Introduction
Lung cancer is the second most common cancer and is
the leading cause of cancer-related death for men and women
worldwide (1–3). Non-small cell lung cancer (NSCLC),
accounting for ~85% cases of lung cancer, represents the most
common histological type of lung cancer (4). The mortality rate of NSCLC is very
high, and the 5-year survival rate is <20% (5), which is due to the lack of reliable
tools for early diagnosis or effective therapy. Therefore,
investigation is required to identify specific molecules that may
contribute to the diagnosis of NSCLC, and serve as prognostic
markers.
Centromere protein F (CENPF), a transient
kinetochore protein, exhibits a cell-cycle dependent localization,
and is completely degraded at the end of cell division (4–6).
Evidence has shown that CENPF is overexpressed in head and neck
squamous cell carcinomas, hepatocellular carcinoma, breast cancer
and prostate cancer, and it may be a prognostic marker for these
cancers (7–12). Forkhead box M1 (FOXM1) is a typical
proliferation-associated factor and plays an important role in
development (13–18). FOXM1 expression is frequently
elevated in numerous malignancies and participates actively in the
development and progression of various human cancers, including
NSCLC (19–21). High expression of FOXM1 is correlated
with shorter disease-free survival of NSCLC patients (22,23). The
synergistic effect of FOXM1 and CENPF has been found in promoting
the growth of prostate cancer and their co-expression predicts poor
survival (24–26). However, the clinical significance of
CENPF in NSCLC is unknown.
In the present study, the expression of CENPF and
FOXM1 in NSCLC was explored by quantitative PCR, western blot
analysis and immunohistochemical staining. The relationship between
protein expression of CENPF and clinicopathological parameters were
investigated to assess the possible prognostic value of CENPF and
FOXM1 expression in NSCLC.
Patients and methods
Patients and clinicopathological
data
The study was approved by the ethics committee of
Shenyang Fifth People Hospital. A total of 75 patients with NSCLC
who underwent surgery in Shenyang Fifth People's Hospital between
2009 and 2011 were enrolled after signed informed consent form was
received. Tumor tissues and adjacent non-tumorous tissues were
obtained from all the patients. Of these samples, 28 pairs of tumor
tissues and adjacent non-tumorous tissues were frozen immediately,
stored at −80°C and used for quantitative PCR analysis. The samples
were formalin-fixed, paraffin-embedded, and cut into 5-µm thick
sections.
Quantitative PCR
Total RNA was isolated from collected tissues with
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. Then, single-stranded cDNA was
generated from 1 µg of total RNA using cDNA synthesis kit (Thermo
Fisher Scientific, Inc.). Quantitative PCR was carried out on ABI
7300 system (Applied Biosystem; Thermo Fisher Scientific, Inc.)
with SYBR-Green qPCR Master Mixes (Thermo Fisher Scientific, Inc.)
as per the manufacturer's instructions. The primers used in the
study were: CENPF, forward, 5′-CTCGTTCCATCCCTGTCATC-3′ and reverse
primer 5′-TCCTGGTCAGATTCTCCTCC-3′; FOXM1, forward,
5′-GAAACGACCGAATCCAGAG-3′ and reverse primer,
5′-GCAGATCGCCACTAAAGAAC-3′; GAPDH, forward,
5′-AATCCCATCACCATCTTC-3′ and reverse primer
5′-AGGCTGTTGTCATACTTC-3′. CENPF and FOXM1 mRNA expression was
calculated using the 2−ΔΔCq method (27).
Western blot analysis
Total protein extracted from collected specimens
(0.5 g per sample) was cut into small sections and homogenized in
RIPA lysis buffer containing protease and phosphatase inhibitors
(Beyotime). After centrifugation at 13,000 × g, at 4°C for 20 min,
the supernatant was recovered.
After separation by 6% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), proteins
were electroblotted onto nitrocellulose membranes (Millipore) and
subjected to western blot analysis, then incubated with primary
antibodies, CENPF (rabbit polyclonal, 1:1,000 dilution, ab5), FOXM1
(rabbit polyclonal, 1:1,000dilution, ab226928) (both from Abcam)
and GAPDH (rabbit monoclonal, 1:1,000dilution, no. 5174; Cell
Signaling Technology), and membranes were incubated with
horseradish peroxidase (HRP)-labeled secondary antibody (goat anti
rabbit, 1:1,000 dilution, A0208; Beyotime). The immunoreactive
signal was detected by the enhanced chemiluminescence kit
(Millipore). Quantification of band intensity was analyzed with
ImageJ software (http://rsb.info.nih.gov/ij/), and normalized to the
intensity of GAPDH.
Immunohistochemical analysis
The sections were deparaffinized in xylene and
hydrated in a graded series of ethanol, then antigen retrieved by
heat exposure in Tris/EDTA buffer (pH 9.0) for 15 min, and blocked
for endogenous peroxidase activity in 3% hydrogen peroxide at room
temperature for 10 min. Followed by blocking with 5% normal
blocking serum, the sections were reacted with anti-CENPF (1:200
dilution, ab5) (28) or anti-FOXM1
(1:250 dilution, ab207298) (both from Abcam) (29) at 4°C overnight. After probing with
HRP-conjugated secondary antibody, immunocomplexes were visualized
using 3,3′-diaminobenzidine (DAB) (both from Long Island Biotech
Co., Ltd.) and lightly counterstained with hematoxylin. The
sections were reviewed and classified by two independent
investigators as previously described (27). The percentage of positive stained
cancer cells was scored as 0, negative; 1, 1–10%; 2, 11–50%
positive; 3, >50% positive. The staining intensity was scored as
0, absent; 1, weak; 2, moderate; 3, strong. The immunoreactive
score (IS) was calculated as follows: IS= percentage × staining
intensity. The protein was considered to be highly expressed when
IS was 3–9, otherwise to be lowly expressed.
Statistical analysis
Statistical analysis was carried out with the
Statistical Package for the Social Sciences software (SPSS Inc.).
Paired Student's t-test was performed to analyze the difference of
mRNA and protein expression between NSCLC tissues and adjacent
non-cancerous tissues. Pearsons correlation analysis was used to
investigate the relationship between mRNA expression of CENPF and
FOXM1 in NSCLC tissues. The Fisher's exact test was used to analyze
the relationship between CENPF expression and the
clinicopathological features. Kaplan-Meier and log rank test was
used to estimate and compare survival. The significance level was
set at 0.05.
Results
Association of CENPF and FOXM1 in
NSCLC tissues
To describe the mRNA expression of CENPF and FOXM1
in NSCLC, we performed quantitative PCR analysis on 28 pairs of
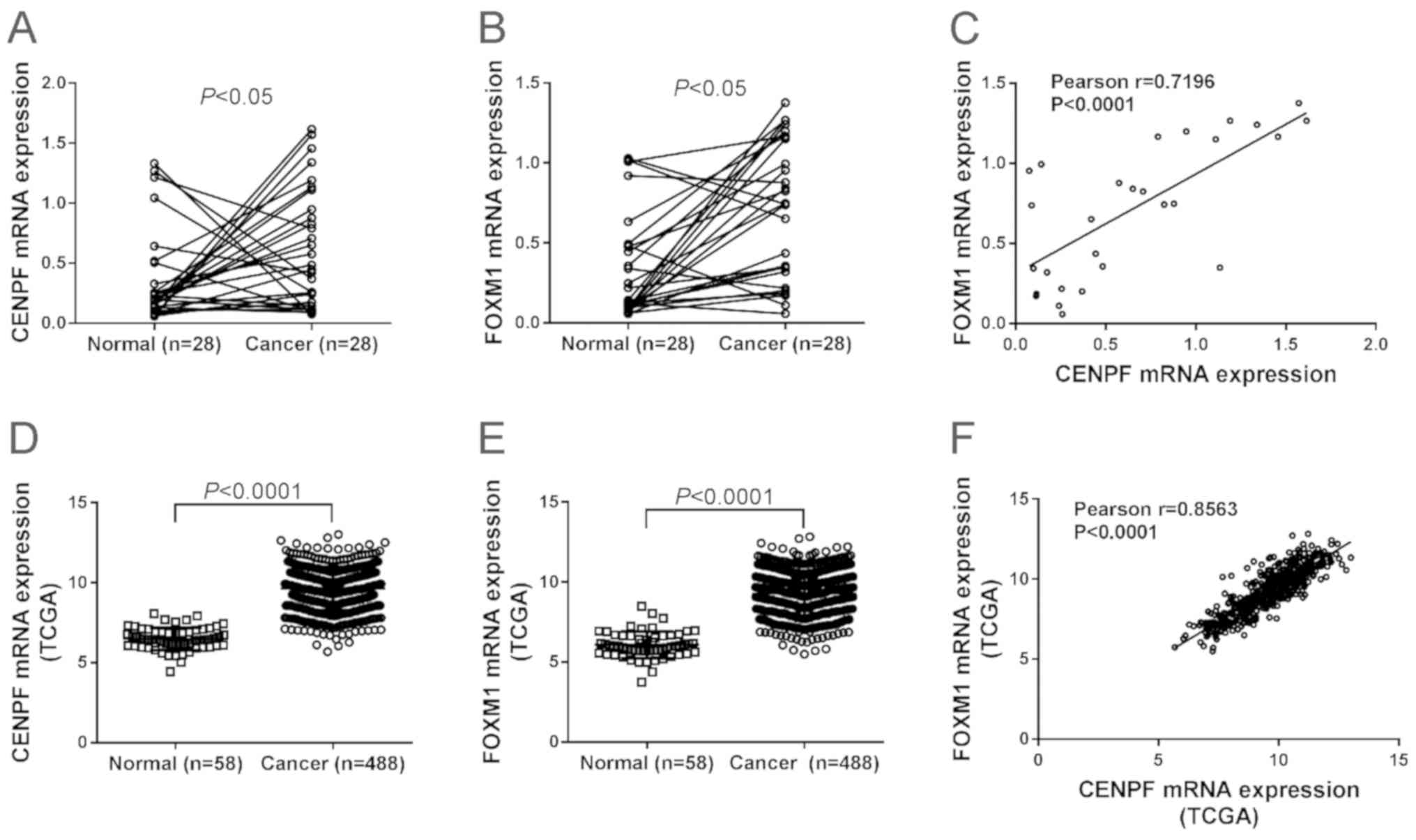
NSCLC tissues and adjacent non-cancerous tissues. Fig. 1A and B shows that 67.9% (19 cases)
and 78.6% (22 cases) of patients showed high mRNA expression of
CENPF and FOXM1, respectively. Paired Student's t-test revealed
that mRNA levels of both genes were significantly elevated in NSCLC
tissues compared to the non-cancerous tissues (P<0.05).
Pearson's r correlation analysis displayed a significant positive
association between CENPF and FOXM1 in NSCLC tissues (P<0.0001)
(Fig. 1C).
mRNA profile data of lung cancer tissues and control
tissues were downloaded from The Cancer Genome Atlas (TCGA)
database. The expression of CENPF (Fig.
1D) and FOXM1 (Fig. 1E) was also
significantly higher in lung cancer tissues than in normal control,
and CENPF expression was positively correlated with FOXM1
expression in lung cancer tissues (Fig.
1F).
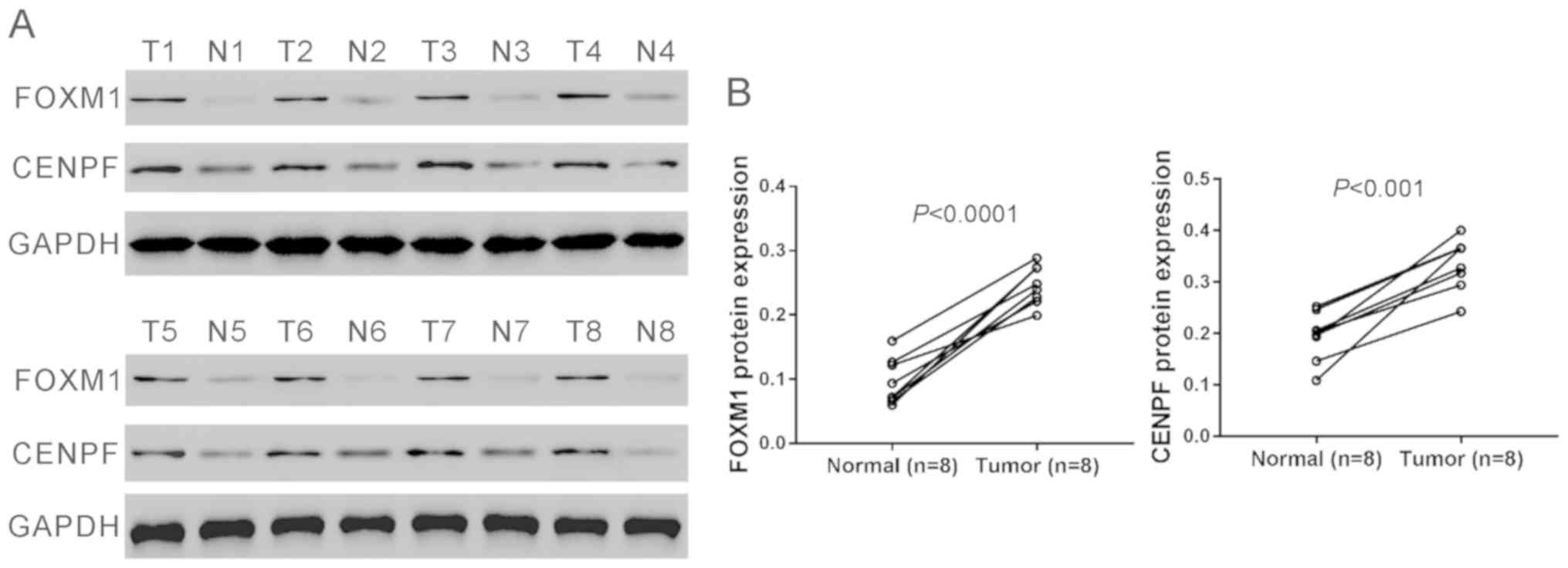
Furthermore, eight pairs of tissue samples were
randomly selected from the above 28 pairs of samples and subjected
to western blot analysis and the results confirmed the elevated
protein levels of CENPF and FOXM1 in NSCLC tissues (Fig. 2A and B).
Elevated expression of CENPF
correlated with clinical parameters of NSCLC
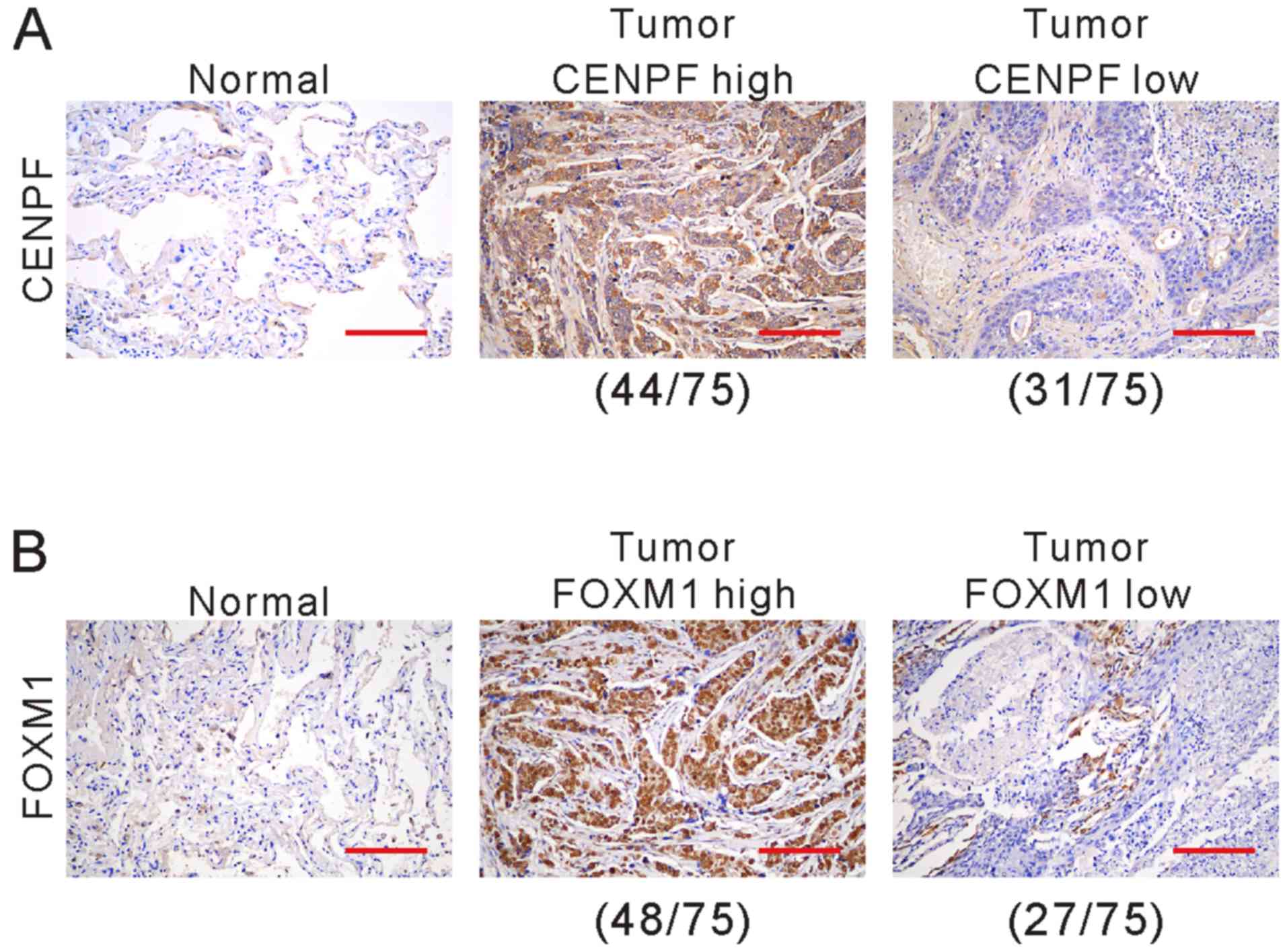
We further detected the protein expression of CENPF
and FOXM1 in cancerous specimens and matched non-cancerous
specimens from 75 NSCLC patients by immunohistochemistry. The
clinical and pathological characteristics of these patients are
listed in Table I. CENPF (Fig. 3A) and FOXM1 (Fig. 3B) expression was observed in
cytoplasm and nucleus. Of the 75 patients, 58.7% (44 cases) and
41.3% (31 cases) showed high and low expression of CENPF,
respectively, while 64.0% (48 cases) and 36.0% (27 cases) showed
high and low expression of FOXM1, respectively.
 | Table I.Clinicopathological characteristics in
NSCLC patients (n=75). |
Table I.
Clinicopathological characteristics in
NSCLC patients (n=75).
| Characteristic | Cases | % |
|---|
| Age (years) |
|
<60 | 37 | 49.3 |
| ≥60 | 38 | 50.7 |
| Sex |
| Male | 42 | 56.0 |
|
Female | 33 |
|
| Smoking status |
|
Smoker | 25 | 33.3 |
|
Non-smoker | 50 | 66.7 |
| Tumor size |
| <5
cm | 31 | 41.3 |
| ≥5
cm | 44 | 58.7 |
| TNM stage |
|
|
| I+II | 30 | 40.0 |
| III | 45 | 60.0 |
| Lymph node
metastasis |
|
Absent | 43 | 57.3 |
|
Present | 32 | 42.7 |
| Pathological
type |
|
Adenocarcinoma | 46 | 61.3 |
| Squamous
cell carcinoma | 29 | 39.7 |
| Vital status (at
follow-up) |
|
Alive | 24 | 32.0 |
| Dead | 51 | 68.0 |
| CENPF expression |
| Low | 31 | 41.3 |
| High | 44 | 58.7 |
| FOXM1 expression |
| Low | 27 | 36.0 |
| High | 48 | 64.0 |
The correlation between CENPF expression and the
clinical parameters of NSCLC was analyzed by Fisher's exact test.
As shown in Table II, CENPF protein
expression was positively correlated with tumor size (P=0.0179),
vital status (P=0.0008) and FOXM1 expression (P=0.0013), which
suggested clinical significance of CENPF in NSCLC.
 | Table II.Correlation of CENPF expression in
NSCLC tissues with different clinicopathological features
(n=75). |
Table II.
Correlation of CENPF expression in
NSCLC tissues with different clinicopathological features
(n=75).
|
| CENPF |
|
|---|
|
|
|
|
|---|
| Characteristic | Low (n=31) | High (n=44) | P-value |
|---|
| Age (years) |
|
| 0.6410 |
|
<60 | 14 | 23 |
|
|
≥60 | 17 | 21 |
|
| Sex |
|
| 0.8163 |
|
Male | 18 | 24 |
|
|
Female | 13 | 20 |
|
| Smoking status |
|
| 0.6210 |
|
Smoker | 9 | 16 |
|
|
Non-smoker | 22 | 28 |
|
| Tumor size |
|
| 0.0179a |
| <5
cm | 18 | 13 |
|
| ≥5
cm | 13 | 31 |
|
| TNM stage |
|
| 0.0991 |
|
I/II | 16 | 14 |
|
|
III | 15 | 30 |
|
| Lymph node
metastasis |
|
| 0.3474 |
|
Absent | 20 | 23 |
|
|
Present | 11 | 21 |
|
| Pathological
Type |
|
| 0.4705 |
|
Adenocarcinoma | 21 | 25 |
|
|
Squamous cell carcinoma | 10 | 19 |
|
| Vital status (at
follow-up) |
|
| 0.0008c |
|
Alive | 17 | 7 |
|
|
Dead | 14 | 37 |
|
| FOXM1
expression |
|
| 0.0013b |
|
Low | 18 | 9 |
|
|
High | 13 | 35 |
|
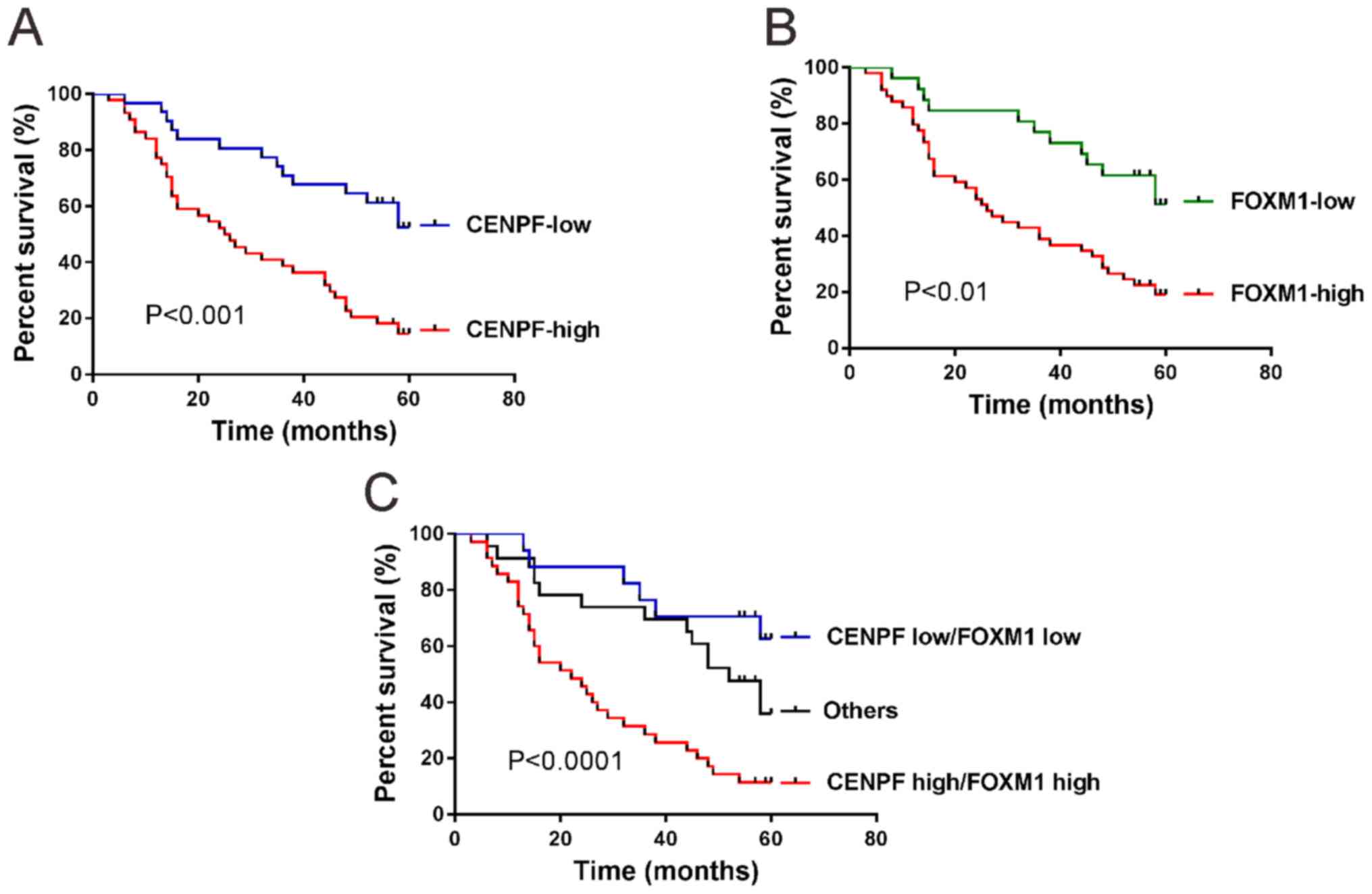
Expression of CENPF is closely related with the poor
prognosis of patients with NSCLC. High expression of CENPF
(P<0.001; Fig. 4A) and high
expression of FOXM1 (P<0.01; Fig.
4B) in NSCLC were correlated with the short survival time of
patients by Kaplan-Meier and log-rank test.
When both CENPF and FOXM1 were analyzed (Fig. 4C), patients whose tumors exhibited
high expression of CENPF and high expression of FOXM1 had the
shortest overall survival time, whereas patients with tumors
displaying low expression of CENPF and low expression of FOXM1 had
the longest overall survival time (P<0.0001).
Finally, a multivariate Cox regression analysis was
performed. CENPF (hazard ratio, 2.694; 95% CI, 1.397–5.195;
P=0.003) was an independent parameter that was associated with
overall survival when compared with tumor size and FOXM1 expression
(Table III).
 | Table III.Multivariate Cox regression of
prognostic parameters for survival in 75 NSCLC patients. |
Table III.
Multivariate Cox regression of
prognostic parameters for survival in 75 NSCLC patients.
|
| Multivariate
analysis |
|---|
|
|
|
|---|
| Prognostic
parameter | HR | 95% CI | P-value |
|---|
| CENPF expression
(low vs. high) | 2.694 | 1.397–5.195 | 0.003a |
| Tumor size (<5
vs. ≥5 cm) | 1.045 | 0.574–1.903 | 0.886 |
| FOXM1 expression
(low vs. high) | 1.751 | 0.911–3.366 | 0.093 |
Discussion
Identification of specific biomarkers is important
for diagnosis, therapy and prognosis of NSCLC. Previous studies
have revealed the potential prognostic values of CENPF in several
human cancers except NSCLC (7–12). In
the present study, we pinpointed that CENPF expression was elevated
in NSCLC tissues at both mRNA and protein levels. Then the protein
expression of CENPF in 75 cases of NSCLC and its association with
overall survival and clinical characteristics were investigated.
The results indicated that there was a significant correlation
between CENPF expression and tumor size, vital status, and overall
survival. Multivariate Cox regression analysis demonstrated that
CENPF expression was an independent prognostic factor of patients
with NSCLC. Our data suggest that CENPF expression may serve as a
novel prognostic marker for NSCLC although further validation data
with larger sample size are required.
FOXM1, a transcription factor, plays a critical role
during development (13–18) and carcinogenesis (19–21).
Previous studies have demonstrated that FOXM1 is an independent
prognostic factor for NSCLC (22,23).
FOXM1 and CENPF colocalized in the nucleus of prostate cancer
cells, and co-expression of FOXM1 and CENPF is a prognostic
indicator for poor survival of prostate cancer (24–26). In
the present study, the findings also demonstrated the value of
diagnosis and prognosis of FOXM1 in NSCLC. Importantly, we found
that CENPF mRNA expression was positively correlated with FOXM1
mRNA expression in NSCLC samples by analyzing TCGA database and our
own samples. CENPF protein expression was positively correlated
with FOXM1 protein expression in NSCLC specimens as indicated by
immunohistochemical staining. In addition, immunohistochemical
staining analysis indicated a similar subcellular localization of
CENPF and FOXM1 in NSCLC specimens. Patients with high expression
of CENPF and FOXM1 had the worst overall survival, whereas patients
with low expression of both proteins had the best overall survival.
Thus, the present study suggests that CENPF and FOXM1 may
co-operate in NSCLC. Aytes et al (24) reported that knockdown of CENPF
decreased the binding of FOXM1 to its target genes as revealed by
chromatin immunoprecipitation analysis. Similar mechanism may exist
in NSCLC cells, which needs to be investigated in the future.
In conclusion, the present study has demonstrated
that CENPF expression in NSCLC is correlated with FOXM1 expression
and worse clinical outcome. These findings suggest that CENPF may
function as a potential prognostic indicator for NSCLC. However,
the present findings are based on a small sample size and further
study with larger number of patients is needed.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
RL and YL wrote the manuscript. RL, XW and XZ
performed PCR and western blot analysis. XZ and HC were responsible
for immunohistochemical staining. YM and YL helped with statistical
analysis. All the authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Shenyang Fifth People's Hospital. Patients who participated in this
research, signed an informed consent and had complete clinical
data. Signed informed consents were obtained from the patients or
the guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang Y, Zheng T and Zhang W: Report of
cancer incidence and mortality in China. 4:1–7. 2018.
|
|
4
|
Bomont P, Maddox P, Shah JV, Desai AB and
Cleveland DW: Unstable microtubule capture at kinetochores depleted
of the centromere-associated protein CENP-F. EMBO J. 24:3927–3939.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Varis A, Salmela AL and Kallio MJ: Cenp-F
(mitosin) is more than a mitotic marker. Chromosoma. 115:288–295.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liao H, Winkfein RJ, Mack G, Rattner JB
and Yen TJ: CENP-F is a protein of the nuclear matrix that
assembles onto kinetochores at late G2 and is rapidly degraded
after mitosis. J Cell Biol. 130:507–518. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Clark GM, Allred DC, Hilsenbeck SG,
Chamness GC, Osborne CK, Jones D and Lee WH: Mitosin (a new
proliferation marker) correlates with clinical outcome in
node-negative breast cancer. Cancer Res. 57:5505–5508.
1997.PubMed/NCBI
|
|
8
|
de la Guardia C, Casiano CA,
Trinidad-Pinedo J and Báez A: CENP-F gene amplification and
overexpression in head and neck squamous cell carcinomas. Head
Neck. 23:104–112. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim HE, Kim DG, Lee KJ, Son JG, Song MY,
Park YM, Kim JJ, Cho SW, Chi SG, Cheong HS, et al: Frequent
amplification of CENPF, GMNN and CDK13 genes in hepatocellular
carcinomas. PLoS One. 7:e432232012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
O'Brien SL, Fagan A, Fox EJ, Millikan RC,
Culhane AC, Brennan DJ, McCann AH, Hegarty S, Moyna S, Duffy MJ, et
al: CENP-F expression is associated with poor prognosis and
chromosomal instability in patients with primary breast cancer. Int
J Cancer. 120:1434–1443. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shahid M, Lee MY, Piplani H, Andres AM,
Zhou B, Yeon A, Kim M, Kim HL and Kim J: Centromere protein F
(CENPF), a microtubule binding protein, modulates cancer metabolism
by regulating pyruvate kinase M2 phosphorylation signaling. Cell
Cycle. 17:2802–2818. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li P, You S, Nguyen C, Wang Y, Kim J,
Sirohi D, Ziembiec A, Luthringer D, Lin SC, Daskivich T, et al:
Genes involved in prostate cancer progression determine MRI
visibility. Theranostics. 8:1752–1765. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang IC, Snyder J, Zhang Y, Lander J,
Nakafuku Y, Lin J, Chen G, Kalin TV, Whitsett JA and Kalinichenko
VV: Foxm1 mediates cross talk between Kras/mitogen-activated
protein kinase and canonical Wnt pathways during development of
respiratory epithelium. Mol Cell Biol. 32:3838–3850. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bolte C, Zhang Y, Wang I-C, Kalin TV,
Molkentin JD and Kalinichenko VV: Expression of Foxm1 transcription
factor in cardiomyocytes is required for myocardial development.
PLoS One. 6:e222172011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ustiyan V, Wert SE, Ikegami M, Wang IC,
Kalin TV, Whitsett JA and Kalinichenko VV: Foxm1 transcription
factor is critical for proliferation and differentiation of Clara
cells during development of conducting airways. Dev Biol.
370:198–212. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim I-M, Ramakrishna S, Gusarova GA, Yoder
HM, Costa RH and Kalinichenko VV: The forkhead box m1 transcription
factor is essential for embryonic development of pulmonary
vasculature. J Biol Chem. 280:22278–22286. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zeng J, Wang L, Li Q, Li W, Björkholm M,
Jia J and Xu D: FoxM1 is up-regulated in gastric cancer and its
inhibition leads to cellular senescence, partially dependent on p27
kip1. J Pathol. 218:419–427. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang C, Chen H, Tan G, Gao W, Cheng L,
Jiang X, Yu L and Tan Y: FOXM1 promotes the epithelial to
mesenchymal transition by stimulating the transcription of Slug in
human breast cancer. Cancer Lett. 340:104–112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Z, Ahmad A, Li Y, Banerjee S, Kong D
and Sarkar FH: Forkhead box M1 transcription factor: A novel target
for cancer therapy. Cancer Treat Rev. 36:151–156. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu M, Dai B, Kang S-H, Ban K, Huang FJ,
Lang FF, Aldape KD, Xie TX, Pelloski CE, Xie K, et al: FoxM1B is
overexpressed in human glioblastomas and critically regulates the
tumorigenicity of glioma cells. Cancer Res. 66:3593–3602. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim I-M, Ackerson T, Ramakrishna S,
Tretiakova M, Wang IC, Kalin TV, Major ML, Gusarova GA, Yoder HM,
Costa RH, et al: The Forkhead Box m1 transcription factor
stimulates the proliferation of tumor cells during development of
lung cancer. Cancer Res. 66:2153–2161. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu N, Jia D, Chen W, Wang H, Liu F, Ge H,
Zhu X, Song Y, Zhang X, Zhang D, et al: FoxM1 is associated with
poor prognosis of non-small cell lung cancer patients through
promoting tumor metastasis. PLoS One. 8:e594122013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang DK, Son CH, Lee SK, Choi PJ, Lee KE
and Roh MS: Forkhead box M1 expression in pulmonary squamous cell
carcinoma: Correlation with clinicopathologic features and its
prognostic significance. Hum Pathol. 40:464–470. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Aytes A, Mitrofanova A, Lefebvre C,
Alvarez MJ, Castillo-Martin M, Zheng T, Eastham JA, Gopalan A,
Pienta KJ, Shen MM, et al: Cross-species regulatory network
analysis identifies a synergistic interaction between FOXM1 and
CENPF that drives prostate cancer malignancy. Cancer Cell.
25:638–651. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lin SC, Kao CY, Lee HJ, Creighton CJ,
Ittmann MM, Tsai SJ, Tsai SY and Tsai MJ: Dysregulation of
miRNAs-COUP-TFII-FOXM1-CENPF axis contributes to the metastasis of
prostate cancer. Nat Commun. 7:114182016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lokody I: Signalling: FOXM1 and CENPF:
co-pilots driving prostate cancer. Nat Rev Cancer. 14:450–451.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee JJ, Maeng CH, Baek SK, Kim GY, Yoo JH,
Choi CW, Kim YH, Kwak YT, Kim DH, Lee YK, et al: The
immunohistochemical overexpression of ribonucleotide reductase
regulatory subunit M1 (RRM1) protein is a predictor of shorter
survival to gemcitabine-based chemotherapy in advanced non-small
cell lung cancer (NSCLC). Lung Cancer. 70:205–210. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mizuno K, Mataki H, Arai T, Okato A,
Kamikawaji K, Kumamoto T, Hiraki T, Hatanaka K, Inoue H and Seki N:
The microRNA expression signature of small cell lung cancer: Tumor
suppressors of miR-27a-5p and miR-34b-3p and their targeted
oncogenes. J Hum Genet. 62:671–678. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ma J, Qi G, Xu J, et al: Overexpression of
forkhead box m1 and urokinase-type plasminogen activator in gastric
cancer is associated with cancer progression and poor prognosis.
Oncol Lett. 14:7288–7296. 2017.PubMed/NCBI
|