Introduction
Esophageal squamous cell carcinoma (ESCC) is a
common malignancy of the digestive system. In 2018, the incidence
of esophageal cancer ranked seventh among malignancies, and there
was a total of 572,034 new cases (1). The epidemiology of ESCC is not only
associated with country, dietary habit, ethnicity and sex, but its
incidence is significantly different within different regions
(2,3). According to the global cancer data
released by the American Cancer Society in 2012, East Asia had the
highest mortality rate of esophageal cancer, and its incidence and
mortality rates ranked fifth (age-standardized incidence rate,
7.74%) and fourth (age-standardized mortality rate, 9.29%),
respectively among malignancies in China (4). Esophageal cancer is predominantly
divided into squamous cell carcinoma and adenocarcinoma, the latter
of which is more common in western countries. However, squamous
cell carcinoma dominates in China, accounting for ~90% of all
reported cases of esophageal cancer (5), and Northern Jiangsu possesses one of
the highest incidences of esophageal cancer in China and in 2006,
the incidences rate of esophageal cancer was 86.45/100,000 in
Chuzhou (Jiangsu, China) (2,6). According to the cancer data report
registered in Huai'an City, esophageal cancer accounts for 32.02%
of all malignancies, and the incidence and mortality of ESCC in the
Huai'an area have been among the highest of all malignancies in the
last 10 years (7). Despite
advancements in the treatment of esophageal cancer, the prognosis
of patients with esophageal cancer remains poor, whereby the 5-year
survival rate is <20% (8).
However, in early stage esophageal cancer, the 5-year survival rate
can be as high as 80–90% (9), and
40% of esophageal cancer is prone to recurrence (10). Due to the concealment and
non-specificity of the early symptoms of esophageal cancer, the
majority of patients are diagnosed and treated in the middle or
advanced stages of disease (11).
ESCC and adenocarcinoma exhibit different pathogeneses, biological
tumor characteristics and prognoses (12). Therefore, adopting the foreign
standard may not be suitable for patients with esophageal cancer in
China, as it would do more harm than good. Further studies are
required in order to develop the diagnosis and treatment plans
according to the characteristics of ESCC in China, in order to
improve diagnosis of early stage esophageal cancer (13).
The continuous development of molecular biology and
an improved understanding of its underlying mechanisms has
subsequently allowed for advancements to the development of tumor
markers for the assessment of tumor treatment efficacy, recurrence
monitoring and prognosis. Song et al (14) demonstrated that eight genes were
associated with ESCC, including six well-known oncology-associated
genes: p53, RB1, CDKN2, PIK3CA, NOTCH1 and NF2L2. Furthermore, to
the best of our knowledge, A Disintegrin And Metalloprotease Domain
29 (ADAM29) and Family With Sequence Similarity 135-member B
(FAM135B) were demonstrated to be associated with ESCC for the
first time.
A member of the ADAMs family, ADAM29 is a type-I
transmembrane protein located in human chromosome 4q34.1 (15), which plays an important role in
regulating cell-to-cell or cell-matrix interactions (16). ADAMs are widely involved in a number
of physiological processes, including sperm-egg fusion, nervous
system development and inflammatory response (17). Furthermore, ADAMs play an important
role in the invasion and metastasis of malignancies, Victor et
al (18) demonstrated that ADAM9
participates in the vascular metastasis process of pancreatic
ductal adenocarcinoma. Besides, ADAM12 promotes the metastasis of
esophageal cancer by influencing the interaction between tumor
cells and the extracellular matrix (19) and ADAM17 participates in Notch and
Wnt signaling pathways to promotes metastasis in gastric cancer
(20). ADAMs are also known as
metalloproteinase depolymerization (21). Zhang et al (22) indicated that ADAM29 was associated
with breast cancer subtypes, and it was also detected in non-small
cell lung cancer (23). In a study
on gastric cancer, it was revealed that ADAM29 influenced gastric
carcinoma proliferation, migration, invasion and motility via
upregulation of the MKN45-C1 and -C2 clones, and the MKN45-SC
control (24). Brim et al
(25) determined that ADAM29 is
associated with colorectal tumors, and it has also been revealed
that the ADAM29 mutation affects the adhesion of melanoma cells to
specific extracellular matrix proteins and promotes tumor invasion
(26). Conversely, the FAM135B gene
is located in human chromosome 8q24.23 (27). A number of studies have demonstrated
that the FAM135B gene is associated with the occurrence and
development of tumor, and can promote the proliferation, migration
and invasion of tumor cells (11,14,28).
To the best of our knowledge, the expression of
ADAM29 and FAM135B proteins in the precancerous stage of esophageal
cancer has not yet been reported. Immunohistochemistry (IHC) was
performed in the present study in order to investigate the
differences and clinical significance of the expression of ADAM29
and FAM135B in the pathological evolution from normal esophageal
epithelium to esophageal cancer.
Materials and methods
Human esophagus specimens
A total of 120 esophageal paraffin mass specimens
were collected from 80 patients with ESCC at the Department of
Pathology of the 82nd Hospital of the People's Liberation Army
(Huai'an, China) between January 2013 to January 2015. There were
40 cases of esophageal intraepithelial neoplasia following biopsy
(20 cases of high-grade esophageal intraepithelial neoplasia and 20
cases of low-grade esophageal intraepithelial neoplasia) and 40
cases of post-surgical ESCC, of which 21 cases were >60 years
old and 19 cases ≤60 years old. A total of 40 cases of normal
mucosal tissue of the corresponding esophageal carcinoma (≥5 cm
from the tumor border) were treated as the control group for ESCC.
The pathological classification were as follows: A total of 19
cases in the G1 stage (29–30), 19 cases in the G2 stage and two cases
in the G3 stage. The clinical stages were as follows: A total of
eight cases in stage I (29–30), 22 cases in stage II: 10 cases in
stage III. Lymph node metastasis was reported in 10 cases and
absent in 30 cases. The tumor size were as follows: >5 cm in two
patients and ≤5 cm in 38 patients. There was an equal sex
distribution among the pathological grades of esophageal cancer.
The American Joint Committee on Cancer (AJCC) 2010 (31) was adopted in order to develop the
standard manual for tumor staging. The inclusion criteria were as
follows: i) Patient had no prior history of cancer; ii) samples
submitted for examination and materials must be sufficient; iii)
radiotherapy and/or chemotherapy had not been performed; iv)
inflammatory diseases were not combined; and v) specimens were
confirmed repeatedly by pathology (confirmed by two or more
specialist pathologists). The present study was approved by the
People's Liberation Army 82nd Hospital Ethics Committee. All
patients, or their families, provided written informed consent
prior to the study start.
Reagents
Rabbit anti-human ADAM29 polyclonal antibody (1:50;
cat. no. ab198875) was purchased from Abcam. Rabbit anti-human
FAM135B polyclonal antibody (1:50; cat. no. 51059) was purchased
from Sigma-Aldrich (Merck KGaA). The secondary antibody kit
[maxvison™2 HRP (Mouse/Rabbit) IHC kit; cat. no. KIT5920;
Ready-to-use antibody] and DAB (cat. no. DAB-2031/2032) dye were
purchased from Fuzhou Maixin Biotech Co, Ltd. The secondary
antibody kit was used to amplify the signal of the primary
antibody, and the dye was used to locate, characterize and quantify
intracellular antigens.
Immunohistochemistry
All postoperative or biopsy specimens were fixed for
24 h with 10% phosphate buffer formalin fixation solution and
embedded in paraffin at 20°C. Paraffin-embedded tissues were cut
into sections (3 µm-thick) for use in routine pathological
examination and immunohistochemical examination. All
immunohistochemical methods were performed using the Elivision
two-step method (32). Briefly, the
following procedures were performed: Specimens were baked at 70°C
for 1 h, before being dewaxed in xylene for 20 min and rehydrated
with graded concentrations of alcohol for 17 min. Slices were
placed in solution of citrate (pH 6.0) and heated in a pressure
cooker (1.03 kpa) for 3 min, 3% H2O2 was
added and incubated at room temperature for 15 min to block
endogenous peroxidase. Sections were then incubated for 60 min at
37°C with primary antibodies (Rabbit anti-human ADAM29 polyclonal
antibody; 1:50; cat. no. ab198875; Abcam) and phosphate buffer was
used as a negative control. Second antibody [maxvisonTM2 HRP
(Mouse/Rabbit) IHC kit; Ready-to-use antibody; cat. no. KIT5920;
Sigma-Aldrich; Merck KGaA] was added and incubated at room
temperature for 20 min. Slices were stained using DAB at room
temperature for 5 min, redyed with hematoxylin at room temperature
for 20 sec and differentiated with 1% hydrochloric acid alcohol for
1 sec at room temperature.
Result judgement
The degree of section coloring was observed using a
high-power optical light microscope (magnification, ×400). A
double-blind method was implemented in order to observe the
esophageal tissue staining. A total of 10 high-power visual fields
on each slice were randomly-selected and counted, and at least 100
cells were observed in each field. The positive expression of
ADAM29 protein was predominantly concentrated in the cell membrane
or cytoplasm, while the positive expression of FAM135B protein was
predominantly concentrated in the nucleus. Esophageal tissue
appeared brown as the positive expression marker of ADAM29 and
FAM135B proteins in the glass slice. The comprehensive score was
obtained, according to the staining intensity of the esophageal
tissue and the proportion of positive cells. The staining intensity
score were as follows: Zero indicated no coloring, 1 indicated
light yellow, 2 indicated yellow-brown and 3 indicated brown.
Stained tissues were counted in five randomly-selected fields using
a high-power optical light microscope (magnification, ×400), and at
least 100 cells were observed in each field. The percentage score
of positive cell slices were as follows: 0, <5% positive cells;
1, 5–25%, 2, 26–50%; 3, 51–75%; and 4, >75%. The product of two
scores were as follows: 0–1 indicated a negative score (−), 2–5
indicated a positive score (+), 6–8 indicated a positive score
(++)/(2+) and ≥9 indicated a strong positive score (+++)/(3+);
low-positive expression group (≥2 and <9) and high-positive
expression group (≥9).
Statistical analysis
All data were analyzed using SPSS software (version
22.0; IBM Corp.). Categorical variables were presented as
frequencies and percentages. Categorical variables were compared
with χ2 tests or Fisher exact tests (Tables I and II). Relevance between categorical
variables were performed using χ2 tests (Table III). P<0.05 was considered to
indicate a statistically significant difference.
 | Table I.Expression of ADAM29 and FAM135B in
different esophageal lesions. |
Table I.
Expression of ADAM29 and FAM135B in
different esophageal lesions.
| Tissue type | Cases, n | Positive rate of
ADAM29, n (%) | Positive rate of
FAM135B, n (%) |
|---|
| Normal squamous
epithelium | 40 | 7 (17.5) | 10 (25.0) |
| Low-grade
intraepithelial neoplasia | 20 | 12 (60.0) | 11 (55.0) |
| High-grade
intraepithelial neoplasia | 20 | 18 (90.0) | 18 (90.0) |
| Esophageal
cancer | 40 | 40 (100.0) | 40 (100.0) |
| χ2 |
| 66.179 | 56.647 |
| P-value |
| <0.001 | <0.001 |
 | Table II.Baseline characteristics of study
populations. |
Table II.
Baseline characteristics of study
populations.
| Characteristic | Cases, n | ADAM29 (score, 3+),
n (%) | D2 | P-value | FAM135B (score,
3+), n (%) | A2 | P-value |
|---|
| Age |
|
|
|
|
|
| 0.115 |
| ≤60
years | 21 | 7 (33.3) | 0.234 | 0.629 | 8 (38.1) | 2.489 |
|
| >60
years | 19 | 5 (26.3) |
|
| 3 (15.8) |
|
|
| Pathological
grade |
|
|
|
|
|
| 0.006 |
| G1 | 19 | 2 (10.5) | 6.903 | 0.024 | 1 (5.3) | 9.636 |
|
| G2 | 19 | 9 (47.4) |
|
| 9 (47.4) |
|
|
| G3 | 2 | 1 (50.0) |
|
| 1 (50) |
|
|
| Clinical stage |
|
|
|
|
|
| 0.015 |
| I | 8 | 0 (0.0) | 7.414 | 0.016 | 0 (0.0) | 7.836 |
|
| II | 22 | 6 (27.3) |
|
| 5 (22.7) |
|
|
|
III | 10 | 6 (60.0) |
|
| 6 (60.0) |
|
|
| Lymph node
metastasis |
|
|
|
|
|
| 0.002 |
| No | 30 | 5 (16.7) | 10.159 | 0.003 | 4 (13.8) | 12.079 |
|
|
Yes | 10 | 7 (70.0) |
|
| 7 (63.6) |
|
|
| Tumor size |
|
|
|
|
|
| 0.515 |
| ≤5
cm | 38 | 11 (22.4) | 0.401 | 0.515 | 11 (22.4) | 0.401 |
|
| >5
cm | 2 | 1 (50.0) |
|
| 1 (50.0) |
|
|
 | Table III.Association between ADAM29 and
FAM135B proteins. |
Table III.
Association between ADAM29 and
FAM135B proteins.
|
| FAM135B |
|---|
|
|
|
|---|
| ADAM29 | Positive | Negative |
|---|
| Positive | 70.0 | 7.0 |
| Negative | 9.0 | 34.0 |
| χ2 | 60.071 |
| P-value | <0.001 |
Results
Expression of ADAM29 protein in
different stages of esophageal lesions
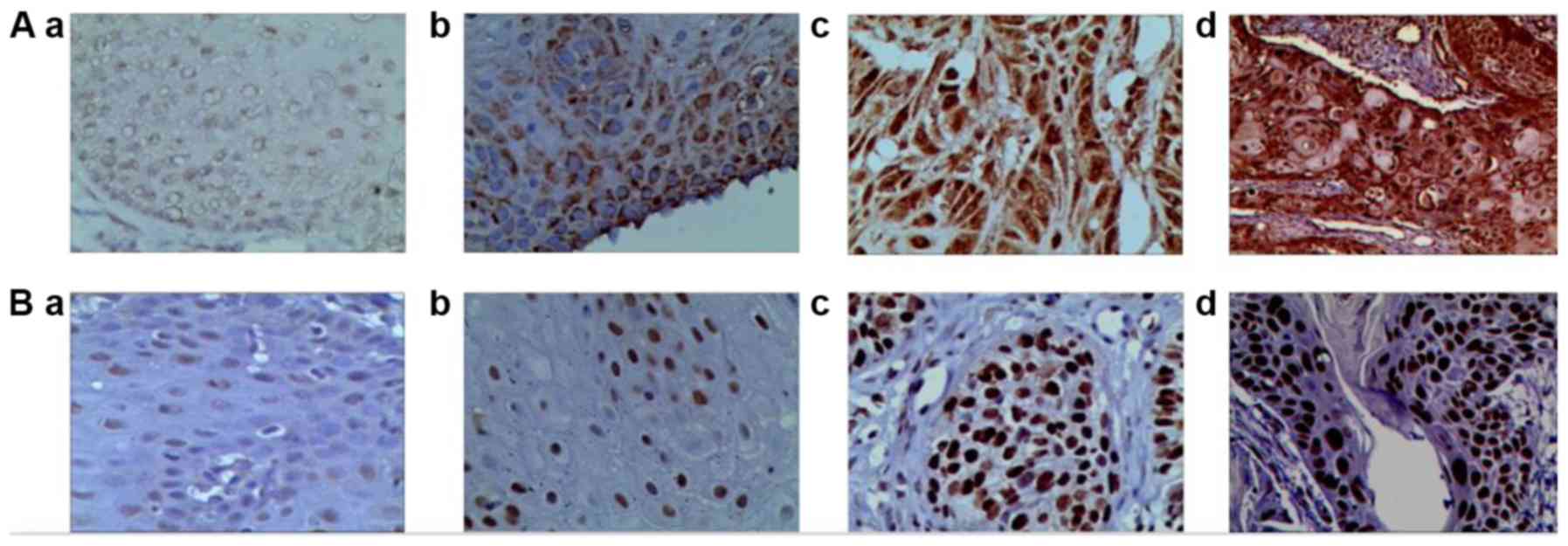
Overall, ADAM29 protein was negatively expressed in
normal esophageal epithelial cells; however, weak positive
expression was occasionally demonstrated in the cytoplasm of basal
cells. Furthermore, the staining was relatively shallow in normal
esophageal epithelial cells, but the majority of ESCC cells
appeared brown. ADAM29 expression in esophageal lesions was
concentrated in the cytoplasm of squamous cell carcinoma (Fig. 1). The expression of ADAM29 protein
gradually enhanced from normal esophageal mucosal epithelium and
intraepithelial esophageal neoplasia, to ESCC. Furthermore, ADAM29
staining gradually deepened and the staining area gradually
increased in these groups (Fig. 1).
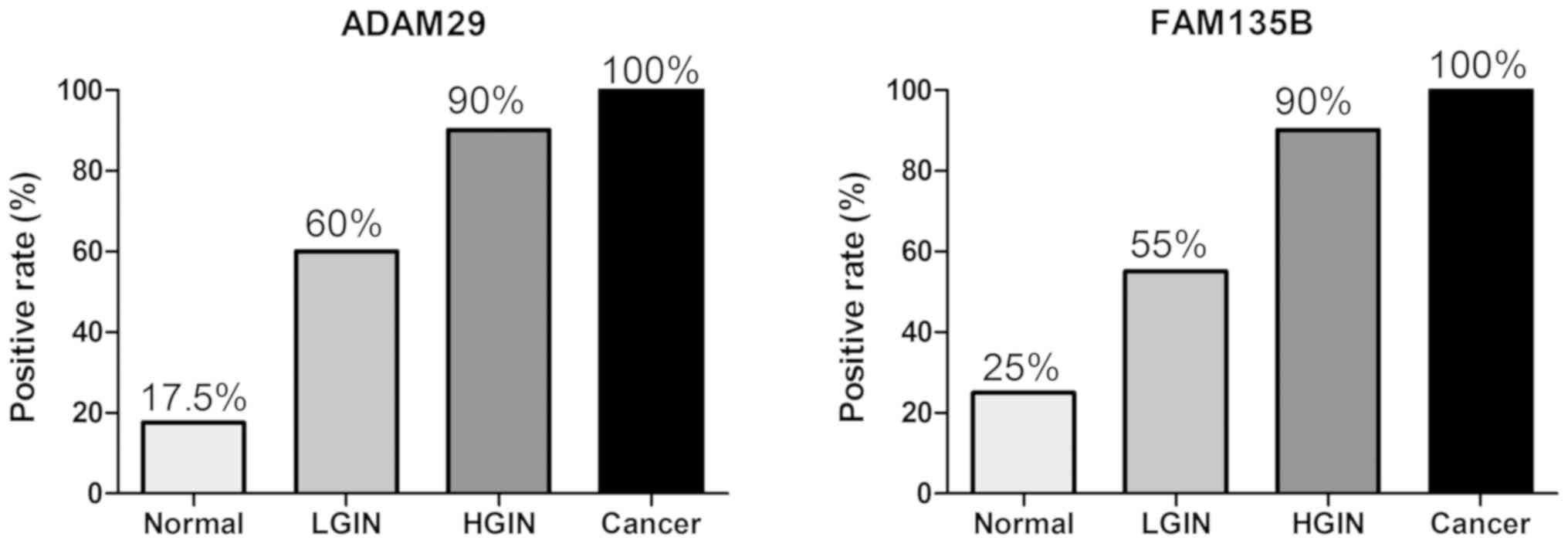
The positive expression rate of ADAM29 was 17.5% in normal
esophageal tissue, 60.0% in low-grade intraepithelial neoplasia
tissue, 90.0% in high-grade intraepithelial neoplasia tissue and
100.0% in ESCC tissue (Fig. 2). The
differences in distribution among the four groups were
statistically significant (Table I;
P<0.05).
Expression of FAM135B protein in
different stages of esophageal lesions
FAM135B protein was expressed in certain normal
epithelial basal cells of the esophagus, and appeared light yellow.
The majority of tumor cells in the esophageal cancer tissues
appeared brown, and the positive expression was concentrated in the
nucleus of squamous cell carcinoma (Fig.
1). The expression of FAM135B protein gradually increased from
the normal esophageal mucosal epithelium and intraepithelial
esophageal neoplasia, to ESCC. Furthermore, FAM135B staining
gradually deepened and the staining area gradually increased in
these groups (Fig. 1). The positive
expression rate of FAM135B was 25% in normal esophageal tissue, 55%
in low-grade intraepithelial neoplasia tissue, 90% in high-grade
intraepithelial neoplasia tissue and 100% in ESCC (Fig. 2). The differences in distribution
among the four groups were statistically significant (Table I; P<0.05).
Association between the expression of
ADAM29 and FAM135B proteins and the severity of esophageal
epithelial cell lesions
The positive rates of ADAM29 and FAM135B proteins
gradually increased from normal esophageal mucosal epithelium and
intraepithelial esophageal neoplasia, to ESCC (Table I; P<0.05).
Association between the expression of
ADAM29 and FAM135B proteins and the clinic-pathological features of
ESCC
Patients with esophageal cancer were divided into
the high expression group (≥9) and the low expression group (≥2 and
<9), according to the product of two scores of percentage
(positive cell slices and staining intensity) as presented in
Table II, the cutoff value was a
score of 9.
Association between the expression of
ADAM29 protein and the clinicopathological features of ESCC
As presented in Table
II, increased ADAM29 expression was significantly associated
with the lymph node metastasis features of ESCC (P=0.003).
Furthermore, high ADAM29 expression was significantly associated
with the pathological stage (P=0.024) and clinical stage (P=0.016).
The case of ADAM29 expression in each pathological stage, two cases
(10.5%) were observed in G1, nine cases (47.4%) in G2 and one case
(50%) in G3. High ADAM29 expression occurred in zero cases (0%) in
clinical stage I, six cases (27.3%) in clinical stage II: six cases
(60%) in clinical stage III. High expression of ADAM29 was not
associated with patient age (P=0.629) and tumor size (P=0.515).
Association between the expression of
FAM135B protein and the clinicopathological features of ESCC
Increased FAM135B expression was significantly
associated with the lymph node metastasis features of ESCC
(P=0.002). Furthermore, high FAM135B expression was significantly
associated with the pathological stage (P=0.006) and clinical stage
(P=0.015). The case of FAM135B expression in each pathological
stage, one case (5.30%) was observed in G1, nine cases (47.4%) in
G2 and one case (50%) in G3. High FAM135B expression occurred in
zero cases (0%) in clinical stage I, five cases (22.7%) in clinical
stage II: six cases (60%) in clinical stage III. High expression of
FAM135B was not associated with patient age (P=0.115) or tumor size
(P=0.515).
Association between ADAM29 and FAM135B
protein expressions
The association between ADAM29 and FAM135B protein
expressions was assessed using χ2 tests which
demonstrated a statistically significant correlation between the
two groups (Table III;
χ2=60.071; P<0.001).
Discussion
The ADAM29 gene is a novel biomarker for esophageal
cancer, and is a member of the ADAMs family of single-pass
transmembrane and secreted metalloendopeptidases. Different members
of the ADAMs family have been detected from cells, transgenic
animals and other experimental systems in previous studies
(14). Currently, >40 species of
ADAMs family have been discovered. Their functional regions include
the polypeptide domain, metalloproteinase domain,
protein-disintegrating domain and cysteine-rich domain, whereby
each domain has its associated functions (33). The ADAMs family plays an important
role in the occurrence, invasion and metastasis of malignancies,
also known as metalloproteinase depolymerization (21,34).
They participate in cell signal transduction, adhesion and
degradation of extracellular matrix (involving membrane fusion,
shedding of cytokines and growth factors), the control of cell
migration and regulation of other biological processes (21). ADAM29 has the ability to change the
surroundings of processes such as cell proliferation, angiogenesis,
apoptosis and invasion, which are strongly influenced by the
surrounding microenvironment of the tumor, thus contributing to
tumor growth and dissemination (35). Furthermore, Noël et al
(35) demonstrated that the ADAM29
gene is involved in the regulation of cell proliferation,
angiogenesis, apoptosis, cell invasion and other microenvironments.
The ADAM29 gene has been reported to be highly expressed in
esophageal cancer compared with the corresponding normal tissue
(14), and overexpressed in breast
cancer (22), lung cancer (23), gastric cancer (24), colorectal cancer (25) and melanoma (26). Mutations of the ADAM29 gene
predominantly occur in somatic cells, in the prolysin and
propeptide domains, and are associated with increased collagen
adhesion of types I and IV compared with wild-type ADAM29 (26). Somatic aberrations are predominantly
involved in the Wnt, cell cycle and Notch signaling pathways
(14). Brim et al
demonstrated that ADAM29 increased the migration ability of cancer
cells and caused chromosomes aberrations, resulting in colorectal
tumor (25). ADAM29 was also
demonstrated to be differently expressed, predominantly in the
cytoplasmic binding domain (15).
Through analysis of the staining degree and range of
ADAM29 in normal esophageal tissues and esophageal lesions, the
present study demonstrated that the expression level of ADAM29 is
positively associated with the severity of esophageal epithelial
cell lesions. This suggests that ADAM29 may be involved in the
evolution process of malignant transformation of esophageal
lesions, and a high increment state of ADAM29 exists in each phase.
The present study further investigated the association between
ADAM29 protein and the clinical characteristics of patients with
esophageal cancer. The expression level of ADAM29 protein of ESCC
patients with lymph node metastasis was demonstrated to be higher
than that of ESCC patients without lymph node metastasis. The
ADAM29 gene is considered to be associated with tumor metastasis
via its regulatory role in the malignant behavior of ESCC.
Furthermore, the results of the present study demonstrated that
ADAM29 protein is increasingly expressed in late clinical stage and
high pathological grade, in patients with ESCC. The positive
expression levels of ADAM29 protein may be associated with
malignant transformation of ESCC, which can predict the likelihood
of transformation from precancerous lesions to esophageal
carcinoma. The results of the present study further indicate that
ADAM29 protein expression is closely associated with the occurrence
and development of esophageal lesions, which corresponds to the
results of previous studies.
The ADAMs family plays an important role in the ErbB
(epidermal growth factor) signaling pathway by hydrolyzing a
variety of Erb ligand types. The ADAMs family can activate the Erb
ligand and cause the shedding of extracellular functional regions
(36,37). This particular role has become an
important target for the development of new drugs in the Erb
signaling pathway. Therefore, the targeted therapy strategy for
ADAMs will become an important breakthrough in anti-ErbB signaling.
Currently, there are no ADAMs inhibitor used in clinical practice,
however; studies on inhibiting ADAM-mediated reactions have entered
clinical trials. For example, ADAM10 and ADAM17 participate in the
Notch signaling pathway (21,37).
Maloum et al (38)
demonstrated that ADAM29 gene expression could be detected in order
to predict the effect of oral administration of fludarabine and
cyclophosphamide on patients with chronic lymphocytic leukemia, and
that ADAM29 could guide individualized treatment.
The FAM135B gene is located in chromosome 8 and has
been reported to be differentially expressed in healthy people with
tuberculosis (27), mental illness
(39) and autism (40). To the best of our knowledge, Song
et al (14) first reported
that the FAM135B gene is a tumor-associated gene for esophageal
cancer, by whole-genome sequencing and other technologies.
Subsequently, a number of studies have indicated that increased
FAM135B expression can enhance the malignant phenotype of ESCC,
reflected by the promotion of cell proliferation, metastasis and
invasion. Furthermore, high FAM135B expression may be associated
with endogenous phosphoglyceraldehyde dehydrogenase (28,41).
This suggests that FAM135B is closely associated with the
occurrence and development of ESCC. Dong et al (42) demonstrated the function and mechanism
behind FAM135B in esophageal cancer, FAMl35B promotes inflammation
by inducing granulin (GRN; a protein-coding gene located on
chromosome 17, q21.32.) precursor secretion and activating the
PI3K/AKT/mTOR signaling pathway; moreover, FAMl35B and GRN form
positive feedback regulation to enhance their respective
cancer-promoting functions. The present study demonstrated that
there was positive expression of FAM135B in various processes of
esophageal precancerous lesions, and occasional or low expression
in normal esophageal tissue. Furthermore, the expression of FAM135B
was positively associated with the degree of malignant lesions. The
results of the present study suggest that FAM135B may be involved
in the evolution of malignant transformation of esophageal lesions.
Furthermore, the present study assessed the clinical
characteristics of patients with esophageal cancer and demonstrated
that the expression of FAM135B was positively associated with
clinical stage and pathological grade. This experiment demonstrated
that high expression of FAM135B could enhance the malignant degree
of esophageal lesions. The present study demonstrated that the
positive expression of FAM135B in ESCC patients with lymph node
metastasis was higher than that of patients without lymph node
metastasis. This suggests that the FAM135B gene may be involved in
the invasion and migration of different types of esophageal tumor,
and in the evolution of normal esophageal tissues into ESCC, which
is closely associated with the occurrence, development and
invasiveness of esophageal cancer. FAM135B may be useful for the
diagnosis of early stage esophageal cancer and precancerous
lesions. The present study assessed the association between the
FAM135B gene and esophageal cancer, however, further studies are
required in order to better understand the underlying molecular
mechanisms involved.
Overall, ADAM29 and FAM135B were significantly
differentially expressed in different esophageal tissues.
Furthermore, the results of the present study demonstrated that
they are closely associated with the occurrence and development of
ESCC. In addition, a significant correlation was demonstrated
between ADAM29 and FAM135B. Both ADAM29 and FAM135B are expected to
be the important biological markers for the diagnosis of early
stage ESCC and precancerous lesions, and for further contribution
to guiding early treatment.
The present study poses a number of limitations due
to the selection of biopsy specimens from some out-patient patients
with esophageal intraepithelial neoplasia, resulting in incomplete
clinical data and a small tissue sample size. First, the present
study failed to perform multiple experimental methods in order to
verify the results obtained. Secondly, the present study failed to
assess the association of ADAM29 and FAM13B expressions with
smoking or alcohol consumption. However, Song et al
(14) demonstrated that ESCC
development is associated with alcohol consumption, through genomic
analyses. Thirdly, some specimens were obtained by biopsy in the
present study; however, post-surgical specimens are considered to
be more credible. It is important to collect more complete
experimental images for further studies, in order to increase the
reliability of the experimental results. The present study
investigated the association between the clinical characteristics
of patients with esophageal cancer and the expression of ADAM29 and
FAM13B proteins in esophageal cancer tissue samples; however,
further analyses on the association between protein expression and
the clinic-pathological features in normal tumor-adjacent tissues
is required in future studies. The present study is only a
retrospective study in some parts of China and the sample size is
relatively small, thus future research requires analyses in
different populations and larger samples.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National ‘863’
Project Foundation of China (grant no. 2014AA020901).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TW performed the experiments, analyzed the data and
drafted the initial manuscript. XL, SH and SJ collected specimens
and performed the experiments. YW designed the study and performed
the quality control. All authors participated in the review of the
final manuscript.
Ethics approval and consent to
participate
The present study was approved by the People's
Liberation Army 82nd Hospital Ethics Committee. All patients
participating in the study (or their family members) have signed
informed consent prior to the implementation of the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018 GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wei KR, Lian QH, Liu J and Liang ZH:
Prevalence of esophageal cancer. Chin J Int Med. 51:156–158.
2012.(In Chinese).
|
|
3
|
Malhotra GK, Yanala U, Ravipati A, Follet
M, Vijayakumar M and Are C: Global trends in esophageal cancer. J
Surg Oncol. 115:564–579. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang Y: Epidemiology of esophageal
cancer. World J Gastroenterol. 19:5598–5606. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhou MG, Wang XF, Hu JP, Li GL, Chen WQ,
Zhang SW, Wan X, Wang LJ, Xiang C, Hu YS and Yang GH: Geographical
distribution of cancer mortality in China, 2004–2005. Zhonghua Yu
Fang Yi Xue Za Zhi. 44:303–308. 2010.(In Chinese). PubMed/NCBI
|
|
7
|
Li QW, Yuan GJ, Du YX, Pan CN and He Y:
Analysis of the prevalence and treatment of esophageal cancer in
huai an area. J Clin Oncol. 17:142–145. 2012.
|
|
8
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang GQ, Jiao GG, Chang FB, Fang WH, Song
JX, Lu N, Lin DM, Xie YQ and Yang L: Long-term results of operation
for 420 patients with early squamous cell esophageal carcinoma
discovered by screening. Ann Thorac Surg. 77:1740–1744. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Aminian A, Panahi N, Mirsharifi R,
Karimian F, Meysamie A, Khorgami Z and Alibakhshi A: Predictors and
outcome of cervical anastomotic leakage after esophageal cancer
surgery. J Cancer Res Ther. 7:448–453. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang GQ, Zhang YM and He S: Present
situation and prospect of diagnosis and treatment of early
esophageal cancer and precancerous lesions in China. Chin Cancer.
18:690–694. 2009.(In Chinese).
|
|
12
|
Siewert JR and Ott K: Are squamous and
adenocarcinomas of the esophagus the same disease. Semin Radiat
Oncol. 17:38–44. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hao J and Shao K: Epidemiological status,
diagnosis and treatment of esophageal cancer in China and China's
future countermeasures. Chin J Cancer. 501–504. 2011.(In
Chinese).
|
|
14
|
Song Y, Li L, Ou Y, Gao Z, Li E, Li X,
Zhang W, Wang J, Xu L, Zhou Y, et al: Identification of genomic
alterations in oesophageal squamous cell cancer. Nature. 509:91–95.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cerretti DP, DuBose RF, Black RA and
Nelson N: Isolation of two novel metalloproteinase-disintegrin
(ADAM) cDNAs that show testis-specific gene expression. Biochem
Biophys Res Commun. 263:810–815. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Edwards DR, Handsley MM and Pennington CJ:
The ADAM metalloproteinases. Mol Aspects Med. 29:258–289. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang F, Xu R, Zhu P, Hu J, Ying B, Zhao S
and Li C: Preliminarily functional analysis of a cloned novel human
gene ADAM29. Sci China C Life Sci. 44:392–399. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Oria VO, Lopatta P, Schmitz T, Preca BT,
Nyström A, Conrad C, Bartsch JW, Kulemann B, Hoeppner J, Maurer J,
et al: ADAM9 contributes to vascular invasion in pancreatic ductal
adenocarcinoma. Mol Oncol. 13:456–479. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Luo ML, Zhou Z, Sun LC, Sun L, Yu L, Sun
L, Liu J, Yang Z, Ran Y, Yao Y and Hu H: An ADAM12 and FAK positive
feedback loop amplifies the interaction signal of tumor cells with
extracellular matrix to promote esophageal cancer metastasis.
Cancer Lett. 422:118–128. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li W, Wang D, Sun X, Zhang Y, Wang L and
Suo J: ADAM17 promotes lymph node metastasis in gastric cancer via
activation of the Notch and Wnt signaling pathways. Int J Mol Med.
43:914–926. 2019.PubMed/NCBI
|
|
21
|
Mochizuki S and Okada Y: ADAMs in cancer
cell proliferation and progression. Cancer Sci. 98:621–628. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang B, Li Y, Li L, Chen M, Zhang C, Zuo
XB, Zhou FS, Liang B, Zhu J, Li P, et al: Association study of
susceptibility loci with specific breast cancer subtypes in Chinese
women. Breast Cancer Res Treat. 146:503–514. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shan Q, Lou X, Xiao T, Zhang J, Sun H, Gao
Y, Cheng S, Wu L, Xu N and Liu S: A cancer/testis antigen
microarray to screen autoantibody biomarkers of non-small cell lung
cancer. Cancer Lett. 328:160–167. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Costa NR, Paulo P, Caffrey T,
Hollingsworth MA and Santos-Silva F: Impact of MUC1 mucin down
regulation in the phenotypic characteristics of MKN45 gastric
carcinoma cell line. PLoS One. 6:e269702011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Brim H, Abu-Asab MS, Nouraie M, Salazar J,
Deleo J, Razjouyan H, Mokarram P, Schaffer AA, Naghibhossaini F and
Ashktorab H: An integrative CGH, MSI and candidate genes
methylation analysis of colorectal tumors. PLoS One. 9:e821852014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wei X, Moncada-Pazos A, Cal S,
Soria-Valles C, Gartner J, Rudloff U, Lin JC; NISC Comparative
Sequencing Program, ; Rosenberg SA, López-Otín C and Samuels Y:
Analysis of the disintegrin-metalloproteinases family reveals
ADAM29 and ADAM7 are often mutated in melanoma. Hum Mutat.
32:E2148–E2175. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Oki NO, Motsinger-Reif AA, Antas PR, Levy
S, Holland SM and Sterling TR: Novel human genetic variants
associated with extrapulmonary tuberculosis A pilot genome wide
association study. BMC Res Notes. 4:282011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou C, Ye M, Ni S, Li Q, Ye D, Li J, Shen
Z and Deng H: DNA methylation biomarkers for head and neck squamous
cell carcinoma. Epigenetics. 13:398–409. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rice TW: Esophageal cancer staging. Korean
J Thorac Cardiovasc Surg. 48:157–163. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rice TW, Rusch VW, Ishwaran H and
Blackstone EH; Worldwide Esophageal Cancer Collaboration, : Cancer
of the esophagus and esophagogastric junction Data-driven staging
for the seventh edition of the American joint committee on
cancer/international union against cancer staging manuals. Cancer.
116:3763–3773. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Edge SB, Byrd DR, Compton CC, et al: AJCC
cancer staging manual. 7th. Springer; New York, NY: 2010
|
|
32
|
Zhou ZJ, Xie JL, Wei P and Zhou XG:
Pathologic subtyping of primary lymphoma of breast and prognostic
analysis. Zhonghua Bing Li Xue Za Zhi. 46:618–622. 2017.(In
Chinese; Abstract available in Chinese from the publisher).
PubMed/NCBI
|
|
33
|
Seals DF and Courtneidge SA: The ADAMs
family of metalloproteases: Multidomain proteins with multiple
functions. Genes Dev. 17:7–30. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rocks N, Paulissen G, El Hour M, Quesada
F, Crahay C, Gueders M, Foidart JM, Noel A and Cataldo D: Emerging
roles of ADAM and ADAMTS metalloproteinases in cancer. Biochimie.
90:369–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Noël A, Gutiérrez-Fernández A, Sounni NE,
Behrendt N, Maquoi E, Lund IK, Cal S, Hoyer-Hansen G and López-Otín
C: New and paradoxical roles of matrix metalloproteinases in the
tumor microenvironment. Front Pharmacol. 3:1402012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lu X, Lu D, Scully M and Kakkar V: ADAM
proteins-therapeutic potential in cancer. Curr Cancer Drug Targets.
8:720–732. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Murphy G: The ADAMs Signalling scissors in
the tumour microenvironment. Nat Rev Cancer. 8:929–941. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Maloum K, Settegrana C, Chapiro E, Cazin
B, Leprêtre S, Delmer A, Leporrier M, Dreyfus B, Tournilhac O, Mahe
B, et al: IGHV gene mutational status and LPL/ADAM29 gene
expression as clinical outcome predictors in CLL patients in
remission following treatment with oral fludarabine plus
cyclophosphamide. Ann Hematol. 88:1215–1221. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Quilter CR, Sargent CA, Bauer J, Bagga MR,
Reiter CP, Hutchinson EL, Southwood OI, Evans G, Mileham A, Griffin
DK and Affara NA: An association and haplotype analysis of porcine
maternal infanticide A model for human puerperal psychosis. Am J
Med Genet B Neuropsychiatr Genet. 159:908–927. 2012. View Article : Google Scholar
|
|
40
|
Tsang KM, Croen LA, Torres AR, Kharrazi M,
Delorenze GN, Windham GC, Yoshida CK, Zerbo O and Weiss LA: A
genome-wide survey of transgenerational genetic effects in autism.
PLoS One. 8:e769782013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lin JQ, Huang ZJ, Chen ZR, et al:
Relationship between FAM135B gene and lymph node metastasis of
esophageal squamous cell carcinoma. Chin J Exp Surg. 6:1170–1171.
2018.(In Chinese).
|
|
42
|
Dong DZ: The Function and Mechanism of
FAM135B in the Deveopment of esophageal squamous cell carcinoma
(unpublished PhD thesis). Peking Union Medical College; 2018
|