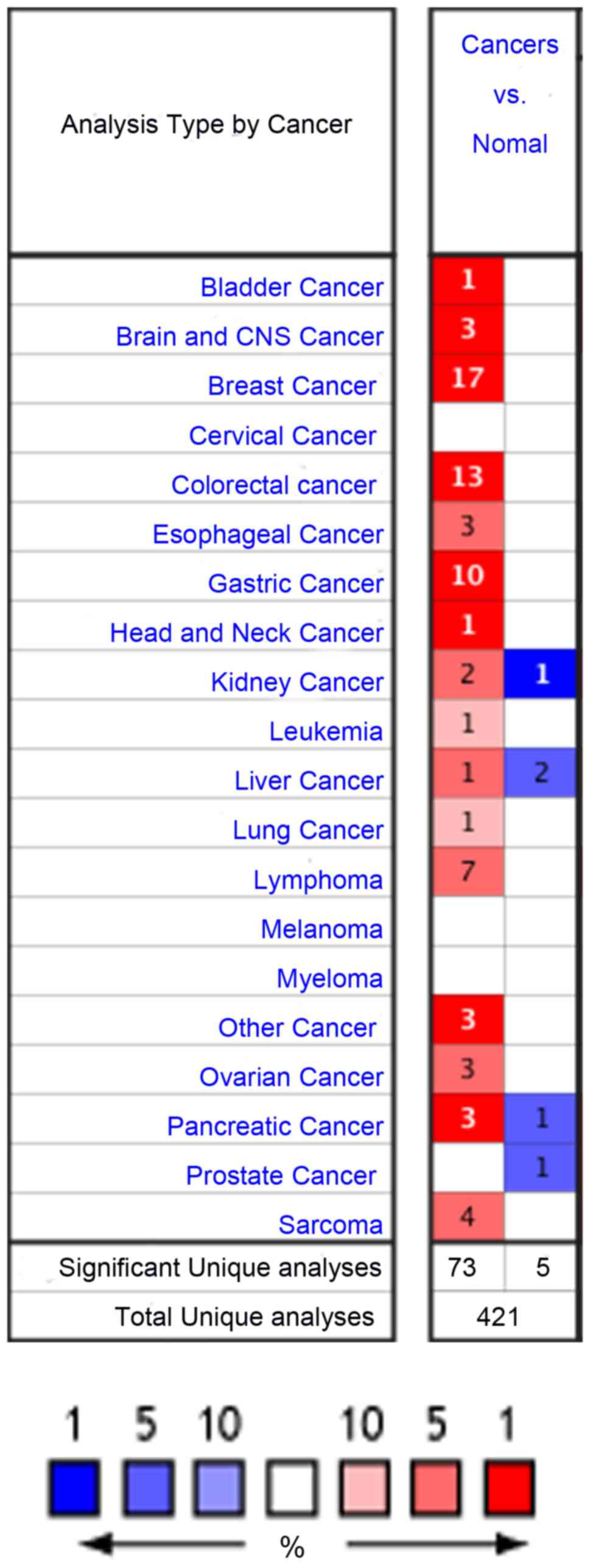
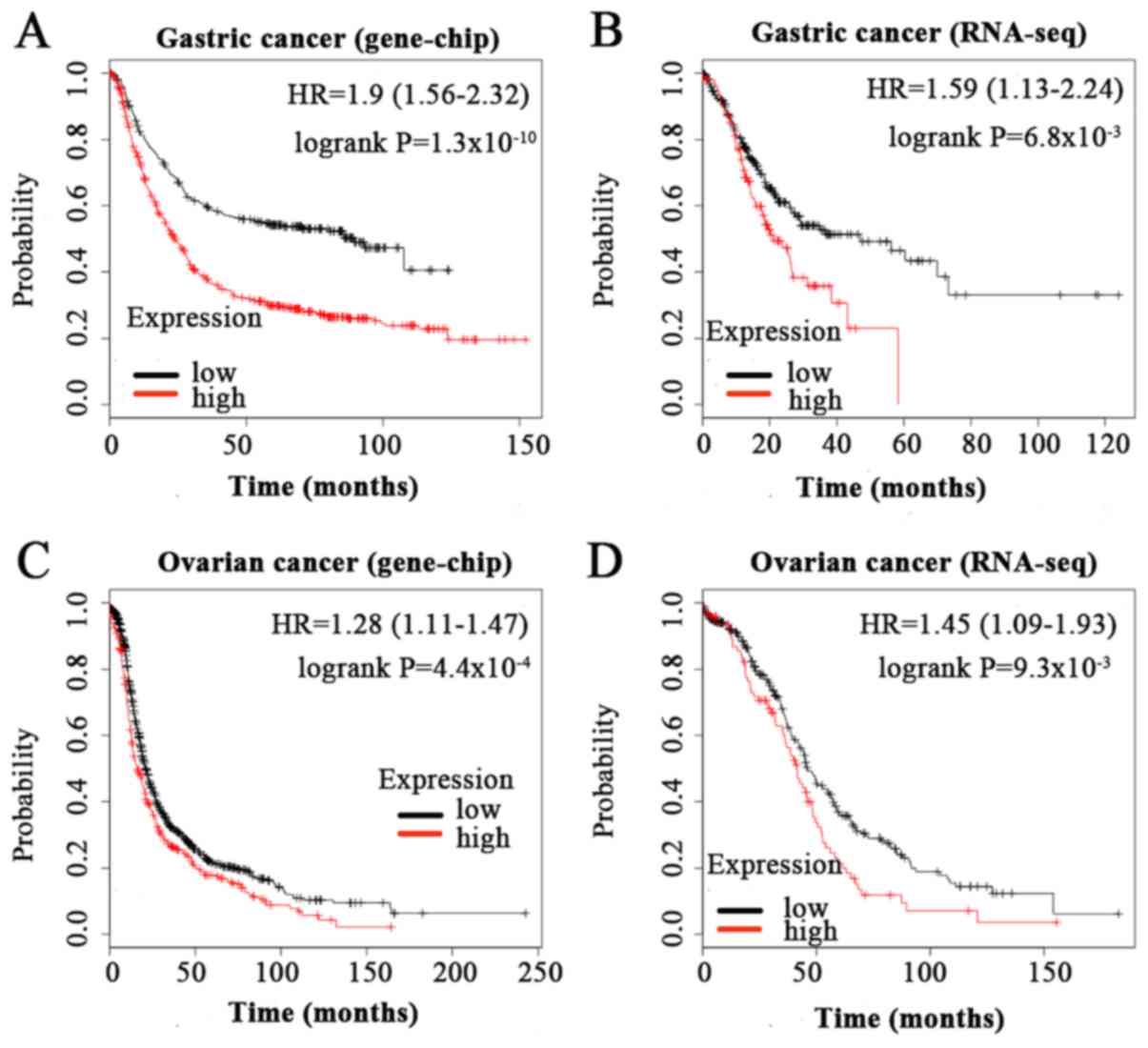
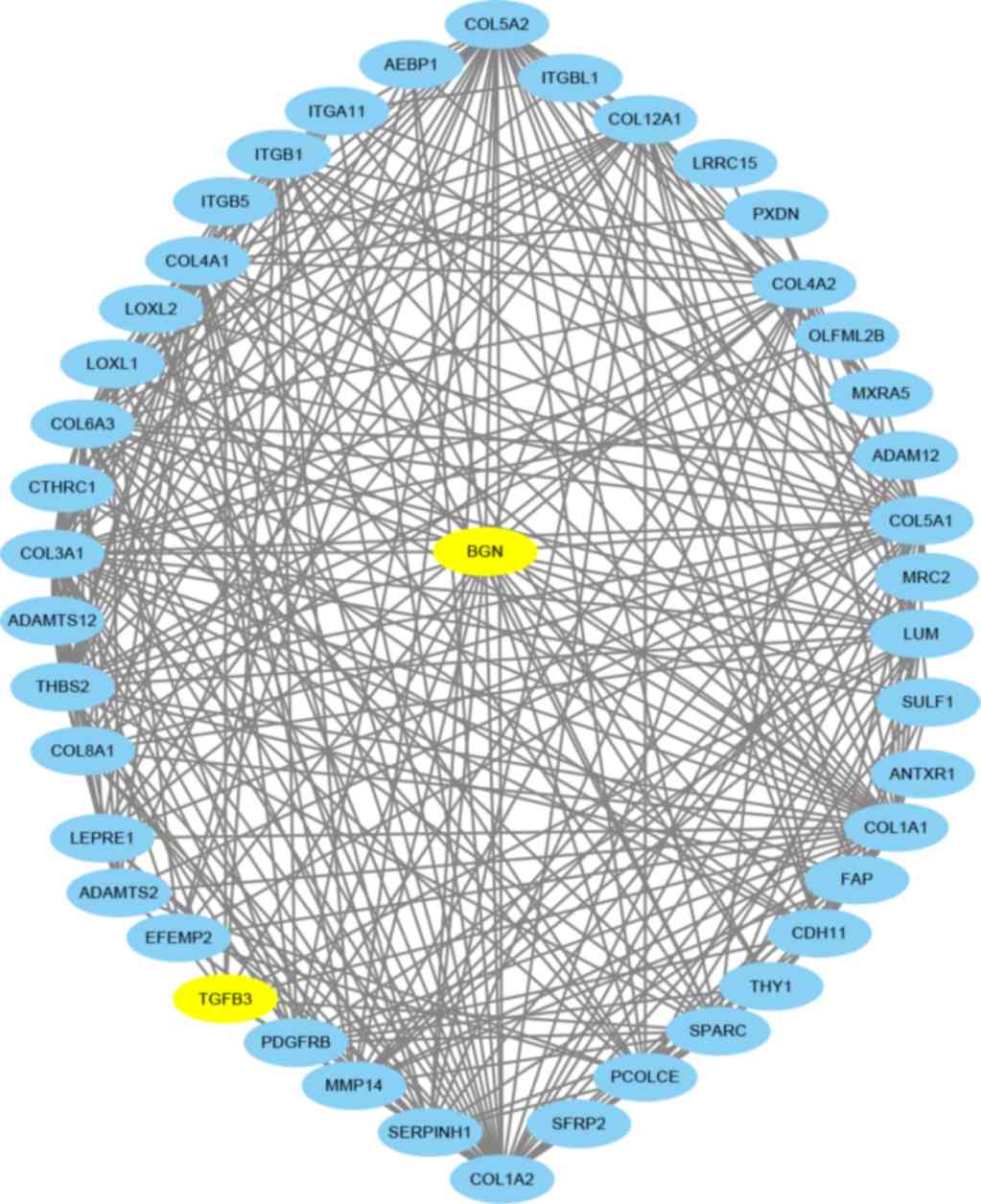
|
1
|
Chaker L, Falla A, van der Lee SJ, Muka T,
Imo D, Jaspers L, Colpani V, Mendis S, Chowdhury R, Bramer WM, et
al: The global impact of non-communicable diseases on
macro-economic productivity: A systematic review. Eur J Epidemiol.
30:357–395. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Patel SA and DeMichele A: Adding adjuvant
systemic treatment after neoadjuvant therapy in breast cancer:
Review of the data. Curr Oncol Rep. 19:562017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Miller KD, Nogueira L, Mariotto AB,
Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL and Siegel
RL: Cancer treatment and survivorship statistics, 2019. CA Cancer J
Clin. 69:363–385. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Geerkens C, Vetter U, Just W, Fedarko NS,
Fisher LW, Young MF, Termine JD, Robey PG, Wöhrle D and Vogel W:
The X-chromosomal human biglycan gene BGN is subject to X
inactivation but is transcribed like an X-Y homologous gene. Hum
Genet. 96:44–52. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
McBride OW, Fisher LW and Young MF:
Localization of PGI (biglycan, BGN) and PGII (decorin, DCN, PG-40)
genes on human chromosomes Xq13-qter and 12q, respectively.
Genomics. 6:219–225. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nastase MV, Young MF and Schaefer L:
Biglycan: A multivalent proteoglycan providing structure and
signals. J Histochem Cytochem. 60:963–975. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fallon JR and McNally EM: Non-Glycanated
biglycan and LTBP4: Leveraging the extracellular matrix for
Duchenne muscular dystrophy therapeutics. Matrix Biol.
68-69:616–627. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kocbek V, Hevir-Kene N, Bersinger NA,
Mueller MD and Rižner TL: Increased levels of biglycan in
endometriomas and peritoneal fluid samples from ovarian
endometriosis patients. Gynecol Endocrinol. 30:520–524. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jacobsen F, Kraft J, Schroeder C,
Hube-Magg C, Kluth M, Lang DS, Simon R, Sauter G, Izbicki JR,
Clauditz TS, et al: Up-regulation of biglycan is associated with
poor prognosis and PTEN deletion in patients with prostate cancer.
Neoplasia. 19:707–715. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Weber CK, Sommer G, Michl P, Fensterer H,
Weimer M, Gansauge F, Leder G, Adler G and Gress TM: Biglycan is
overexpressed in pancreatic cancer and induces G1-arrest in
pancreatic cancer cell lines. Gastroenterology. 121:657–667. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu L, Duan YT, Li JF, Su LP, Yan M, Zhu
ZG, Liu BY and Yang QM: Biglycan enhances gastric cancer invasion
by activating FAK signaling pathway. Oncotarget. 5:1885–1896. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xing X, Gu X, Ma T and Ye H: Biglycan
up-regulated vascular endothelial growth factor (VEGF) expression
and promoted angiogenesis in colon cancer. Tumour Biol.
36:1773–1780. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang B, Li GX, Zhang SG, Wang Q, Wen YG,
Tang HM, Zhou CZ, Xing AY, Fan JW, Yan DW, et al: Biglycan
expression correlates with aggressiveness and poor prognosis of
gastric cancer. Exp Biol Med (Maywood). 236:1247–1253. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Aprile G, Avellini C, Reni M, Mazzer M,
Foltran L, Rossi D, Cereda S, Iaiza E, Fasola G and Piga A:
Biglycan expression and clinical outcome in patients with
pancreatic adenocarcinoma. Tumor Biol. 34:131–137. 2013. View Article : Google Scholar
|
|
17
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Anaya J: OncoLnc: Linking TCGA survival
data to mRNAs, miRNAs, and lncRNAs. PeerJ Computer Sci. 2:e672016.
View Article : Google Scholar
|
|
19
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Györffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Győrffy B, Lánczky A and Szállási Z:
Implementing an online tool for genome-wide validation of
survival-associated biomarkers in ovarian-cancer using microarray
data from 1287 patients. Endocr Relat Cancer. 19:197–208. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Győrffy B, Surowiak P, Budczies J and
Lánczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8:e822412013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Szász AM, Lánczky A, Nagy Á, Förster S,
Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A and Győrffy B:
Cross-validation of survival associated biomarkers in gastric
cancer using transcriptomic data of 1,065 patients. Oncotarget.
7:49322–49333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen X, Cheung ST, So S, Fan ST, Barry C,
Higgins J, Lai KM, Ji J, Dudoit S, Ng IO, et al: Gene expression
patterns in human liver cancers. Mol Biol Cell. 13:1929–1939. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Higgins JP, Shinghal R, Gill H, Reese JH,
Terris M, Cohen RJ, Fero M, Pollack JR, van de Rijn M and Brooks
JD: Gene expression patterns in renal cell carcinoma assessed by
complementary DNA microarray. Am J Pathol. 162:925–932. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Iacobuzio-Donahue CA, Maitra A, Olsen M,
Lowe AW, van Heek NT, Rosty C, Walter K, Sato N, Parker A, Ashfaq
R, et al: Exploration of global gene expression patterns in
pancreatic adenocarcinoma using cDNA microarrays. Am J Pathol.
162:1151–1162. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Buchholz M, Braun M, Heidenblut A, Kestler
HA, Klöppel G, Schmiegel W, Hahn SA, Lüttges J and Gress TM:
Transcriptome analysis of microdissected pancreatic intraepithelial
neoplastic lesions. Oncogene. 24:6626–6636. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Segara D, Biankin AV, Kench JG, Langusch
CC, Dawson AC, Skalicky DA, Gotley DC, Coleman MJ, Sutherland RL
and Henshall SM: Expression of HOXB2, a retinoic acid signaling
target in pancreatic cancer and pancreatic intraepithelial
neoplasia. Clin Cancer Res. 11:3587–3596. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Badea L, Herlea V, Dima SO, Dumitrascu T
and Popescu I: Combined gene expression analysis of whole-tissue
and microdissected pancreatic ductal adenocarcinoma identifies
genes specifically overexpressed in tumor epithelia.
Hepatogastroenterology. 55:2016–2027. 2008.PubMed/NCBI
|
|
31
|
Mas VR, Maluf DG, Archer KJ, Yanek K, Kong
X, Kulik L, Freise CE, Olthoff KM, Ghobrial RM, McIver P and Fisher
R: Genes involved in viral carcinogenesis and tumor initiation in
hepatitis C virus-induced hepatocellular carcinoma. Mol Med.
15:85–94. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yusenko MV, Kuiper RP, Boethe T, Ljungberg
B, van Kessel AG and Kovacs G: High-resolution DNA copy number and
gene expression analyses distinguish chromophobe renal cell
carcinomas and renal oncocytomas. BMC Cancer. 9:1522009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Roessler S, Jia HL, Budhu A, Forgues M, Ye
QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX and Wang XW: A
unique metastasis gene signature enables prediction of tumor
relapse in early-stage hepatocellular carcinoma patients. Cancer
Res. 70:10202–10212. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Franzmann E: Carcinoma. Encyclopedia of
Behavioral Medicine. Gellman MD and Turner JR: Springer New York;
New York, NY: pp. 329–330. 2013, View Article : Google Scholar
|
|
35
|
Blaveri E, Simko JP, Korkola JE, Brewer
JL, Baehner F, Mehta K, Devries S, Koppie T, Pejavar S, Carroll P
and Waldman FM: Bladder cancer outcome and subtype classification
by gene expression. Clin Cancer Res. 11:4044–4055. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bredel M, Bredel C, Juric D, Harsh GR,
Vogel H, Recht LD and Sikic BI: Functional network analysis reveals
extended gliomagenesis pathway maps and three novel MYC-interacting
genes in human gliomas. Cancer Res. 65:8679–8689. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ma XJ, Dahiya S, Richardson E, Erlander M
and Sgroi DC: Gene expression profiling of the tumor
microenvironment during breast cancer progression. Breast Cancer
Res. 11:R72009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Curtis C, Shah SP, Chin SF, Turashvili G,
Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et
al: The genomic and transcriptomic architecture of 2,000 breast
tumours reveals novel subgroups. Nature. 486:346–352. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Karnoub AE, Dash AB, Vo AP, Sullivan A,
Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R and Weinberg
RA: Mesenchymal stem cells within tumour stroma promote breast
cancer metastasis. Nature. 449:557–563. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhao H, Langerod A, Ji Y, Nowels KW,
Nesland JM, Tibshirani R, Bukholm IK, Kåresen R, Botstein D,
Børresen-Dale AL and Jeffrey SS: Different gene expression patterns
in invasive lobular and ductal carcinomas of the breast. Mol Biol
Cell. 15:2523–2536. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cancer Genome Atlas Network: Comprehensive
molecular portraits of human breast tumours. Nature. 490:61–70.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ciriello G, Gatza ML, Beck AH, Wilkerson
MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, et
al: Comprehensive molecular portraits of invasive lobular breast
cancer. Cell. 163:506–519. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kaiser S, Park YK, Franklin JL, Halberg
RB, Yu M, Jessen WJ, Freudenberg J, Chen X, Haigis K, Jegga AG, et
al: Transcriptional recapitulation and subversion of embryonic
colon development by mouse colon tumor models and human colon
cancer. Genome Biol. 8:R1312007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Skrzypczak M, Goryca K, Rubel T, Paziewska
A, Mikula M, Jarosz D, Pachlewski J, Oledzki J and Ostrowski J:
Modeling oncogenic signaling in colon tumors by multidirectional
analyses of microarray data directed for maximization of analytical
reliability. PLoS One. 5(pii): e130912010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Graudens E, Boulanger V, Mollard C,
Mariage-Samson R, Barlet X, Grémy G, Couillault C, Lajémi M,
Piatier-Tonneau D, Zaborski P, et al: Deciphering cellular states
of innate tumor drug responses. Genome Biol. 7:R192006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Cancer Genome Atlas Network, ; Muzny DM,
Bainbridge MN, Chang K, Dinh HH, Drummond JA, Fowler G, Kovar CL,
Lewis LR, Morgan MB, et al: Comprehensive molecular
characterization of human colon and rectal cancer. Nature.
487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gaedcke J, Grade M, Jung K, Camps J, Jo P,
Emons G, Gehoff A, Sax U, Schirmer M, Becker H, et al: Mutated KRAS
results in overexpression of DUSP4, a MAP-kinase phosphatase, and
SMYD3, a histone methyltransferase, in rectal carcinomas. Genes
Chromosomes Cancer. 49:1024–1034. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hong Y, Downey T, Eu KW, Koh PK and Cheah
PY: A ‘metastasis-prone’ signature for early-stage mismatch-repair
proficient sporadic colorectal cancer patients and its implications
for possible therapeutics. Clin Exp Metastasis. 27:83–90. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Su H, Hu N, Yang HH, Wang C, Takikita M,
Wang QH, Giffen C, Clifford R, Hewitt SM, Shou JZ, et al: Global
gene expression profiling and validation in esophageal squamous
cell carcinoma and its association with clinical phenotypes. Clin
Cancer Res. 17:2955–2966. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Hu N, Clifford RJ, Yang HH, Wang C,
Goldstein AM, Ding T, Taylor PR and Lee MP: Genome wide analysis of
DNA copy number neutral loss of heterozygosity (CNNLOH) and its
relation to gene expression in esophageal squamous cell carcinoma.
BMC Genomics. 11:5762010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hao Y, Triadafilopoulos G, Sahbaie P,
Young HS, Omary MB and Lowe AW: Gene expression profiling reveals
stromal genes expressed in common between Barrett's esophagus and
adenocarcinoma. Gastroenterology. 131:925–933. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen X, Leung SY, Yuen ST, Chu KM, Ji J,
Li R, Chan AS, Law S, Troyanskaya OG, Wong J, et al: Variation in
gene expression patterns in human gastric cancers. Mol Biol Cell.
14:3208–3215. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Cho JY, Lim JY, Cheong JH, Park YY, Yoon
SL, Kim SM, Kim SB, Kim H, Hong SW, Park YN, et al: Gene expression
signature-based prognostic risk score in gastric cancer. Clin
Cancer Res. 17:1850–1857. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang Q, Wen YG, Li DP, Xia J, Zhou CZ, Yan
DW, Tang HM and Peng ZH: Upregulated INHBA expression is associated
with poor survival in gastric cancer. Med Oncol. 29:77–83. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
D'Errico M, de Rinaldis E, Blasi MF, Viti
V, Falchetti M, Calcagnile A, Sera F, Saieva C, Ottini L, Palli D,
et al: Genome-wide expression profile of sporadic gastric cancers
with microsatellite instability. Eur J Cancer. 45:461–469. 2009.
View Article : Google Scholar
|
|
57
|
Cui J, Chen Y, Chou WC, Sun L, Chen L, Suo
J, Ni Z, Zhang M, Kong X, Hoffman LL, et al: An integrated
transcriptomic and computational analysis for biomarker
identification in gastric cancer. Nucleic Acids Res. 39:1197–1207.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Frierson HF Jr, El-Naggar AK, Welsh JB,
Sapinoso LM, Su AI, Cheng J, Saku T, Moskaluk CA and Hampton GM:
Large scale molecular analysis identifies genes with altered
expression in salivary adenoid cystic carcinoma. Am J Pathol.
161:1315–1323. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Talbot SG, Estilo C, Maghami E, Sarkaria
IS, Pham DK, O-Charoenrat P, Socci ND, Ngai I, Carlson D, Ghossein
R, et al: Gene expression profiling allows distinction between
primary and metastatic squamous cell carcinomas in the lung. Cancer
Res. 65:3063–3071. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Welsh JB, Zarrinkar PP, Sapinoso LM, Kern
SG, Behling CA, Monk BJ, Lockhart DJ, Burger RA and Hampton GM:
Analysis of gene expression profiles in normal and neoplastic
ovarian tissue samples identifies candidate molecular markers of
epithelial ovarian cancer. Proc Natl Acad Sci USA. 98:1176–1181.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Bonome T, Levine DA, Shih J, Randonovich
M, Pise-Masison CA, Bogomolniy F, Ozbun L, Brady J, Barrett JC,
Boyd J and Birrer MJ: A gene signature predicting for survival in
suboptimally debulked patients with ovarian cancer. Cancer Res.
68:5478–5486. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Cancer Genome Atlas Research Network, .
Integrated genomic analyses of ovarian carcinoma. Nature.
474:609–615. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Riker AI, Enkemann SA, Fodstad O, Liu S,
Ren S, Morris C, Xi Y, Howell P, Metge B, Samant RS, et al: The
gene expression profiles of primary and metastatic melanoma yields
a transition point of tumor progression and metastasis. BMC Med
Genomics. 1:132008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Appunni S, Anand V, Khandelwal M, Gupta N,
Rubens M and Sharma A: Small leucine rich proteoglycans (decorin,
biglycan and lumican) in cancer. Clin Chim Acta. 491:1–7. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Schaefer L, Babelova A, Kiss E, Hausser
HJ, Baliova M, Krzyzankova M, Marsche G, Young MF, Mihalik D, Götte
M, et al: The matrix component biglycan is proinflammatory and
signals through Toll-like receptors 4 and 2 in macrophages. J Clin
Invest. 115:2223–2233. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Hildebrand A, Romaris M, Rasmussen LM,
Heinegård D, Twardzik DR, Border WA and Ruoslahti E: Interaction of
the small interstitial proteoglycans biglycan, decorin and
fibromodulin with transforming growth factor beta. Biochem J.
302:527–534. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Bischof AG, Yüksel D, Mammoto T, Mammoto
A, Krause S and Ingber DE: Breast cancer normalization induced by
embryonic mesenchyme is mediated by extracellular matrix biglycan.
Integr Biol (Camb). 5:1045–1056. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Liu B, Xu T, Xu X, Cui Y and Xing X:
Biglycan promotes the chemotherapy resistance of colon cancer by
activating NF-kappaB signal transduction. Mol Cell Biochem.
449:285–294. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Yang Z, Wen Y, Qian L and Hospital Z:
Up-regulation of biglycan is associated with malignant phenotype of
nonsmall cell lung cancer. J Med Res. 45:41–46. 2016.
|
|
72
|
Ferlay J, Colombet M, Soerjomataram I,
Mathers C, Parkin DM, Piñeros M, Znaor A and Bray F: Estimating the
global cancer incidence and mortality in 2018: GLOBOCAN sources and
methods. Int J Cancer. 144:1941–1953. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Appunni S, Anand V, Khandelwal M, Seth A,
Mathur S and Sharma A: Altered expression of small leucine-rich
proteoglycans (Decorin, Biglycan and Lumican): Plausible diagnostic
marker in urothelial carcinoma of bladder. Tumour Biol.
39:10104283176991122017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Liu JY, Jiang L, Liu JJ, He T, Cui YH,
Qian F and Yu PW: AEBP1 promotes epithelial-mesenchymal transition
of gastric cancer cells by activating the NF-κB pathway and
predicts poor outcome of the patients. Sci Rep. 8:119552018.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Kasurinen A, Gramolelli S, Hagström J,
Laitinen A, Kokkola A, Miki Y, Lehti K, Yashiro M, Ojala PM,
Böckelman C and Haglund C: High tissue MMP14 expression predicts
worse survival in gastric cancer, particularly with a low PROX1.
Cancer Med. 8:6995–7005. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Liu J, Liu Z, Zhang X, Gong T and Yao D:
Bioinformatic exploration of OLFML2B overexpression in gastric
cancer base on multiple analyzing tools. BMC Cancer. 19:2272019.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Wang G, Shi B, Fu Y, Zhao S, Qu K, Guo Q,
Li K and She J: Hypomethylated gene NRP1 is co-expressed with
PDGFRB and associated with poor overall survival in gastric cancer
patients. Biomed Pharmacother. 111:1334–1341. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Liao P, Li W, Liu R, Teer JK, Xu B, Zhang
W, Li X, Mcleod HL and He Y: Genome-scale analysis identifies
SERPINE1 and SPARC as diagnostic and prognostic biomarkers in
gastric cancer. Onco Targets Ther. 11:6969–6980. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wang H, Duan XL, Qi XL, Meng L, Xu YS, Wu
T and Dai PG: Concurrent Hypermethylation of SFRP2 and DKK2
Activates the Wnt/β-catenin pathway and is associated with poor
prognosis in patients with gastric cancer. Mol Cells. 40:45–53.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Ao R, Guan L, Wang Y and Wang JN:
Silencing of COL1A2, COL6A3, and THBS2 inhibits gastric cancer cell
proliferation, migration, and invasion while promoting apoptosis
through the PI3k-Akt signaling pathway. J Cell Biochem.
119:4420–4434. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhuo C, Li X, Zhuang H, Tian S, Cui H,
Jiang R, Liu C, Tao R and Lin X: Elevated THBS2, COL1A2, and SPP1
expression levels as predictors of gastric cancer prognosis. Cell
Physiol Biochem. 40:1316–1324. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Hao S, Lv J, Yang Q, Wang A, Li Z, Guo Y
and Zhang G: Identification of key genes and circular RNAs in human
gastric cancer. Med Sci Monit. 25:2488–2504. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Suzuki S, Dobashi Y, Hatakeyama Y, Tajiri
R, Fujimura T, Heldin CH and Ooi A: Clinicopathological
significance of platelet-derived growth factor (PDGF)-B and
vascular endothelial growth factor-A expression, PDGF receptor-β
phosphorylation, and microvessel density in gastric cancer. BMC
Cancer. 10:6592010. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Mazzoccoli G, Pazienza V, Panza A, Valvano
MR, Benegiamo G, Vinciguerra M, Andriulli A and Piepoli A: ARNTL2
and SERPINE1: Potential biomarkers for tumor aggressiveness in
colorectal cancer. J Cancer Res Clin Oncol. 138:501–511. 2012.
View Article : Google Scholar : PubMed/NCBI
|