Introduction
Thyroid cancer (TC), as a common malignant thyroid
tumor, can be divided into papilla carcinoma, follicular carcinoma,
myeloid carcinoma and undifferentiated carcinoma through its
pathological classification. According to Chinese scholars, the
incidence of TC ranks seventh among female malignant tumors. Most
TC patients have no obvious symptoms, the most common manifestation
is thyroid nodules, often found in physical examination, and a very
small number of patients with cervical lymph node enlargement
(1). In recent years, TC treatments
have made significant progress, such as resection of thyroid,
thyroid stimulating hormone suppression therapy and drug targeting
therapy, radiation therapy, have been proven beneficial for
patients with TC treatments (2), but
its incidence is still rising steadily year by year, as there is no
clear molecular explantion for the pathogenesis of TC. Therefore,
it is very important for the treatment of TC to explore the
pathogenesis of TC from the molecular mechanism perspective.
microRNA (miRNA) is a small molecule intrinsic
non-coding RNA. In recent 10 years, research on miRNA in cell
differentiation, proliferation and apoptosis has been paid
increased attention. miRNA, as a post-transcriptional regulator, is
involved in the regulation of gene deletion, mutation or
amplification of various mRNAs, which may lead to misexpression,
which is related to the occurrence and development of many diseases
(3). miRNA, as a tumor inhibitor or
promoter, is widely involved in the occurrence and development of
many malignant tumors (4). For
example, Wang et al suggested that miR-205 can be used as a
tumor inhibitor in triple-negative breast cancer, which can inhibit
the growth, migration and invasion of cancer cells by targeting
HMGB1-RAGE pathway (5). Wu et
al (6) found that miR-501-3p is
misexpressed in colorectal cancer. miR-501-3p may be used as a new
type of miRNA to promote the development of colorectal cancer by
targeting the regulation of APC to promote the development of
colorectal cancer. miR-205 is generally used as a tumor inhibitor,
has low expression in tumors and affects the occurrence and
development of various malignant tumors (7–9). Yang
et al (10) reported that
miR-205 negative regulation of PAR2 promotes invasion and
metastasis of colorectal cancer. Pang and Yue (11) found that the low expression of
miR-205 in cervical cancer was related to the degree of tumor
differentiation and clinical stage. IGF1R was targeted to inhibit
the invasion and metastasis of cervical cancer. Although some
progress has been made in tumor research, the role and mechanism of
miR-205 in tumors, especially the mechanism of miR-205 in TC, the
effect of miR-205 on the proliferation and migration of TC cells
has not been reported, and the relationship between miR-205 and TC
is still unclear and needs to be further studied.
This study explored the expression level of miR-205
in TC, and its effect on the proliferation and migration of TC
cells and its regulatory mechanism, so as to provide a theoretical
basis for explaining the pathogenesis and molecular therapy of
TC.
Materials and methods
Tissue specimens
The 25 pairs of TC and paracancer tissues collected
in this study were tissue specimens from January 2017 to December
2018, removed after surgical treatment in the department of breast
and breast surgery in The Second Affiliated Hospital of Qiqihar
Medical University (Qiqihar, China). Among them, 15 were female and
10 were male, with an average age of 42.25±8.73 years. Inclusion
criteria: The specimens received surgical treatment in the
hospital, and were confirmed to be TC by histopathological
examination, and all were primary tumor lesions. Exclusion
criteria: Patients who had received preoperative radiation and
chemotherapy, patients who had secondary surgery, and patients with
other site metastasis. The adjacent tissues were 1–2 cm away from
the tumor, and were non-tumor tissues by histopathological
examination. Each specimen was stored in liquid nitrogen for 10 min
in vitro for subsequent analysis. All patients and their
families agreed to participate in the experiment and signed the
informed consent form. This study was approved by the Ethics
Committee of The Second Affiliated Hospital of Qiqihar Medical
University.
Cell lines and cell culture
The cell lines (SW579, B-CPAP, TPC-1, WRO, Htori-3)
were from the Cell bank of the Chinese Academy of Sciences
(Shanghai). The cell lines were cultured with DMEM (Corning, Inc.)
culture, and 10% fetal bovine serum (Thermo Fisher Scientific,
Inc.) and 1% streptomycin (Corning, Inc.) were added. The cell
culture conditions were: CO2 concentration 5% at
37°C.
Real-time fluorescence qRT-PCR
Total RNA was extracted by TRIzol kit (Shanghai
Pufei). The concentration and quality of RNA were determined by
spectrophotometer, and cDNA was obtained by reverse transcription.
Specific reaction system (20 µl): The total RNA was 2 µg, 2X miRNA
RT Buffer 10 µl, miRNA RT Enzyme Mix 2 µl, and RNase-Free
H2O was added to 20 µl. U6 (Gene) was the internal
reference gene, according to the fluorescence quantitative PCR
specification (Takara). Establishment of PCR reaction system: SYBR
Premix Ex Taq (Takara) 10 µl, forward primer 0.4 µl, reverse primer
0.4 µl, cDNA 2 µl, sterilized distilled water 7.2 µl, reaction
conditions: Pre-denaturation at 95°C for 30 sec, pre-denaturation
at 95°C for 5 sec, at 60°C for 30 sec, 40 cycles, the relative
quantitative analysis of mRNA was carried out using the method of
2-Thiophans (Table I).
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Primer | Primer sequences |
|---|
| CCNB2 | F:
5′-CAACCCACCAAAACAACA-3′ |
|
| R:
5′-AGAGCAAGGCATCAGAAA-3′ |
| miR-205 | F:
5′-GCTCCTTCATTCCACCGG-3′ |
|
| R:
5′-CAGTGCAGGGTCCGAGGT-3′ |
| U6 | F:
5′-CTCGCTTCGGCACAGT-3′ |
|
| R:
5′-ACGCTTCACGATTGCT-3′ |
Cell transfection and group
miR-205 mimic (5′-UCCUUCAUUCCACCGGAGUCUG-3′) and
CCNB2 overexpression plasmid (5′-CCGUUUCCCAGACUACCUU-3′) were from
Guangzhou Ruibo Biology Company and inoculated into 6-well culture
plate (Corning, Inc.). The cells were transferred to 50–70% of the
cells. According to the specification of Lipofectamine®
3000 transfection kit (Invitrogen; Thermo Fisher Scientific, Inc.),
the 10 µl transfection reagent was diluted with 250 µl serum-free
medium and added to each well of the culture plate for 48 h for
subsequent experiments. The TC cell line model of miR-205
overexpression was constructed by transferring miR-205 mimic, into
the cell line, that is, miR-205 mimic group, and SW579 cells were
transferred into negative virus (5′-TCTCCGAACGTGTCACGT-3′), which
was the negative control (NC).
Cell counting kit-8 (CCK-8)
The logarithmic cells were digested by trypsin to
make cell suspension. Cells (2,000) were inoculated into a 96-well
plate, one group for every 5 compound wells. CCK-8 solution
(Corning, Inc.) Ten microliters was added to each well and
incubated for 1 h. The absorbance of 450 nm was measured by enzyme
labeling instrument (Tecan Infinite) on the 1st day, 2nd day, 3rd
day, 4th day and 5th day, respectively.
Transwell transfer experiment
A 24-well plate was placed in a Transwell chamber
(Corning, Inc.), and 100 µl of serum-free medium was added to the
upper layer to prepare a serum-free cell suspension (concentration
of 2 to 105/ml) and a 600 µl of 30% FBS-containing medium was added
to the lower layer and incubated for 24 h at 37°C in an incubator.
The chamber was carefully removed, 4% paraformaldehyde was fixed at
room temperature for 30 min, and stained with 0.1% crystal purple
(Shanghai Health and Industry Co., Ltd.) at room temperature for 10
min. The solution was cleaned and placed under a microscope
(Olympus) after drying.
Double luciferase test
Target prediction database TargetScan (http://www. Targetscan.org), DIANA (http://athena-innovation.gr) and MiRDB (http://mirdb.org) were used to predict the target gene
of miR-205. Double luciferase kit (Promega) was used to detect
wild-type and mutant CCNB2, which were constructed by Shanghai
Jikai Gene Co., Ltd. The logarithmic SW579 cells were inoculated
into a 96-well plate. Twenty-four hours later, both miR-205 mimic
group and NC group were transfected with wild-type and mutant
CCNB23′UTR plasmid, and the fluorescence intensity was detected by
fluorescence detector (Glomax20/20; Promega). Fluorescein detection
reagent II (100 µl) was placed in 1.5 ml centrifuge tube. A
bioluminescence detector (GloMax) was used to read the luciferase
activity Firely luciferase (FLUC) of firefly by pre-reading for 2
sec, detecting 10 sec per well, adding 20 µl cell lysate, and fully
mixing it with 20 µl cell lysate. Then 1X Stop&Glo preparation
100 µl was added, fully mixed, to read the luciferase activity
Renilla luciferase (RLUC) of sea kidney on the luminous
instrument. The relative fluorescence intensity was calculated by
the ratio of RLUC/FLUC.
Western blot
Logarithmic cells were collected so that the lysate
was in contact with the cells. The total cell protein was
extracted. After the protein concentration was detected by BCA
method (Biyuntian), the gel was made. The protein was separated by
SDS-PAGE electrophoresis at 4°C and 300 mA constant current for 90
min. the protein was transferred to PVDF membrane (Millipore) by
wet transfer method. PVDF membrane was left at room temperature
with 5% skim milk for 1 h. Then the diluted first antibody (mouse
anti-human CCNB2 monoclonal antibody, Abcam, ab18250, dilution
ratio 1200; mouse anti-human GAPDH, SC-32233; Santa Cruz
Biotechnology, Inc.) was incubated for 1 h, and the second antibody
(sheep anti-rabbit IgG; Santa Cruz Biotechnology, Inc., sc-2004,
dilution ratio 1–2,000) was incubated. The chemiluminescence
solution containing ECL substrate (Thermo Fisher Scientific, Inc.)
was added to PVDF film and exposed to GelPro Analyzer (Media
Cybernetics, Silver Spring) for band analysis.
Statistical analysis
All the data were analyzed by SPSS 24 (IBM, Corp.)
and GraphPad 5.0 (GraphPad Software, Inc.). The data were obtained
from more than three independent experiments and expressed as mean
± standard deviation. t-test was used to compare the measurement
data, and the Chi-square test is used to compare the counting data.
P<0.05 was considered as statistically significant.
Results
Expression of miR-205 in TC tissues
and cells
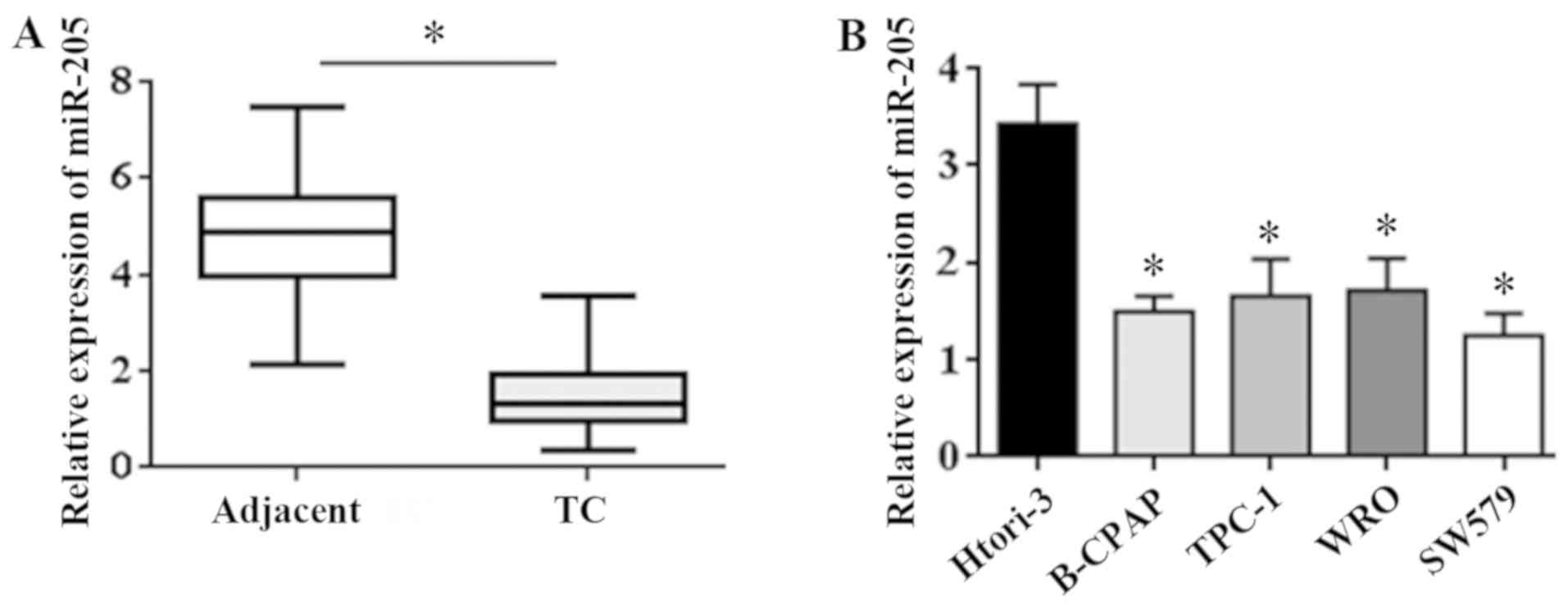
The expression of miR-205 in the tissue was detected
by qRT-PCR. The results showed that the expression of miR-205 in
the cancer tissues was significantly lower than that in the
adjacent tissues (t=3.47, P=0.031) (Fig.
1A). The expression of miR-205 in 4 TC cell lines (SW579,
B-CPAP, TPC-1, WRO) and 1 human normal thyroid cell line Htori-3
was detected by qRT-PCR assay. The results showed that the
expression level of miR-205 in TC cell line was lower than that in
the normal cell line (t=5.41, P=0.016) (Fig. 1B). The above results show that
miR-205 is significantly downregulated in TC tissue and cells.
Overexpression of miR-205 affects the
proliferation and migration of SW579 cells
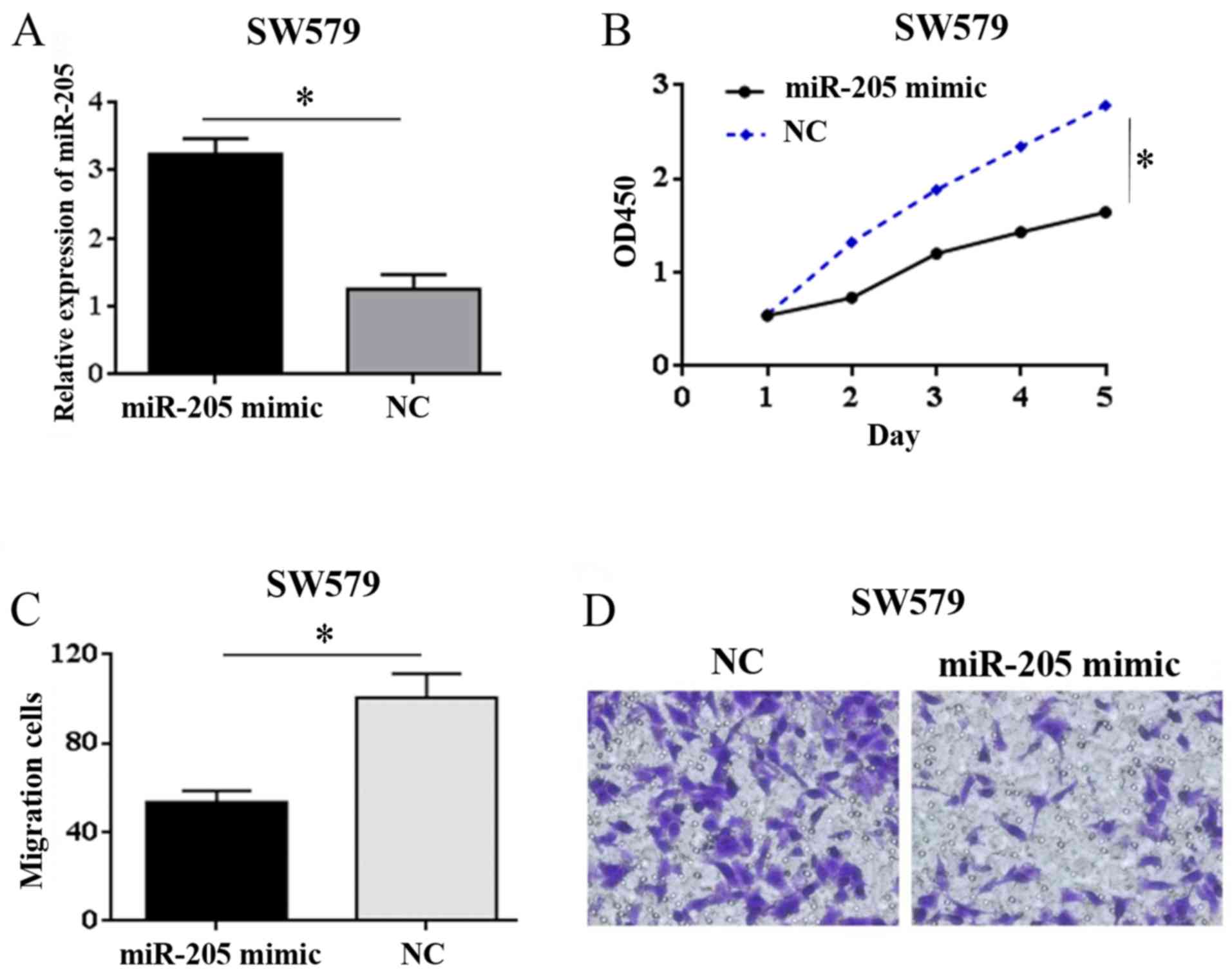
Results of qRT-PCR test showed that expression of
miR-205 in miR-205 mimic group was significantly higher than that
in NC group (t=3.92, P=0.035) (Fig.
2A). It can be concluded that the SW579 cell model with
overexpression of miR-205 was successfully constructed.
The results of CCK-8 assay showed that in SW579
cells, compared with NC group, the proliferation ability of miR-205
mimic group was significantly weakened (t=4.12, P=0.035) (Fig. 2B). The results of Transwell migration
test showed that the cell mobility in miR-205 mimic group was
significantly lower than that in NC group (t=4.47, P=0.027)
(Fig. 2C and D). These results
showed that overexpression of miR-205 inhibited the proliferation
and migration of SW579 cells.
Target relationship between CCNB2 and
miR-205
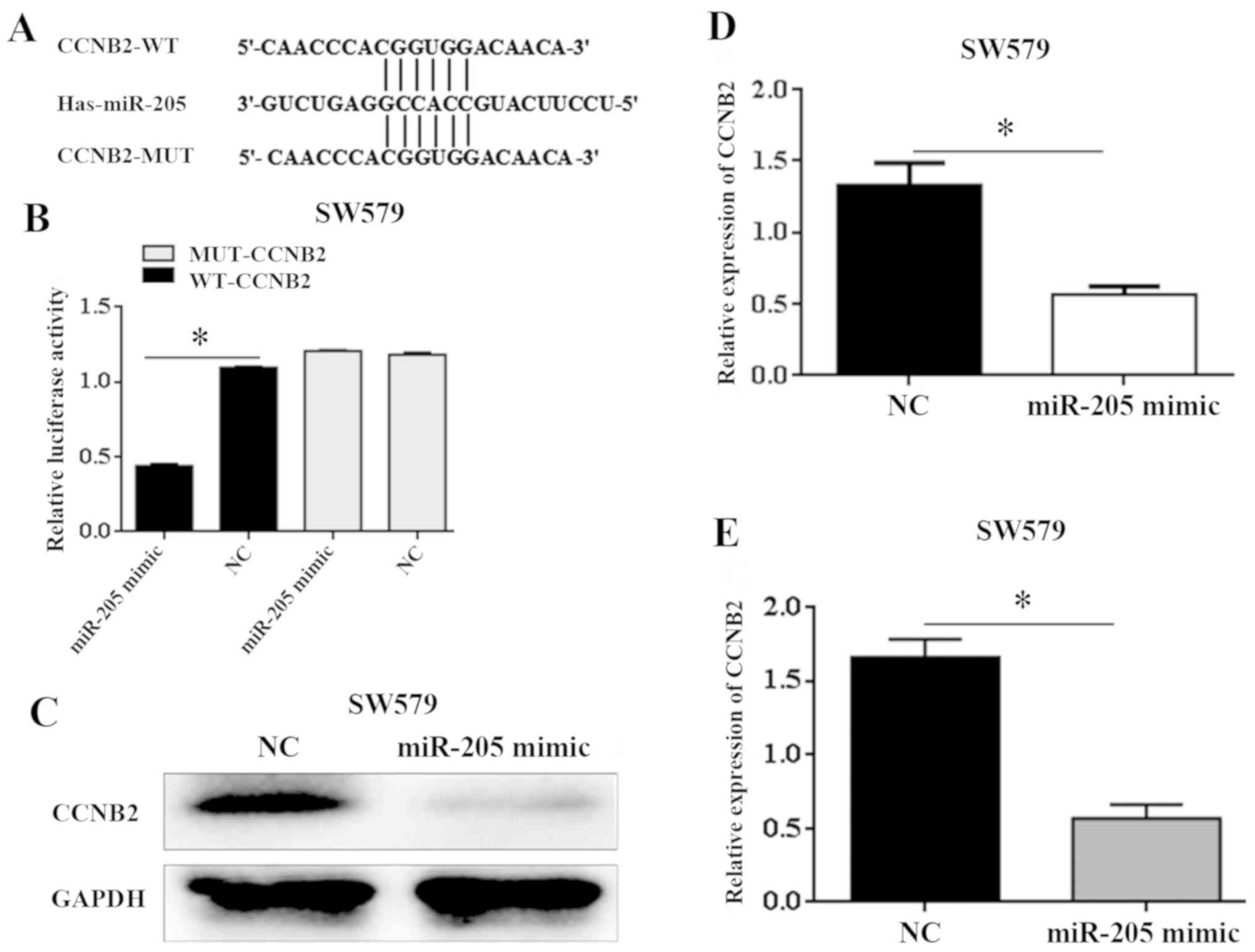
miR-205 can be combined with CCNB23′UTR through
target prediction database TargetScan, DIANA and MiRDB, prediction
miR-205. Double luciferase assay was used to further verify that
the wild-type and mutant sequences of CCNB2 combined with miR-205
as shown in Fig. 3A. Overexpression
of miR-205 significantly decreased the fluorescence activity of
wild-type 3′UTR containing CCNB2, but had no significant effect on
the fluorescence activity of 3′UTR containing CCNB2 mutant (t=4.63,
P=0.024) (Fig. 3B). The results of
western blot and qRT-PCR experiments show that after upregulation
of miR-205 in SW579 cells, CCNB2 expression level decreased
significantly compared with NC group (t=3.55, P=0.029; T=2.86,
P=0.043) (Fig. 3C-E). Expression of
miR205 and CCNB2 may be negatively regulated. Therefore, this
result confirms that CCNB2 is the downstream target of miR-205.
Effect of upregulation of CCNB2 on
proliferation and migration of SW579 cells regulated by
miR-205
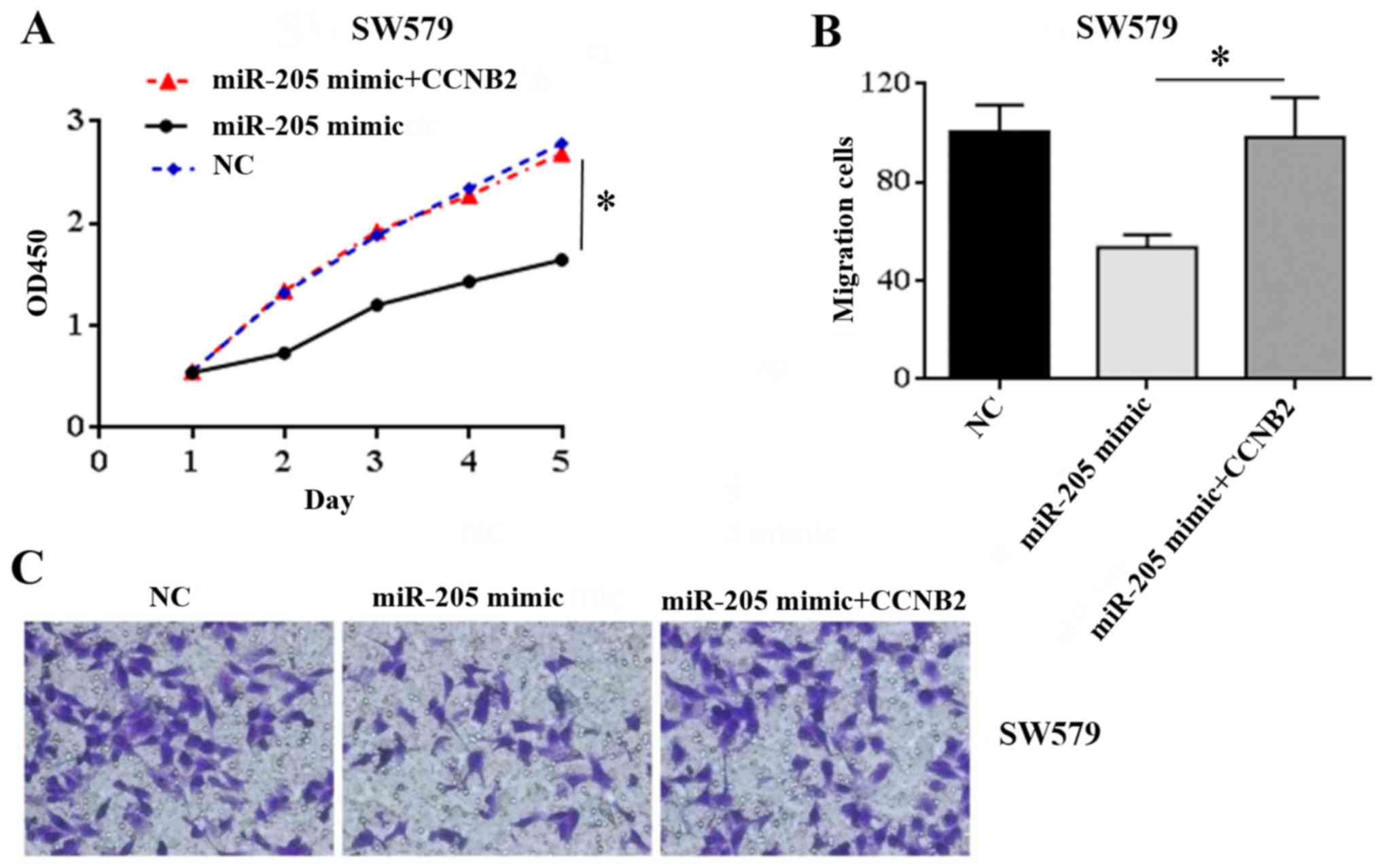
In order to further explore whether miR-205
regulates the proliferation and migration of TC cells by acting on
its downstream target CCNB2, this study continued to transfect
SW579 cells with CCNB2 overexpressed plasmid, and used CCK-8 and
Transwell experiments to observe whether the upregulation of CCNB2
could reverse the proliferation and migration of TC cells caused by
miR-205. CCK-8 experimental results showed that in SW579 cells, the
proliferation capacity of miR-205 mimic+CCNB2 group was
significantly higher than that of miR-205 mimic group (t=3.70,
P=0.031), while there was no significant difference compared with
NC group (t=1.54, P=0.122) (Fig.
4A). The results of Transwell assay showed that the cell
migration ability of miR-205 mimic CCNB2 group was significantly
higher than that of miR-205 mimic group (t=4.12, P=0.022), but
there was no significant difference between miR-205 mimic CCNB2
group and NC group (t=0.39, P=0.765) (Fig. 4B and C). These results suggest that
upregulation of CCNB2 can reverse the inhibitory effect of miR-205
on the proliferation and migration of TC cells.
Discussion
TC is a common malignant tumor in clinic. Although
most of TC are differentiated tumors and the malignant degree is
low, the incidence of TC has shown an upward trend in recent years.
Therefore, it is of great significance to explore the molecules
that can affect the occurrence of TC. miRNA is a kind of non-coding
RNA, non-coding protein, which is often used as upstream regulatory
molecule to affect the biological process of many tumors and plays
an important role in the occurrence and development of tumors
(12–14). In TC, many studies have also reported
that miRNA is involved in the regulation of its biological process.
Jiao et al (15) found that
ZEB1 overexpression reverses the inhibitory effect of miR-873
overexpression on the proliferation and invasion of TC cells, and
miR-873 may play a tumor inhibitory role in the development of TC
by inhibiting ZEB1. Guo and Zhang (16) suggest that miR-30a plays a role in
inhibiting tumor growth in TC by directly targeting the E2F7 gene.
miR-30a may be a new therapeutic target for TC. miR-205 is
misexpressed in many tumors, Dai et al (17) reported that miR-205 is often
underexpressed in glioma tissues. miR-205 can inhibit the growth,
invasion and reverse the EMT process of glioma by downregulating
its target gene HOXD9. In pancreatic cancer, the expression of
miR-205 is also downregulated. miR-205 can inhibit the
proliferation and migration of tumor cells by targeting RUNX2 in
pancreatic cancer (18). Lu et
al (19) confirmed that the
expression of miR-205 in HCC cells decreased, which could inhibit
the migration and invasion of HCC cells. miR-205 may become a
therapeutic target for HCC. However, the expression and role of
miR-205 in TC has not been reported. In this study, by detecting
the expression of miR-205 in TC and paracancerous tissues, we found
that the expression of miR-205 in cancer tissues was lower.
Moreover, expression of miR-205 in cancer cells was also confirmed
in 4 TC cell lines (SW579, B-CPAP, TPC-1, WRO) and the normal
thyroid cell line Htori-3, which suggested that miR-205 has low
expression in TC tumors. It may be working as a tumor suppressor
gene. In order to further study the effect of miR-205 on the
proliferation and migration of TC cells, a TC cell line model
overexpressing miR-205 was constructed in SW579 cells by miR-205
mimic to verify the effect of miR-205 on the biological function of
SW579 cells. The results of CCK-8 and Transwell migration also
confirmed that overexpression of miR-205 inhibited the
proliferation and migration of SW579 cells. These results suggest
that miR-205 may be involved in the regulation of cell
proliferation and migration of TC as a tumor inhibitor in TC. This
is also consistent with the results of Dai et al (17) and Lu et al (19), which fully indicates that miR-205 may
inhibit the proliferation and migration of TC cells. Previous
studies have shown that miR-205 is usually an upstream regulator.
The studies of Dai et al (17) and Lu et al (19) also confirmed that miR-205 plays a
role in tumor by regulating its target genes. However, the
mechanism of miR-205 inhibiting the proliferation and migration of
TC cells is not clear.
Therefore, this study continued to verify the target
gene of miR-205 through database exploration and double fluorescein
reporter enzyme assay, and reveal the mechanism of miR-205
regulating the biological function of TC cells. CCNB2 is an
important member of the cell cycle family regulatory network. It
can prevent damaged cells from entering mitotic phase and maintain
the correct replication of genetic material and genome stability
(20,21). The normal growth and development of
cells need the organic cooperation of cell cycle family members,
orderly regulation, and abnormal expression of CCNB2, may appear as
cell cycle regulation disorder, also inducing cell malignant
transformation. In recent years, CCNB2 abnormal expression in many
malignant tumors has been used as downstream molecules, which
affects the development process of tumors, such as non-small cell
lung cancer, bladder cancer and gastric cancer (22–24).
Shubbar et al (25) found
that ccnb2 overexpression affects the prognosis of breast cancer
patients. During follow-up, ccnb2 overexpression was found to be
associated with poor prognosis. Li et al reported that CCNB2
is also a risk factor for the prognosis of HCC patients. CCNB2 can
promote the proliferation and migration of HCC cells (26). In conclusion, CCNB2, as a member of
the cell cycle family, plays an important role in the orderly
regulation of the cell cycle. Previous studies have confirmed that
CCNB2 may play a carcinogenic role in many malignant tumors. In
this study, it was found by public database prediction analysis
that miR-205 binds to CCNB2 3′UTR. Therefore, the relationship
between the expression of miR-205 and CCNB2 in TC cells and
targeted regulation were also explored. In this study, the results
of double luciferase assay confirmed that CCNB2 may be a target
gene of miR-205. QRT-PCR and western blot experiments also found
that overexpression of miR-205 could inhibit the expression of
CCNB2, and the expression of miR-205 was negatively correlated with
the expression of CCNB2, which may be a negative regulatory
relationship. Furthermore, it was found that the proliferation and
migration ability of SW579 cells in miR-205 mimic CCNB2 group was
significantly higher than that in miR-205 mimic group, but there
was no significant difference compared with NC group, which
indicated that the upregulation of CCNB2 expression could
counteract the inhibitory effect of miR-205 mimic on the
proliferation and migration of TC cells. Based on the above
results, it can be suggested that miR-205 may play a role in the
proliferation and migration of TC cells by targeting the regulation
of CCNB2.
In conclusion, this study found that miR-205 was
differentially expressed in TC tissues and cells, and miR-205
inhibits the proliferation and migration of TC cells. Further study
on the mechanism showed that miR-205 regulated the proliferation
and migration of TC cells by inhibiting expression of CCNB2, which
provided a theoretical basis for exploring the pathogenesis of TC
and a new target for the treatment of TC.
Acknowledgements
Not applicable.
Funding
This study was supported by Qiqihar Science and
Technology Project (no. SFGG-201707).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XW wrote the manuscript. XW, YL, HZ, KJ, CZ and HL
conceived and designed the study. XW, YL, HZ, KJ, CZ, QM, ZW and CF
were responsible for the collection and analysis. XW, YL, HZ, KJ,
CZ, HL and QM interpreted the data and drafted the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
The Second Affiliated Hospital of Qiqihar Medical University
(Qiqihar, China). All patients and their families agreed to
participate in the experiment and signed the informed consent
form.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cabanillas ME, McFadden DG and Durante C:
Thyroid cancer. Lancet. 388:2783–2795. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kong YW, Ferland-McCollough D, Jackson TJ
and Bushell M: microRNAs in cancer management. Lancet Oncol.
13:e249–e258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang L, Kang FB, Wang J, Yang C and He DW:
Downregulation of miR-205 contributes to epithelial-mesenchymal
transition and invasion in triple-negative breast cancer by
targeting HMGB1-RAGE signaling pathway. Anticancer Drugs.
30:225–232. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu F, Xing T, Gao X and Liu F: miR-501-3p
promotes colorectal cancer progression via activation of
Wnt/β-catenin signaling. Int J Oncol. 55:671–683. 2019.PubMed/NCBI
|
|
7
|
Yao L, Shi W and Gu J: Micro-RNA 205-5p is
involved in the progression of gastric cancer and targets
phosphatase and tensin homolog (PTEN) in SGC-7901 human gastric
cancer cells. Med Sci Monit. 25:6367–6377. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hu A, Zhang Y, Zhao X, Li J and Ying Y:
CBX1 is a direct target of miR-205-5p and contributes to the
progression of pituitary tumor. Pharmazie. 74:154–156.
2019.PubMed/NCBI
|
|
9
|
Chen S, Jin L, Nie S, Han L, Lu N and Zhou
Y: miR-205 inhibits neuroblastoma growth by targeting
cAMP-responsive element-binding protein 1. Oncol Res. 26:445–455.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang X, Yang L, Ma Y, Zhao X and Wang H:
MicroRNA-205 mediates proteinase-activated receptor 2
(PAR2)-promoted cancer cell migration. Cancer Invest. 35:601–609.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pang H and Yue X: MiR-205 serves as a
prognostic factor and suppresses proliferation and invasion by
targeting insulin-like growth factor receptor 1 in human cervical
cancer. Tumour Biol. Jun 27–2017.(Epub ahead of print). doi
org/10.1177/1010428317701308. View Article : Google Scholar
|
|
12
|
Liu Y, Qian XM, He QC and Weng JK: MiR-421
inhibition protects H9c2 cells against
hypoxia/reoxygenation-induced oxidative stress and apoptosis by
targeting Sirt3. Perfusion. Aug 30–2019.(Epub ahead of print). doi:
10.1177/0267659119870725. View Article : Google Scholar
|
|
13
|
Mei JW, Yang ZY, Xiang HG, Bao R, Ye YY,
Ren T, Wang XF and Shu YJ: MicroRNA-1275 inhibits cell migration
and invasion in gastric cancer by regulating vimentin and
E-cadherin via JAZF1. BMC Cancer. 19:7402019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ye K, Xu C and Hui T: MiR-34b inhibits the
proliferation and promotes apoptosis in colon cancer cells by
targeting Wnt/β-catenin signaling pathway. Biosci Rep.
39:BSR201917992019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiao D, Guo F and Fu Q: MicroRNA 873
inhibits the progression of thyroid cancer by directly targeting
ZEB1. Mol Med Rep. 20:1986–1993. 2019.PubMed/NCBI
|
|
16
|
Guo H and Zhang L: MicroRNA-30a suppresses
papillary thyroid cancer cell proliferation, migration and invasion
by directly targeting E2F7. Exp Ther Med. 18:209–215.
2019.PubMed/NCBI
|
|
17
|
Dai B, Zhou G, Hu Z, Zhu G, Mao B, Su H
and Jia Q: MiR-205 suppresses epithelial-mesenchymal transition and
inhibits tumor growth of human glioma through downregulation of
HOXD9. Biosci Rep. 39:392019. View Article : Google Scholar
|
|
18
|
Zhuang L, Guo J, Yao Y and Li Z: miR-205
targets runt-related transcription factor 2 to inhibit human
pancreatic cancer progression. Oncol Lett. 17:843–848.
2019.PubMed/NCBI
|
|
19
|
Lu J, Lin Y, Li F, Ye H, Zhou R, Jin Y, Li
B, Xiong X and Cheng N: MiR-205 suppresses tumor growth, invasion,
and epithelial-mesenchymal transition by targeting SEMA4C in
hepatocellular carcinoma. FASEB J. May 25–2018.(Epub ahead of
print). doi: 10.1096/fj.201800113R. View Article : Google Scholar
|
|
20
|
Ballew O and Lacefield S: The DNA damage
checkpoint and the spindle position checkpoint: Guardians of
meiotic commitment. Curr Genet. 65:1135–1140. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hochegger H, Takeda S and Hunt T:
Cyclin-dependent kinases and cell-cycle transitions: Does one fit
all? Nat Rev Mol Cell Biol. 9:910–916. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qian X, Song X, He Y, Yang Z, Sun T, Wang
J, Zhu G, Xing W and You C: CCNB2 overexpression is a poor
prognostic biomarker in Chinese NSCLC patients. Biomed
Pharmacother. 74:222–227. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lei CY, Wang W, Zhu YT, Fang WY and Tan
WL: The decrease of cyclin B2 expression inhibits invasion and
metastasis of bladder cancer. Urol Oncol. 34:237.e1–237.e10. 2016.
View Article : Google Scholar
|
|
24
|
Shi Q, Wang W, Jia Z, Chen P, Ma K and
Zhou C: ISL1, a novel regulator of CCNB1, CCNB2 and c-MYC genes,
promotes gastric cancer cell proliferation and tumor growth.
Oncotarget. 7:36489–36500. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shubbar E, Kovács A, Hajizadeh S, Parris
TZ, Nemes S, Gunnarsdóttir K, Einbeigi Z, Karlsson P and Helou K:
Elevated cyclin B2 expression in invasive breast carcinoma is
associated with unfavorable clinical outcome. BMC Cancer. 13:12013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li R, Jiang X, Zhang Y, Wang S, Chen X, Yu
X, Ma J and Huang X: Cyclin B2 overexpression in human
hepatocellular carcinoma is associated with poor prognosis. Arch
Med Res. 50:10–17. 2019. View Article : Google Scholar : PubMed/NCBI
|