Introduction
MicroRNAs (miRNAs) are small single-stranded
non-coding RNAs, consisting of no more than 25 nucleotides (nts)
cleaved from hairpin pre-miRNA precursors (1). miRNAs serve important roles in the
epigenetic regulation of protein expression, are encoded by their
respective genes and are highly conserved in all multicellular
eukaryotes and certain unicellular eukaryotes (2). Generally, miRNAs recognize and bind to
complementary 3′-untranslated regions of target mRNAs, which can
induce degradation or transcriptional repression of the target
(3). However, it has also been
revealed that miRNAs can upregulate the translation of target genes
by interacting with regulatory complexes (4). A single miRNA has the capacity to
regulate the expression of multiple target mRNAs and one gene can
be modulated by multiple miRNAs (5).
A number of miRNAs have been identified in multiple eukaryotic
organisms; a total of 2,588 human genome miRNAs are included in the
miRBase database (6). It is estimated
that >60% of all mRNAs are modulated by miRNAs at the
post-transcriptional level (7).
Previous studies have identified that miRNAs serve critical roles
in numerous biological processes, including development, cell
differentiation, proliferation and apoptosis (8). Furthermore, miRNA-associated dysfunction
is associated with a number of diseases, including Alzheimer's
disease (9), cardiovascular diseases
(10) and numerous cancer types
(11). Additionally, the modulation
of miRNAs has been demonstrated to provide therapeutic benefits.
For example, miRNAs have been demonstrated to be useful in cancer
therapy (12).
Grape seed proanthocyanidins (GSPs), biologically
active components that make up 70–95% of the proanthocyanidins,
have been reported to modulate the expression of certain miRNAs
that serve important roles in cancer, glucose homeostasis and lipid
homeostasis (13,14). Previous studies have revealed that
GSPs exhibit inhibitory effects in various cancer types, including
pancreatic cancer (PC) (15–17). However, to the best of our knowledge,
the molecular mechanism of this anticancer effect remains unknown.
In our previous study transcriptome analysis of the response of PC
cells to treatment with GSPs was performed using RNA-seq (18). This indicated that the expression
levels of multiple genes associated with the proliferation of PC
cells were altered in GSPs-treated cells. However, to the best of
our knowledge, it remains to be investigated whether treatment with
GSPs can regulate the expression of miRNAs in PC cells.
To determine how treatment with GSPs regulates the
expression of miRNA in PC cells the current study generated PC cell
samples (SS3, SS12 and SS24) treated with 20 µg/ml GSPs for 3, 12
and 24 h, respectively. Subsequently, miRNA-seq experiments were
performed. Compared with the miRNA data obtained from control cell
samples (SC3, SC12 and SC24), numerous differentially expressed
(DE) miRNAs were identified in SS3, SS12 and SS24. In addition, a
number of target genes of the DE miRNAs were identified. Analysis
using the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) databases revealed that multiple target genes were
enriched in functional pathways associated with the proliferation
of cancer cells. This suggested that treatment with GSPs may
exhibit inhibitory effects on cancer cells through the regulation
of miRNAs.
Materials and methods
Cell culture and reagents
The human PC cell line PANC-1 was obtained from
Procell (Wuhan, China) (http://www.procell.com.cn/) and cultured in monolayers
in Dulbecco's modified Eagle's medium (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA)
in a humidified incubator at 37°C and 5% CO2. The GSP
extract, obtained from JF-NATURAL (Tianjin, China; catalog no.
J011003), contained monomeric (9.5%), dimeric (12.8%), trimeric
(76.7%) and oligomeric (1%) procyanidins. The 100 µg GSPs were
dissolved in 100 µl dimethylsulfoxide (DMSO; Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) for 10 min at room temperature prior to
addition to cell culture media. The maximum concentration of DMSO
in the media did not exceed 0.1%. PANC-1 cells were treated with 20
µg/ml GSP for 3, 12 and 24 h at 37°C, and the treated cells were
used to prepare for SS3, SS12 and S24 samples, respectively.
Additionally, control cell samples were treated with DMSO for 3, 12
and 24 h at 37°C, and then SC3, SC12 and SC24 samples were prepared
accordingly.
Cell viability assay
GSP-treated PANC-1 cells were plated in 96-well cell
culture plates at 5×103 cells/well and incubated for 24
h at 37°C. Subsequently, 50 µl of MTT solution (5 mg/ml;
Sigma-Aldrich; Merck KGaA) was added to each well and the cells
were incubated for a further 4 h at 37°C. Following 3 min
centrifugation (5500 × g) at 4°C, the supernatant was removed from
each well. The formazan crystals produced from MTT in each well
were dissolved in 150 µl DMSO and the optical density values were
measured at 490 nm.
Flow cytometry
GSP-induced apoptosis in PC cells was determined by
flow cytometry using the Annexin V-fluorescein isothiocyanate
(FITC) Apoptosis Detection kit (BD Biosciences, Franklin Lakes, NJ,
USA). Following treatment with GSPs for 48 h, 2×105
cells were harvested, washed twice with PBS and incubated with
Annexin V-FITC and propidium iodide for 10 min in the dark at room
temperature. The stained cells were then detected and analyzed by
the MoFlo XDP flow cytometer (Beckman Coulter, Inc., Brea, CA, USA)
and Cell Quest 3.3 software (BD Biosciences, Franklin Lakes, NJ,
USA).
RNA extraction and small RNA
sequencing
Total RNA was extracted from PC cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol. Subsequently, RQ1 DNase (Promega
Corporation, Madison, WI, USA) was used to remove DNA. The quality
and quantity of the purified RNA was monitored by absorbance at 260
and 280 nm, and the A260: A280 ratio, using a SmartSpec Plus
Spectrophotometer (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
RNA integrity was further verified by electrophoresis of a 1.5%
agarose gel.
Total RNA (3 µg) from each sample was used for small
RNA cDNA library preparation with a Balancer NGS Library
Preparation kit (Gnomegen, San Diego, CA, USA), according to the
manufacturer's protocol. The whole library was subjected to 10%
native polyacrylamide gel electrophoresis and the bands
corresponding to miRNA insertion were cut and eluted. Following
ethanol precipitation and washing, the purified libraries were
quantified using the QubitFluorometer (Invitrogen; Thermo Fisher
Scientific, Inc.). These small RNA libraries were applied to
NextSeq 500 (Illumina, Inc., San Diego, CA, USA) 76 nt pair-end
sequencing.
Identification of conserved and novel
miRNA
To obtain reliable clean reads, raw reads were
processed by the FASTX-Toolkit (version 0.0.13)(http://hannonlab.cshl.edu/fastx_toolkit/). During this
procedure, adaptor sequences and low quality tags were discarded.
Based on the length of the mature miRNA and adapter lengths, RNAs
<16 or >30 nts in length were also excluded from further
analysis. The high-quality clean reads were subsequently searched
against the Rfam database (version 12.0) (http://rfam.xfam.org/) using Bowtie (19). Matches to ribosomal RNAs and transfer
RNAs were excluded. Subsequently, the remaining unique sequences
were aligned against the miRBase database (20) using Bowtie, with one mismatch allowed.
The matched small RNA sequences were considered to be conserved
miRNAs and the unaligned sequences were potential candidates for
novel miRNAs. To identify novel miRNAs, the unique sequences were
aligned to the human reference genome sequence (GRCH38) using the
miRDeep algorithm (21).
Bioinformatics analysis
To investigate the expression profiles of identified
miRNAs, the frequency of miRNA counts were normalized to
transcripts per million (TPM) using the following formula:
Normalized expression=actual read count/total read count
×106. DE miRNAs were analyzed using the edgeR (v3.22)
package of Bioconductor software (22). A fold change (FC) ≤2 or <0.5 and
P≤0.01 indicated a statistically significant DE miRNA. The miRanda
algorithm (23) was used on human
miRNA and transcript sequences of miRBase and TargetScan (7) for the prediction of putative miRNA
targets.
To predict the gene function and calculate the
frequency distribution of functional categories, GO and KEGG
analysis were employed using the DAVID bioinformatics database
(24). Networks were constructed by
calculating the Pearson's correlation coefficient (PCC) for the
expression levels of DE miRNAs and target genes. Cytoscape (version
3.0.2) was used to display the co-expression network (25).
Validation of the target genes by
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR)
To validate the RNA-seq data, RT-qPCR was performed
for selected target genes and normalization was achieved with the
human reference gene GAPDH. The primers used are presented in
Table I. The same RNA samples for
RNA-seq were used for RT-qPCR, and RNA extraction was performed
with the same protocol and materials. In each pooled sample, l µg
of total RNA was reverse transcribed using the PrimeScript™ RT
Reagent kit (Takara Bio, Inc., Otsu, Japan), according to the
manufacturer's protocol. qPCR was performed with the S1000 Thermal
Cycler (Bio-Rad Laboratories, Inc.) and Bestar SYBR Green RT-PCR
Master mix (DBI Bioscience, Shanghai, China). The following
thermocycling conditions were used: 95°C for 10 min, 38 cycles of
95°C for 15 sec and 60°C for 1 min. PCR amplifications were
performed in triplicate for each sample, and the results were
quantified by 2−∆∆Cq methods (26).
 | Table I.Genes and primers used for reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Genes and primers used for reverse
transcription-quantitative polymerase chain reaction.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| CDK6 |
GCAGCTTCTCTTCTTCTGGAAT |
TGTGGAGGATTGCTATCTGGAG |
| MSH6 |
GTCCTATGTGTCGCCCAGTA |
TTCCTGCTCCTCTTCCTCAC |
| EGFR |
GTGTGCCCTGTAACCTGAC |
GTGACTGAACATAACTGTAGGC |
| DNMT1 |
CCGTGGATGAGGACCTGTAC |
CCTGCCGTTGCTCTTCTTG |
| GAPDH |
GGTCGGAGTCAACGGATTTG |
GGAAGATGGTGATGGGATTTC |
Statistical analysis
All data are presented as the mean ± standard
deviation. For comparisons between two groups, statistically
significant differences between means were identified by Student's
t-test. For multiple comparisons, the significance was determined
by one-way analysis of variance followed by a Tukey's post hoc
test. P<0.05 was considered to indicate a statistically
significant difference.
Online data deposition
The datasets obtained in the current study were
deposited in the National Center for Biotechnology Information Gene
Expression Omnibus database under the accession number GSE107409.
The transcriptome database (GSE85610) generated in our previous
study (18) was also deposited.
Results
GSPs repressed the viability of PC
cells in vitro
Using flow cytometry, the current study identified
that treatment with 0, 20, 40 or 60 µg/ml GSP significantly induced
apoptosis of the PC cell line PANC-1 in a dose-dependent manner
(Fig. 1A and B). In addition, the
effect of treatment with GSPs on the viability of PC cells was
investigated by MTT assay. A significant dose-dependent decrease in
the viability of PC cells was identified 24 h after treatment
(Fig. 1C). The aforementioned
apoptosis and cell viability results were in accordance with
previous studies (15–17). Furthermore, it was identified that
treatment with GSPs was associated with necrosis in a small number
of of PC cells in a dose-dependent manner (Fig. 1A).
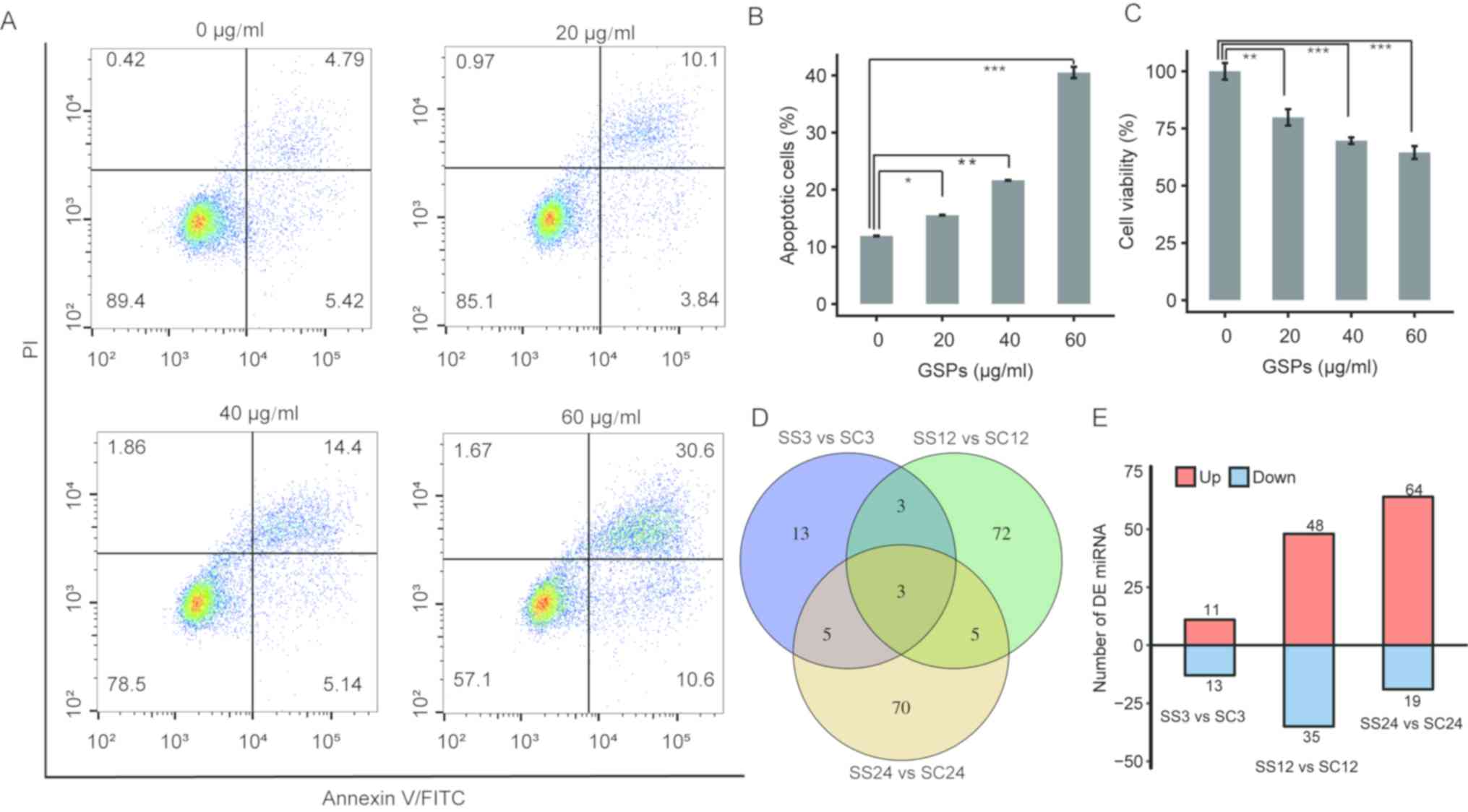 | Figure 1.Determination of DE miRNAs in PC cells
following treatment with GSPs. (A) GSPs induce apoptosis of the PC
cell line PANC-1. Cells were treated with different concentrations
of GSPs and harvested at 24 h post-treatment for assessment of
apoptosis using Annexin V-fluorescein isothiocyanate staining
coupled with flow cytometry. The upper left, upper right and lower
left quadrants represent necrotic, late apoptotic and early
apoptotic events, respectively. (B) Total percentage of apoptotic
PC cells in each treatment group are quantified with data presented
as the mean ± SD of two independent experiments. (C) Treatment with
GSPs reduces the viability of PC cells. Viability of the PC cells
was determined by MTT assay. The data are reported as the
percentage relative to control cells and presented as the mean ± SD
of six replicates. (D) Venn diagrams of the DE miRNAs identified in
the SS3 vs. SC3, SS12 vs. SC12 and SS24 vs. SC24 comparisons. (E)
The number of upregulated and downregulated DE miRNAs in the three
comparisons. Data are presented as the mean ± SD. Statistical
analysis was performed using one-way analysis of variance followed
by Tukey's post hoc test. *P<0.05, **P<0.01, ***P<0.001.
DE, differentially expressed; miRNA, microRNA; GSP, grape seed
proanthocyanidin; PC, pancreatic cancer; SD, standard deviation;
PI, propidium iodide; PE, phycoerythrin. |
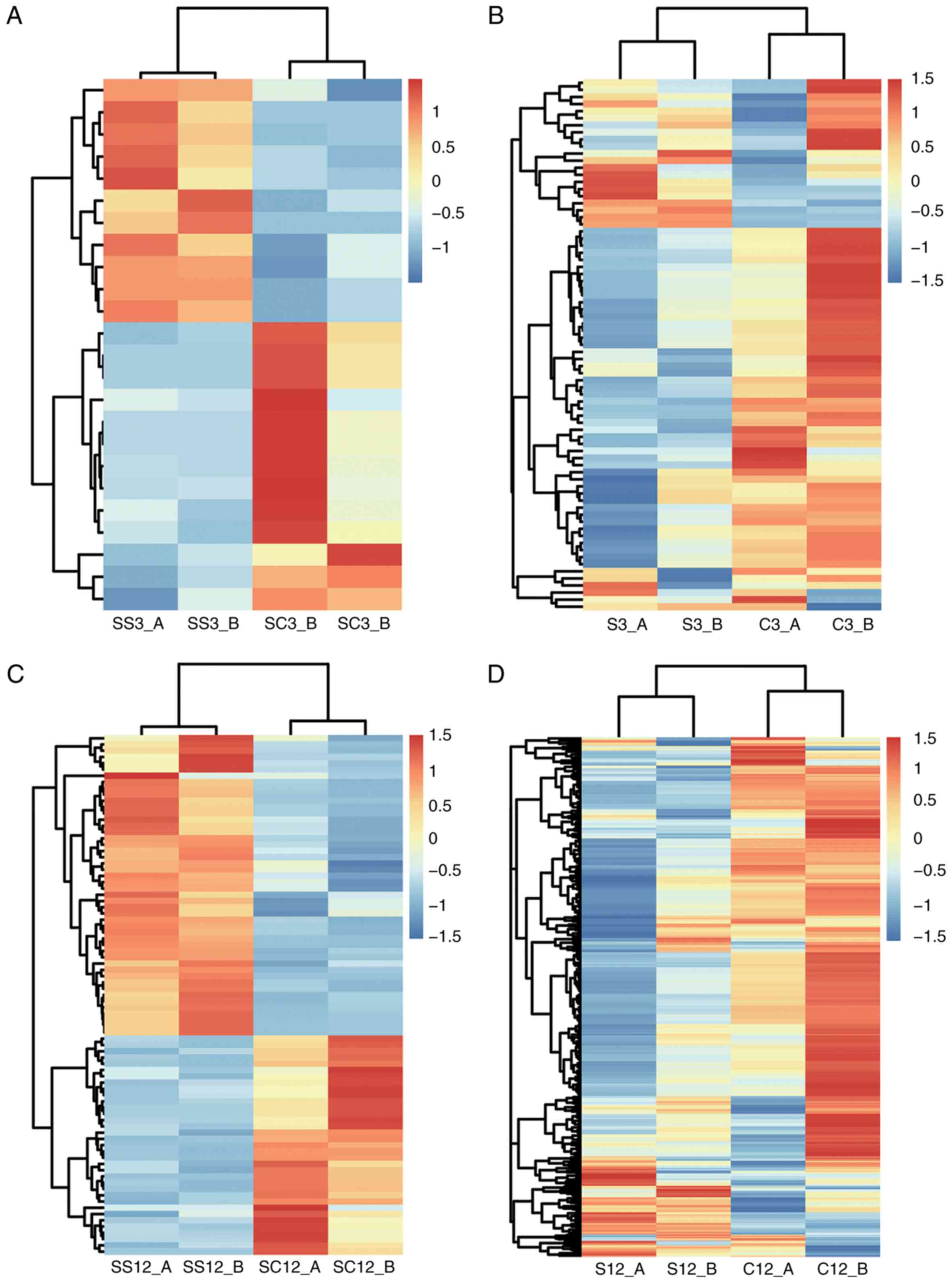
miRNA expression profiles in PC cells
treated with GSPs
To avoid high levels of necrotic cells and
investigate how GSPs modulate the expression of miRNAs in PC cells,
PANC-1 cells were treated with 20 µg/ml GSP for 3, 12 and 24 h, and
the miRNA samples termed SS3, SS12 and SS24 were prepared,
respectively. Cells treated with DMSO for 3, 12 and 24 h served as
controls, and the control miRNA samples termed SC3, SC12 and SC24
were prepared, respectively. Two biological replicates were
generated at each time point, therefore, a total of 12 small RNA
libraries (SS3-A, SS3-B, SC3-A, SC3-B; SS12-A, SS12-B, SC12-A,
SC12-B; SS24-A, SS24-B, SC24-A and SC24-B) were constructed for
miRNA-seq.
Using Illumina NextSeq 500, >89.38 million reads
were generated, corresponding to ~7.44 million sequence reads per
sample and ~70% of all reads were successfully mapped against the
current human reference genome (Table
II). In addition, the proportion of clean reads mapped to the
Rfam database for each sample is presented in Table III. In total, >79.6% of reads
were matched to Rfam and multiple mapped reads were >84.2%.
 | Table II.Small RNA-Seq results. |
Table II.
Small RNA-Seq results.
| Sample | Raw data | Clean reads | Total mapped
reads | Unique mapped
reads |
|---|
| SC3-A | 8764786 | 6564553 | 6344960 | 4373569 |
| SC3-B | 5900986 | 4283482 | 4141642 | 2896531 |
| SC12-A | 7603914 | 4836317 | 4456413 | 2834055 |
| SC12-B | 4918096 | 3034397 | 2614249 | 1657090 |
| SC24-A | 8437896 | 6022757 | 5777249 | 4000517 |
| SC24-B | 7878026 | 5703819 | 5467646 | 3774752 |
| SS3-A | 7470866 | 5732424 | 5538865 | 3835541 |
| SS3-B | 8821086 | 7070260 | 6855861 | 4833465 |
| SS12-A | 6258112 | 5002508 | 4760228 | 3293693 |
| SS12-B | 8683604 | 6627072 | 6366737 | 4431579 |
| SS24-A | 8985184 | 7399980 | 7002763 | 4752743 |
| SS24-B | 5657588 | 3457776 | 3328007 | 2009410 |
 | Table III.Clean reads mapped to Rfam. |
Table III.
Clean reads mapped to Rfam.
| Sample | Input reads | Total mapped
reads | Unique mapped
reads | Multiple mapped
reads |
|---|
| SC3-A | 6564553 | 5412203 | 808375 | 4603828 |
| SC3-B | 4283482 | 3538961 | 529044 | 3009917 |
| SC12-A | 4836317 | 3663218 | 567185 | 3096033 |
| SC12-B | 3034397 | 2082789 | 350653 | 1732136 |
| SC24-A | 6022757 | 4818716 | 780206 | 4038510 |
| SC24-B | 5703819 | 4574160 | 726961 | 3847199 |
| SS3-A | 5732424 | 4752397 | 718256 | 4034141 |
| SS3-B | 7070260 | 5883811 | 906572 | 4977239 |
| SS12-A | 5002508 | 4013582 | 644253 | 3369329 |
| SS12-B | 6627072 | 5256520 | 846982 | 4409538 |
| SS24-A | 7399980 | 5708723 | 930190 | 4778533 |
| SS24-B | 3457776 | 2669152 | 423987 | 2245165 |
The clean reads were mapped to miRBase and it was
identified that ~39.3% reads were matched to mature miRNAs
(Table IV). The read counts were
normalized to TPM and a total of 2,578 miRNAs were revealed,
consisting of 502 novel miRNAs (data not shown).
 | Table IV.Clean reads mapped against mature
microRNAs of miRBase. |
Table IV.
Clean reads mapped against mature
microRNAs of miRBase.
| Sample | Input reads | Total mapped
reads | Unique mapped
reads | Multiple mapped
reads |
|---|
| SC3-A | 6564553 | 2762812 | 2629156 | 133656 |
| SC3-B | 4283482 | 1781052 | 1694783 | 86269 |
| SC12-A | 4836317 | 1710715 | 1618013 | 92702 |
| SC12-B | 3034397 | 1032329 | 980273 | 52056 |
| SC24-A | 6022757 | 2302503 | 2190377 | 112126 |
| SC24-B | 5703819 | 2230508 | 2123120 | 107388 |
| SS3-A | 5732424 | 2420577 | 2299253 | 121324 |
| SS3-B | 7070260 | 3110970 | 2944227 | 166743 |
| SS12-A | 5002508 | 1922143 | 1828317 | 93826 |
| SS12-B | 6627072 | 2534886 | 2396862 | 138024 |
| SS24-A | 7399980 | 2816014 | 2673813 | 142201 |
| SS24-B | 3457776 | 1236899 | 1166154 | 70745 |
Determination of DE miRNAs
DE miRNAs were determined using edgeR (22) and the following criteria: P≤0.01 and
FC ≥2 or ≤0.5. A total of 24, 83 and 83 DE miRNAs were identified
in SS3 vs. SC3, SS12 vs. SC12 and SS24 vs. SC24, respectively
(Fig. 1D and E). The highest and
lowest FC values were 2×108.59 and 2×10−8.12,
respectively (data not shown). These findings indicated that
treatment with GSPs is associated with the expression levels of
numerous miRNAs in PANC-1 cells. In addition, only three common DE
miRNAs were identified between all three comparisons, suggesting
that treatment with GSPs can regulate the expression levels of
different miRNAs depending on the duration of treatment.
Prediction of DE miRNA target
genes
miRNAs commonly exert their functions by binding to
complementary target sites in the mRNAs of their target genes.
Using the miRanda algorithm (23) on
human miRNA and transcript sequences of miRBase and TargetScan
(7), 74, 598 and 1,204 target genes
were identified for the DE miRNAs in SS3 vs. SC3, SS12 vs. SC12 and
SS24 vs. SC24, respectively (Fig. 2).
This suggests that the DE miRNAs have the capacity to regulate the
expression levels of multiple genes.
In our previous study, PANC-1 cells were also
treated with 20 µg/ml GSPs for 3, 12 and 24 h, and 12 cDNA
libraries (S3-A, S3-B, C3-A, C3-B; S12-A, S12-B, C12-A, C12-B;
S24-A, S24-B, C24-A and C24-B) were constructed for RNA-seq. The
current study analyzed the expression levels of the predicted
target genes using the transcriptome database generated in our
previous study (GSE85610). This revealed that all predicted target
genes of the DE miRNAs were downregulated or upregulated following
treatment with GSPs, suggesting that GSPs may modulate gene
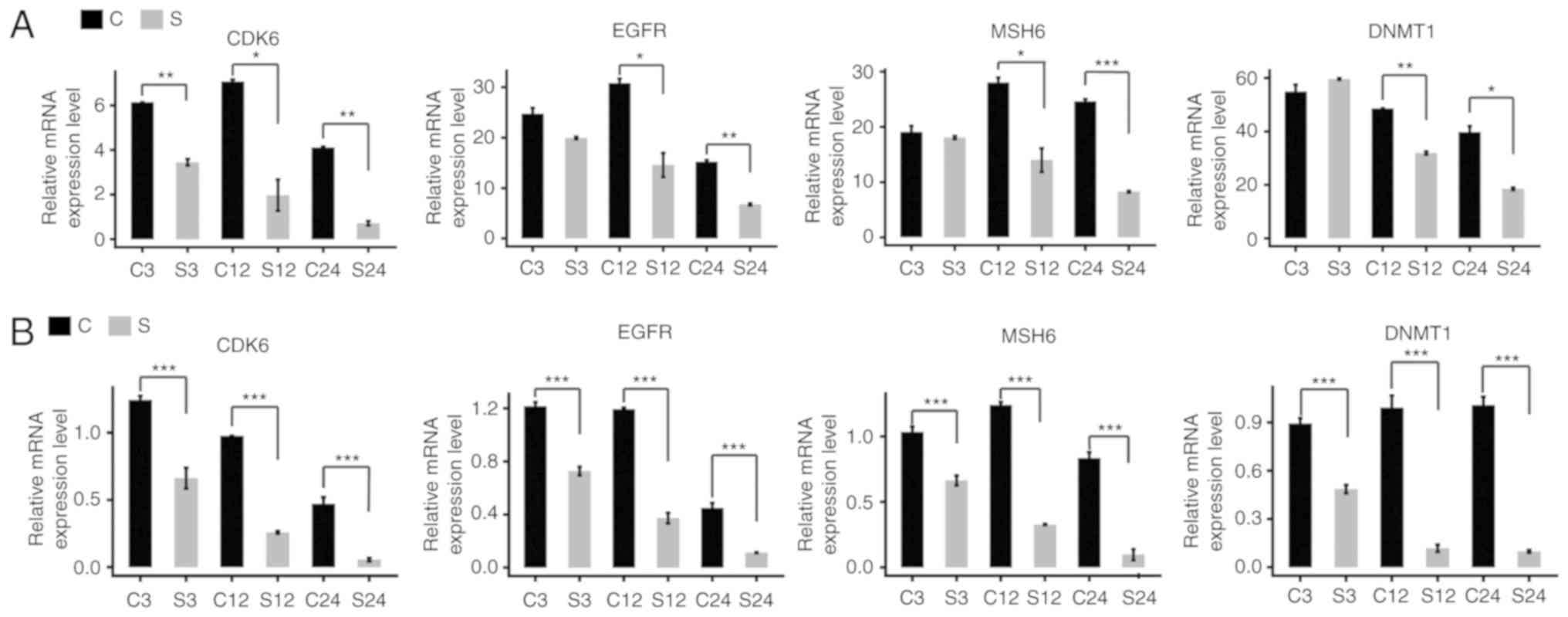
expression by the regulation of miRNAs. The expression levels
determined by RT-qPCR of certain target genes, including MutS
homolog 6, epidermal growth factor receptor, cyclin dependent
kinase 6 and DNA methyltransferase 1, were in accordance with the
RNA-seq results (Fig. 3).
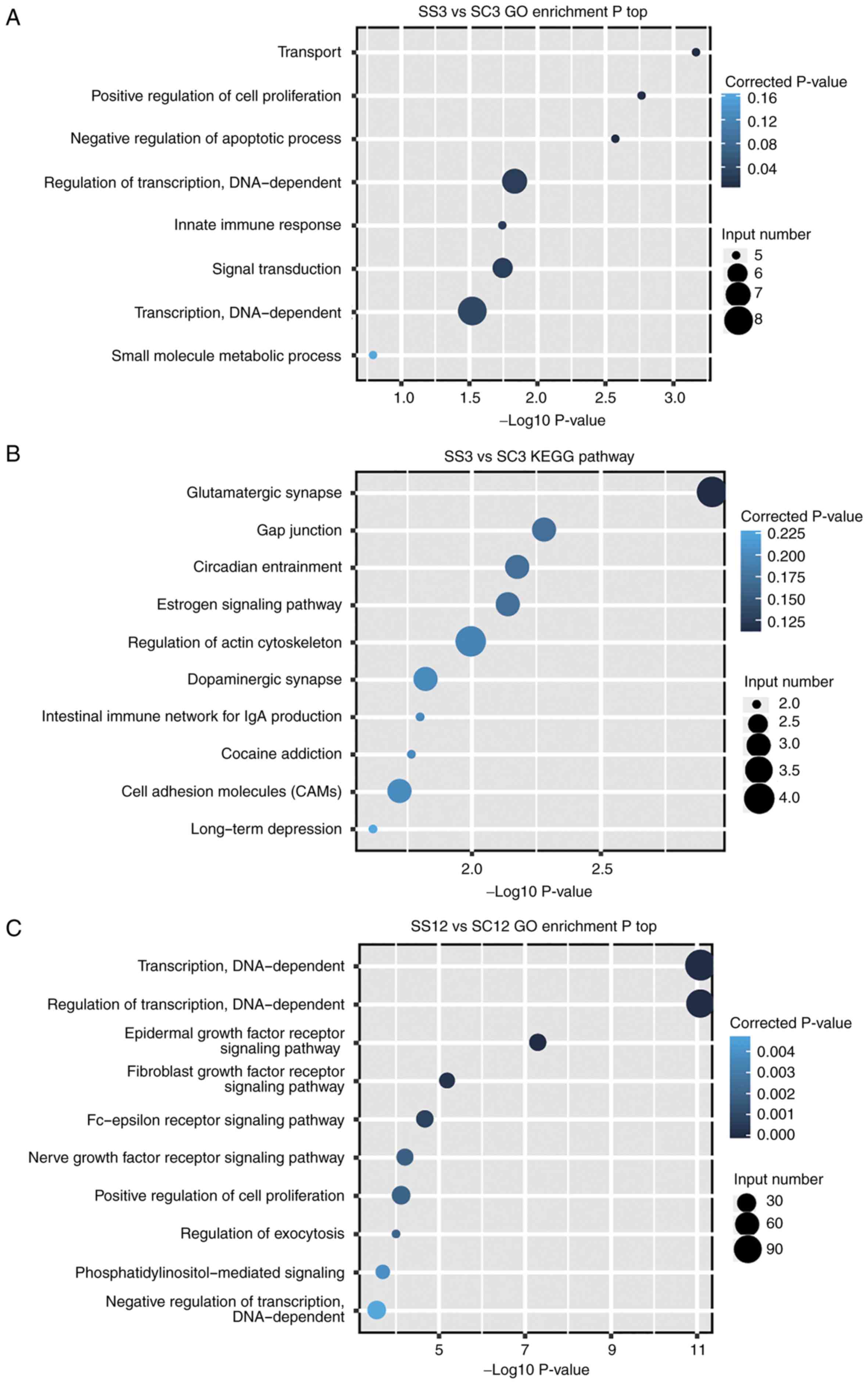
Functional analysis of target
genes
To identify the pathways associated with the target
genes of the DE miRNAs, GO enrichment analysis was first conducted
(Fig. 4). It was demonstrated that 7,
77 and 124 significant functional terms were identified for the
target genes of the DE miRNAs in SS3 vs. SC3, SS12 vs. SC12 and
SS24 vs. SC24, respectively (P<0.05; Fig. 4A, C and E). Multiple target genes were
enriched in ‘positive regulation of cell proliferation’ (GO:
0008284) and ‘negative regulation of apoptotic process’ (GO:
0043066), which indicates that the DE miRNAs induced by treatment
with GSPs may serve a regulatory role in the proliferation and
apoptosis of PANC-1 cells. Targeting factors associated with cell
cycle is a potential approach for cancer therapy; therefore, GSPs
may exhibit inhibitory effects on the proliferation of cancer
cells.
In addition, KEGG enrichment analysis was performed.
It was revealed that 15, 42 and 47 significant functional terms
were identified for the target genes of the DE miRNAs in SS3 vs.
SC3, SS12 vs. SC12 and SS24 vs. SC24, respectively (P<0.05).
Although ‘microRNAs in cancer’ (ID: hsa05206) and ‘pancreatic
cancer’ (ID: hsa05212) were not presented in Fig 4 (only top10 terms were presented),
numerous target genes were enriched in them and ‘pathways in
cancer’ (ID: hsa05200). This suggests that treatment with GSPs may
exhibit anticancer effects by disrupting the functional pathways of
PC cells.
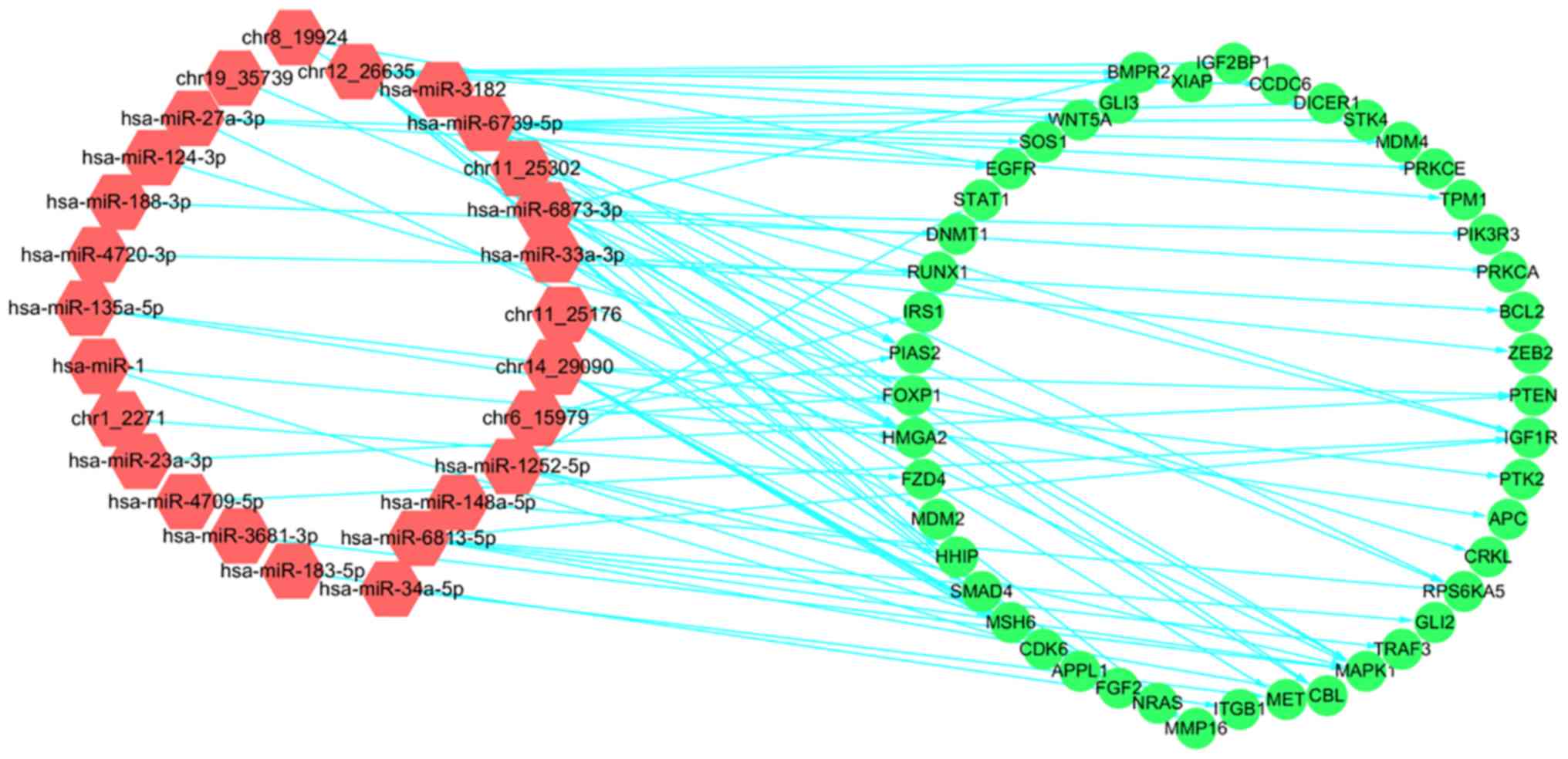
Integrative analysis of DE miRNA and
mRNA expression
The aforementioned results revealed that the target
genes of certain DE miRNAs were associated with the regulation of
cancer. To further investigate this, a co-expression network was
constructed according to PCC values that indicated the strength of
the correlation between the expression levels of the DE miRNAs and
the target genes that were enriched in ‘pathways in cancer’ (ID:
hsa05200), ‘microRNAs in cancer’ (ID: hsa05206) and ‘pancreatic
cancer’ (ID: hsa05212) (PPC ≥0.6, P<0.01; Fig. 5). In total, 36 DE miRNAs and 66 target
genes were analyzed, and 26 DE miRNAs were identified to be
correlated with 46 target genes. The negative co-expression
correlations revealed between DE miRNAs and target genes
demonstrated that treatment with GSPs may serve an anticancer role
by regulating the expression of miRNAs.
Discussion
Phytochemicals, naturally occurring bioactive
compounds, are important constituents of fruits, vegetables and
legumes that provide a rich source of dietary micronutrients.
Proanthocyanidins, the most common polyphenols, are the most
abundant phytochemicals in the human diet and have been
demonstrated to benefit human health (27). For example, proanthocyanidins have
been identified to improve the symptoms of metabolic disorders,
including insulin resistance, obesity, diabetes and inflammation
(28). In addition, proanthocyanidins
have been implicated in miRNA-based anticancer therapies (14), which are effective due to the ability
of small RNAs to influence cell behavior.
Proanthocyanidin extracts from a number of plants
exhibit different characteristic compositions and health effects
(29). GSPs, a group of
proanthocyanidins that primarily contain dimers, trimers and other
oligomers of catechin and epicatechin, and their galloylated esters
(30), have widely been investigated
and used. Previous studies have revealed that GSPs can inhibit the
proliferation of cancer cells by upregulating or downregulating
miRNA expression (13,31,32).
Furthermore, it has been demonstrated that GSPs are more effective
in modulating miRNA expression compared with other proanthocyanidin
extracts (13). In our previous
study, it was identified that treatment with GSPs inhibited the
proliferation of PC cells and certain molecular mechanisms were
examined by transcriptome analysis. In the current study, 2,076
conserved miRNAs were identified and 502 novel miRNAs were revealed
in PC cells. Following treatment with GSPs, the expression levels
of numerous miRNAs markedly changed, which demonstrates that GSPs
may serve a role in miRNA-based therapies. Additionally, multiple
target genes of the DE miRNAs were associated with the
proliferation of cancer cells, suggesting that treatment with GSPs
may inhibit the proliferation of cancer cells by regulating the
expression of miRNAs.
Notably, in SS3 vs. SC3, SS12 vs. SC12 and SS24 vs.
SC24, a total of 24, 83 and 83 DE miRNAs were revealed,
respectively, with only a small number of overlapping DE miRNAs.
Between SS3 vs. SC3 and SS12 vs. SC12 only six overlapping DE
miRNAs were identified, while only eight common DE miRNAs were
revealed in SS3 vs. SC3 and SS24 vs. SC24. Furthermore between SS12
vs. SC12 and SS24 vs. SC24 only eight common DE miRNAs were
identified, which was <10% of the total number of DE miRNAs.
These results indicate that the molecular mechanism underlying the
regulation of miRNA expression varies at different treatment time
points. This suggests that the miRNA expression response to
treatment with GSPs is complicated and further investigation is
required.
Acknowledgements
Not applicable.
Funding
The current study was supported by the National
Natural Science Foundation of China (grant nos. 31360409, 31471667
and 31660489), Program for Young and Middle-aged Leading Talents of
China Xinjiang Production and Construction Corps (grant no.
2017CB009) and the Selection and Cultivation Project of ‘Talents of
Xinjiang Production and Construction Corps’ (Grant no.
00608017).
Availability of data and materials
The data sets analyzed during this study are
available from the NCBI public repository under the accession
number GSE107409 and GSE85610.
Authors' contributions
LZ, WW and ZH designed and managed the study. WW,
LZ, HW and ZH drafted and revised the manuscript. YZ, YW, GM and ZH
performed the analysis. DG, YX and MT participated in sample
collection and carried out experiments. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kato M and Slack FJ: microRNAs: Small
molecules with big roles-C. Elegans to human cancer. Biol Cell.
100:71–81. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vasudevan S, Tong Y and Steitz JA:
Switching from repression to activation: MicroRNAs can up-regulate
translation. Science. 318:1931–1934. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li Y, Qiu C, Tu J, Geng B, Yang J, Jiang T
and Cui Q: HMDD v2.0: A database for experimentally supported human
microRNA and disease associations. Nucleic Acids Res.
42:D1070–D1074. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kozomara A and Griffiths-Jones S: miRBase:
Integrating microRNA annotation and deep-sequencing data. Nucleic
Acids Res. 39:D152–D157. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang R and Su B: Small but influential:
The role of microRNAs on gene regulatory network and 3′UTR
evolution. J Genet Genomics. 36:1–6. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tan L, Yu JT, Tan MS, Liu QY, Wang HF,
Zhang W, Jiang T and Tan L: Genome-wide serum microRNA expression
profiling identifies serum biomarkers for Alzheimer's disease. J
Alzheimers Dis. 40:1017–1027. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dangwal S and Thum T: microRNA
therapeutics in cardiovascular disease models. Annu Rev Pharmacol
Toxicol. 54:185–203. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cheng CJ, Bahal R, Babar IA, Pincus Z,
Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM,
et al: MicroRNA silencing for cancer therapy targeted to the tumour
microenvironment. Nature. 518:107–110. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Arola-Arnal A and Blade C:
Proanthocyanidins modulate microRNA expression in human HepG2
cells. PLoS One. 6:e259822011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bansode RR, Khatiwada JR, Losso JN and
Williams LL: Targeting microRNA in cancer using plant-based
proanthocyanidins. Diseases. 4:E212016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chung YC, Huang CC, Chen CH, Chiang HC,
Chen KB, Chen YJ, Liu CL, Chuang LT, Liu M and Hsu CP: Grape-seed
procyanidins inhibit the in vitro growth and invasion of pancreatic
carcinoma cells. Pancreas. 41:447–454. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Prasad R, Vaid M and Katiyar SK: Grape
proanthocyanidin inhibit pancreatic cancer cell growth in vitro and
in vivo through induction of apoptosis and by targeting the
PI3K/Akt pathway. PLoS One. 7:e430642012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Prasad R and Katiyar SK: Grape seed
proanthocyanidins inhibit migration potential of pancreatic cancer
cells by promoting mesenchymal-to-epithelial transition and
targeting NF-κB. Cancer Lett. 334:118–126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang WH, Zhan LL, Guo DQ, Xiang YJ, Zhang
Y, Tian MX and and Han ZJ: Transcriptome analysis of pancreatic
cancer cell response to treatment with grape seed
proanthocyanidins. Oncol Lett. 17:1741–1749. 2019.
|
|
19
|
Langmead B, Trapnell C, Pop M and Salzberg
SL: Ultrafast and memory-efficient alignment of short DNA sequences
to the human genome. Genome Biol. 10:R252009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kozomara A and Griffiths-Jones S: miRBase:
Annotating high confidence microRNAs using deep sequencing data.
Nucleic Acids Res. 42:D68–D73. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Friedlander MR, Chen W, Adamidi C,
Maaskola J, Einspanier R, Knespel S and Rajewsky N: Discovering
microRNAs from deep sequencing data using miRDeep. Nat Biotechnol.
26:407–415. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Krek A, Grun D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee Y: Cancer chemopreventive potential of
procyanidin. Toxicol Res. 33:273–282. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Serrano J, Puupponen-Pimia R, Dauer A,
Aura AM and Saura-Calixto F: Tannins: Current knowledge of food
sources, intake, bioavailability and biological effects. Mol Nutr
Food Res. 53 Suppl 2:S310–S329. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kidd PM: Bioavailability and activity of
phytosome complexes from botanical polyphenols: The silymarin,
curcumin, green tea, and grape seed extracts. Altern Med Rev.
14:226–246. 2009.PubMed/NCBI
|
|
30
|
Bagchi D, Swaroop A, Preuss HG and Bagchi
M: Free radical scavenging, antioxidant and cancer chemoprevention
by grape seed proanthocyanidin: An overview. Mutat Res. 768:69–73.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ma J, Fang B, Ma C, Pang H, Zeng F and Xia
J: Proanthocyanidins inhibit pancreatic cancer AsPC-1 cell growth
and migration through up-regulation of let-7a. Nan Fang Yi Ke Da
Xue Xue Bao. 35:1110–1115. 2015.(In Chinese). PubMed/NCBI
|
|
32
|
Prasad R and Katiyar SK: Down-regulation
of miRNA-106b inhibits growth of melanoma cells by promoting
G1-phase cell cycle arrest and reactivation of p21/WAF1/Cip1
protein. Oncotarget. 5:10636–10649. 2014. View Article : Google Scholar : PubMed/NCBI
|