics (519m group), an adenovirus vector expressing HUR (HUR
group) or an adenovirus vector expressing HUR and miR-519 mimics
(HUR+519m group). (D) HUR, BAX and BCL-2 protein levels were
measured by western blotting in MCF-7 cells co-transfected with
HUR-specific siRNAs (si-HUR) and miR-519 inhibitors (519i). (E)
Colony formation and (F) proliferation of MCF-7 cells
co-transfected with HUR-specific siRNAs (si-HUR) and miR-519
inhibitors (519i). *P<0.05, **P<0.01 vs. control group.
&P<0.05 vs. 519m or 519i group. n=5. 519i,
miR-519 inhibitors; 519m, miR-519 mimics; HUR, human antigen R; NC,
negative control; NCi, negative control inhibitors; si, small
interfering. |
Discussion
Numerous factors can lead to breast cancer,
including lifestyle (obesity, inactivity, alcohol) (22) and BRCA gene mutation (23). Aberrant expression of miRs, including
miR-148a-3p (24), miR-124-3p
(25) and miR-301a-3p (26), can also contribute to the
pathogenesis of breast cancer. The present study focused on the
effects of miR-519 on breast cancer cell proliferation. The results
suggested that miR-519 may serve a crucial role in breast cancer
cell proliferation.
The miR-519 family includes miR-519a-3p,
miR-519b-3p, miR-519b-3p, miR-519c-3p, miR-519a-5p and miR-519b-5p.
miR-519 robustly inhibits HeLa cell proliferation and induces HeLa
cell senescence (27). miR-519
suppresses nasopharyngeal carcinoma cell proliferation (20). The present results demonstrated that
the relative expression levels of miR-519a-3p, miR-519b-3p and
miR-519c-3p were lower in primary breast cancer tissues compared
with in adjacent normal tissues. The present study demonstrated
that miR-519 overexpression could inhibit MCF-7 cell proliferation,
whereas miR-519 downregulation could promote MCF-7 cell
proliferation. These results were consistent with those from a
previous study (19,20).
miRs exert their biological function by regulating
their target genes. It has been reported that miR-519 can regulate
two prominent subsets of genes. One subset of target genes encodes
proteins involved in DNA maintenance, including deoxyuridine
5′-triphosphate nucleotidohydrolase, exonuclease 1, replication
protein A2 and DNA polymerase epsilon 4, accessory subunit. The
other subset of target genes encodes proteins that control
intracellular calcium levels, including ATPase secretory pathway
Ca2+ transporting 1ATP2C1 and ORAI calcium release-activated
calcium modulator 1 (27). The
present study demonstrated that HUR was a target of miR-519. The
results from luciferase assay confirmed that miR-519 could directly
bind to HUR mRNA and negatively regulate HUR protein expression.
HUR is a RNA-binding protein that can bind and stabilize AU rich
element-mRNAs (28). HUR is
abundantly expressed in cancer cells and associated-malignant
phenotypes (29). HUR has been
implicated in numerous cellular events, including proliferation,
senescence, differentiation, apoptosis, and stress and immune
responses. In turn, HUR can promote certain processes, including
cancer pathogenesis and inflammation (30). HUR can exert its effects in two ways.
Firstly, HUR acts as a positive effector for gene expression by
binding and stabilizing their mRNA, for example cyclooxygenase 2,
p21 and cyclin D1 (31–33). Secondly, HUR can decrease the
translation efficiency of mRNA, including of TNF-α and p27
(34,35). The results from the present study
demonstrated that HUR protein level was increased in primary breast
cancer tissues compared with adjacent normal tissues, and that this
effect was accompanied by increased BCL-2 expression and decreased
BAX expression. Furthermore, the present study demonstrated that
HUR could promote BCL-2 expression and inhibit BAX expression. HUR
overexpression could rescue the effects of miR-519 mimics on MCF-7
cell proliferation and on BCL-2 and BAX expression, whereas HUR
silencing could reverse the effects of miR-519 inhibitors on MCF-7
cell proliferation and on BCL-2 and BAX expression. Previous
studies suggested that HUR could be a positive regulator of BCL-2
by promoting BCL-2 mRNA stability and translation efficiency
(36,37). BCL-2 can promote the survival and
proliferation of cells and therefore participate in the
pathogenesis of cancer and tumor growth (38). BAX is a pro-apoptotic effector that
can be inhibited by BCL-2 (39).
Both proteins belong to the BCL-2 family, although they have
opposite effects on cell apoptosis. BCL-2 overexpression can
inhibit BAX insertion into the mitochondrial outer membrane but
spontaneously increase BAX localization to the mitochondria
(40). Taken together, the findings
from the present study suggested that miR-519 may regulate breast
cancer cell proliferation by targeting HUR, which may be considered
as a potential therapeutic target in breast cancer.
Acknowledgements
Not applicable.
Funding
This study was supported by The Fourth Special Fund
Construction Project of Medical Science (grant no. 2015B2001).
Availability of data and materials
The dataset used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LR, YL and QZ designed the experiments. LR, LF and
BT performed the experiments. AZ and HY analysed the data. LR and
YL wrote and revised the manuscript. All authors approved the final
version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethical Review
Committee of Hebei Medical University (Shijiazhuang, China).
Written informed consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests
References
|
1
|
Miller KD, Siegel RL, Lin CC, Mariotto AB,
Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A: Cancer
treatment and survivorship statistics, 2016. CA Cancer J Clin.
66:271–289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Akram M, Iqbal M, Daniyal M and Khan AU:
Awareness and current knowledge of breast cancer. Biol Res.
50:332017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Aumeeruddy MZ and Mahomoodally MF:
Combating breast cancer using combination therapy with 3
phytochemicals: Piperine, sulforaphane, and thymoquinone. Cancer.
125:1600–1611. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shamsi U, Khan S, Usman S, Soomro S and
Azam I: A multicenter matched case control study of breast cancer
risk factors among women in Karachi, Pakistan. Asian Pac J Cancer
Prev. 14:183–188. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zainal NZ, Nik-Jaafar NR, Baharudin A,
Sabki ZA and Ng CG: Prevalence of depression in breast cancer
survivors: A systematic review of observational studies. Asian Pac
J Cancer Prev. 14:2649–2656. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shaukat U, Ismail M and Mehmood N:
Epidemiology, major risk factors and genetic predisposition for
breast cancer in the Pakistani population. Asian Pac J Cancer Prev.
14:5625–5629. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Han G, Qiu N, Luo K, Liang H and Li H:
Downregulation of miroRNA-141 mediates acquired resistance to
trastuzumab and is associated with poor outcome in breast cancer by
upregulating the expression of ERBB4. J Cell Biochem. Feb
11–2019.(Epub ahead of print).
|
|
9
|
Wu H, Tao J, Li X, Zhang T, Zhao L, Wang
Y, Zhang L, Xiong J, Zeng Z, Zhan N, et al: MicroRNA-206 prevents
the pathogenesis of hepatocellular carcinoma by modulating
expression of met proto-oncogene and cyclin-dependent kinase 6 in
mice. Hepatology. 66:1952–1967. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun KK, Shen XJ, Yang D, Gan MQ, Liu G,
Zhang YF, Hua P, Wang HD and Wu XY: MicroRNA-31 triggers G2/M cell
cycle arrest, enhances the chemosensitivity and inhibits migration
and invasion of human gastric cancer cells by downregulating the
expression of zeste homolog 2 (ZH2). Arch Biochem Biophys.
663:269–275. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu L, Feng X, Hao X, Wang P, Zhang Y,
Zheng X, Li L, Ren S, Zhang M and Xu M: CircSETD3
(Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting
hepatocellular carcinoma growth. J Exp Clin Cancer Res. 38:982019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang Y, Ishak Gabra MB, Hanse EA, Lowman
XH, Tran TQ, Li H, Milman N, Liu J, Reid MA, Locasale JW, et al:
miR-135 suppresses glycolysis and promotes pancreatic cancer cell
adaptation to metabolic stress by targeting phosphofructokinase-1.
Nat Commun. 10:8092019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ramalho S, Andrade LAA, Filho CC, Natal
RA, Pavanello M, Ferracini AC, Sallum LF, Sarian LO and Derchain S:
Role of discoidin domain receptor 2 (DDR2) and microRNA-182 in
survival of women with high-grade serous ovarian cancer. Tumour
Biol. 41:10104283188239882019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ding Z, Zhu J, Zeng Y, Du W, Zhang Y, Tang
H, Zheng Y, Qin H, Liu Z and Huang JA: The regulation of Neuropilin
1 expression by miR-338-3p promotes non-small cell lung cancer via
changes in EGFR signaling. Mol Carcinog. 58:1019–1032. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ren L, Chen H, Song J, Chen X, Lin C,
Zhang X, Hou N, Pan J, Zhou Z, Wang L, et al: miR-454-3p-mediated
Wnt/β-catenin signaling antagonists suppression promotes breast
cancer metastasis. Theranostics. 9:449–465. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liolios T, Kastora SL and Colombo G:
MicroRNAs in female malignancies. Cancer Inform.
18:11769351198287462019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu Y, Shi W, Tang T, Wang Y, Yin X, Chen
Y, Zhang Y, Xing Y, Shen Y, Xia T, et al: miR-29a contributes to
breast cancer cells epithelial-mesenchymal transition, migration,
and invasion via down-regulating histone H4K20 trimethylation
through directly targeting SUV420H2. Cell Death Dis. 10:1762019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang J, Yang X, He X, Ma W, Wang J, Zhou
Q, Li M and Yu S: MicroRNA-449b-5p suppresses the growth and
invasion of breast cancer cells via inhibiting CREPT-mediated
Wnt/beta-catenin signaling. Chem Biol Interact. 302:74–82. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Abdelmohsen K, Srikantan S, Kuwano Y and
Gorospe M: miR-519 reduces cell proliferation by lowering
RNA-binding protein HuR levels. Proc Natl Acad Sci USA.
10:20297–20302. 2008. View Article : Google Scholar
|
|
20
|
Yu G, Zhang T, Jing Y, Bao Q, Tang Q and
Zhang Y: miR-519 suppresses nasopharyngeal carcinoma cell
proliferation by targeting oncogene URG4/URGCP. Life Sci.
175:47–51. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Michelle H..Howell A and Evans DG: Can
diet and lifestyle prevent breast cancer: What is the evidence. Am
Soc Clin Oncol Educ Bool. e66–e73. 2015.
|
|
23
|
Baretta Z, Mocellin S, Goldin E, Olopade
OI and Huo D: Effect of BRCA germline mutations on breast cancer
prognosis: A systematic review and meta-analysis.
Medicine(Baltimore). 95:e49752016.PubMed/NCBI
|
|
24
|
Lacerda JZ, Ferreira LC, Lopes BC,
Aristizábal-Pachón AF, Bajgelman MC, Borin TF and Zuccari DAPC:
Therapeutic potential of melatonin in the regulation of miR-148a-3p
and angiogenic factors in breast cancer. Microrna. 8:237–247. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hu D, Li M, Su J, Miao K and Qiu X:
Dual-targeting of miR-124-3p and ABCC4 promotes sensitivity to
adriamycin in breast cancer cells. Genet Test Mol Biomarkers.
23:156–165. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Oztemur Islakoglu Y, Noyan S and Gur
Dedeoglu B: hsa-miR-301a- and SOX10-dependent miRNA-TF-mRNA
regulatory circuits in breast cancer. Turk J Biol. 42:103–112.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Abdelmohsen K, Srikantan S, Tominaga K,
Kang MJ, Yaniv Y, Martindale JL, Yang X, Park SS, Becker KG,
Subramanian M, et al: Growth inhibition by miR-519 via multiple
p21-inducing pathways. Mol Cell Biol. 32:2530–2548. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lv Z, He K, Shi L, Shi K, Jiang T and Chen
Y: Interaction between C2ORF68 and HuR in human colorectal cancer.
Oncol Rep. 41:1918–1928. 2019.PubMed/NCBI
|
|
29
|
Kakuguchi W, Nomura T, Kitamura T,
Otsuguro S, Matsushita K, Sakaitani M, Maenaka K and Tei K:
Suramin, screened from an approved drug library, inhibits HuR
functions and attenuates malignant phenotype of oral cancer cells.
Cancer Med. 7:6269–6280. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Grammatikakis I, Abdelmohsen K and Gorospe
M: Posttranslational control of HuR function. Wiley Interdiscip Rev
RNA. 8:Jun 16–2017.(Epub ahead of print). doi: 10.1002/wrna.1372.
View Article : Google Scholar
|
|
31
|
Ko CY, Wang WL, Li CF, Jeng YM, Chu YY,
Wang HY, Tseng JT and Wang JM: IL-18-induced interaction between
IMP3 and HuR contributes to COX-2 mRNA stabilization in acute
myeloid leukemia. J Leukoc Biol; 99. pp. 131–141. 2016, PubMed/NCBI
|
|
32
|
Lafarga V, Cuadrado A, Lopez de Silanes I,
Bengoechea R, Fernandez-Capetillo O and Nebreda AR: p38
mitogen-activated protein kinase- and HuR-dependent stabilization
of p21(Cip1) mRNA mediates the G(1)/S checkpoint. Mol Cell Biol.
9:4341–4351. 2009. View Article : Google Scholar
|
|
33
|
Ghosh U and Adhya S: Posttranscriptional
regulation of cyclin D1 by ARE-binding proteins AUF1 and HuR in
cycling myoblasts. J Biosci. 43:685–691. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhu Z, Zhao Y, Li J, Tao L, Shi P, Wei Z,
Sheng X, Shen D, Liu Z, Zhou L, et al: Cryptotanshinone, a novel
tumor angiogenesis inhibitor, destabilizes tumor necrosis factor-α
mRNA via decreasing nuclear-cytoplasmic translocation of
RNA-binding protein HuR. Mol Carcinog. 55:1399–1410. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mukherjee J, Ohba S, See WL, Phillips JJ,
Molinaro AM and Pieper RO: PKM2 uses control of HuR localization to
regulate p27 and cell cycle progression in human glioblastoma
cells. Int J Cancer. 139:99–111. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ghisolfi L, Calastretti A, Franzi S, Canti
G, Donnini M, Capaccioli S, Nicolin A and Bevilacqua A: B cell
lymphoma (Bcl)-2 protein is the major determinant in bcl-2
adenine-uridine-rich element turnover overcoming HuR activity. J
Biol Chem. 284:20946–20955. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ishimaru D, Ramalingam S, Sengupta TK,
Bandyopadhyay S, Dellis S, Tholanikunnel BG, Fernandes DJ and
Spicer EK: Regulation of Bcl-2 expression by HuR in HL60 leukemia
cells and A431 carcinoma cells. Mol Cancer Res. 7:1354–1366. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ruefli-Brasse A and Reed JC: Therapeutics
targeting Bcl-2 in hematological malignancies. Biochem J.
474:3643–3657. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pan LL, Wang AY, Huang YQ, Luo Y and Ling
M: Mangiferin induces apoptosis by regulating Bcl-2 and Bax
expression in the CNE2 nasopharyngeal carcinoma cell line. Asian
Pac J Cancer Prev. 15:7065–7068. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Teijido O and Dejean L: Upregulation of
Bcl2 inhibits apoptosis-driven BAX insertion but favors BAX
relocalization in mitochondria. FEBS Lett. 584:3305–3310. 2010.
View Article : Google Scholar : PubMed/NCBI
|