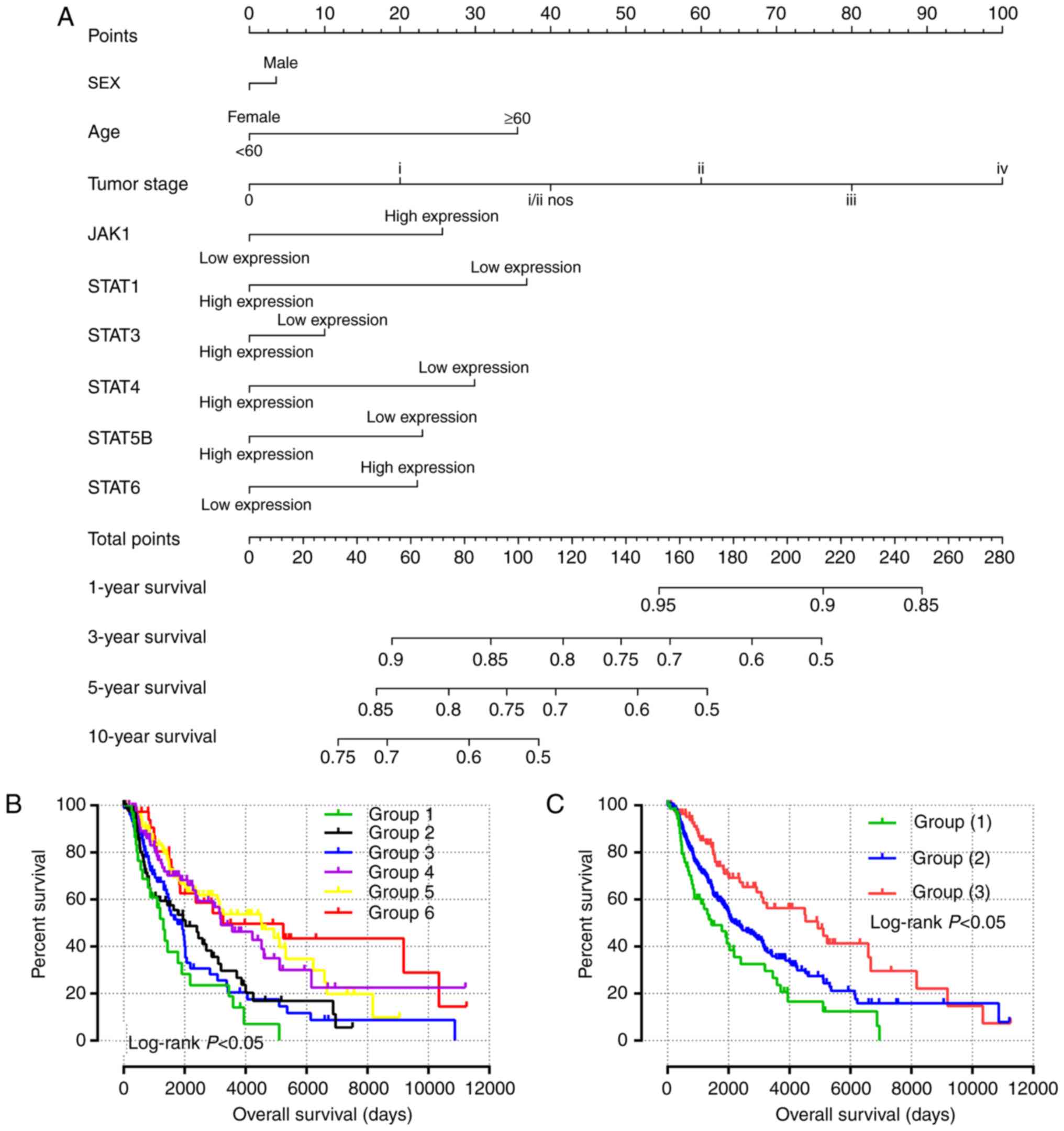
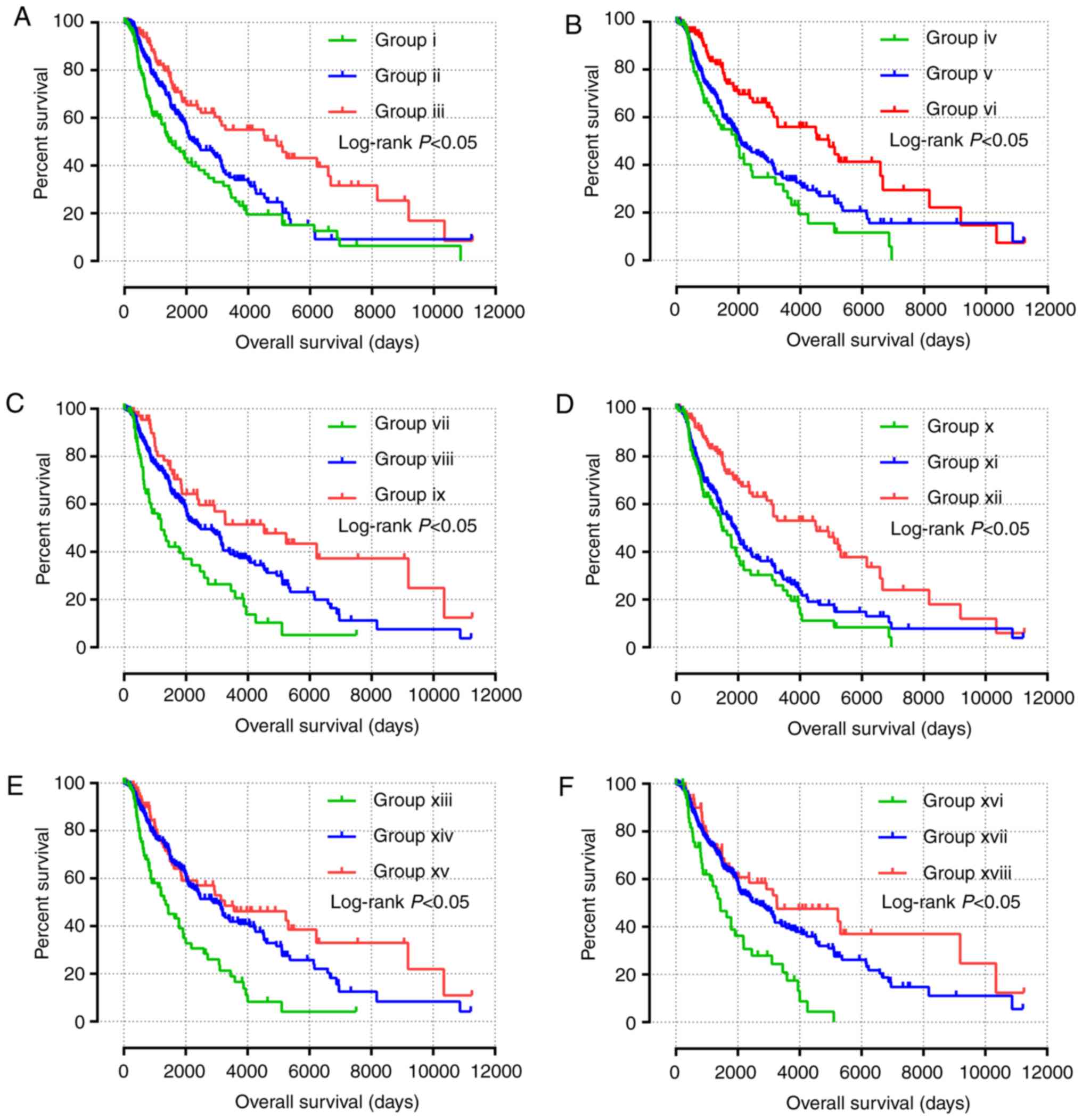
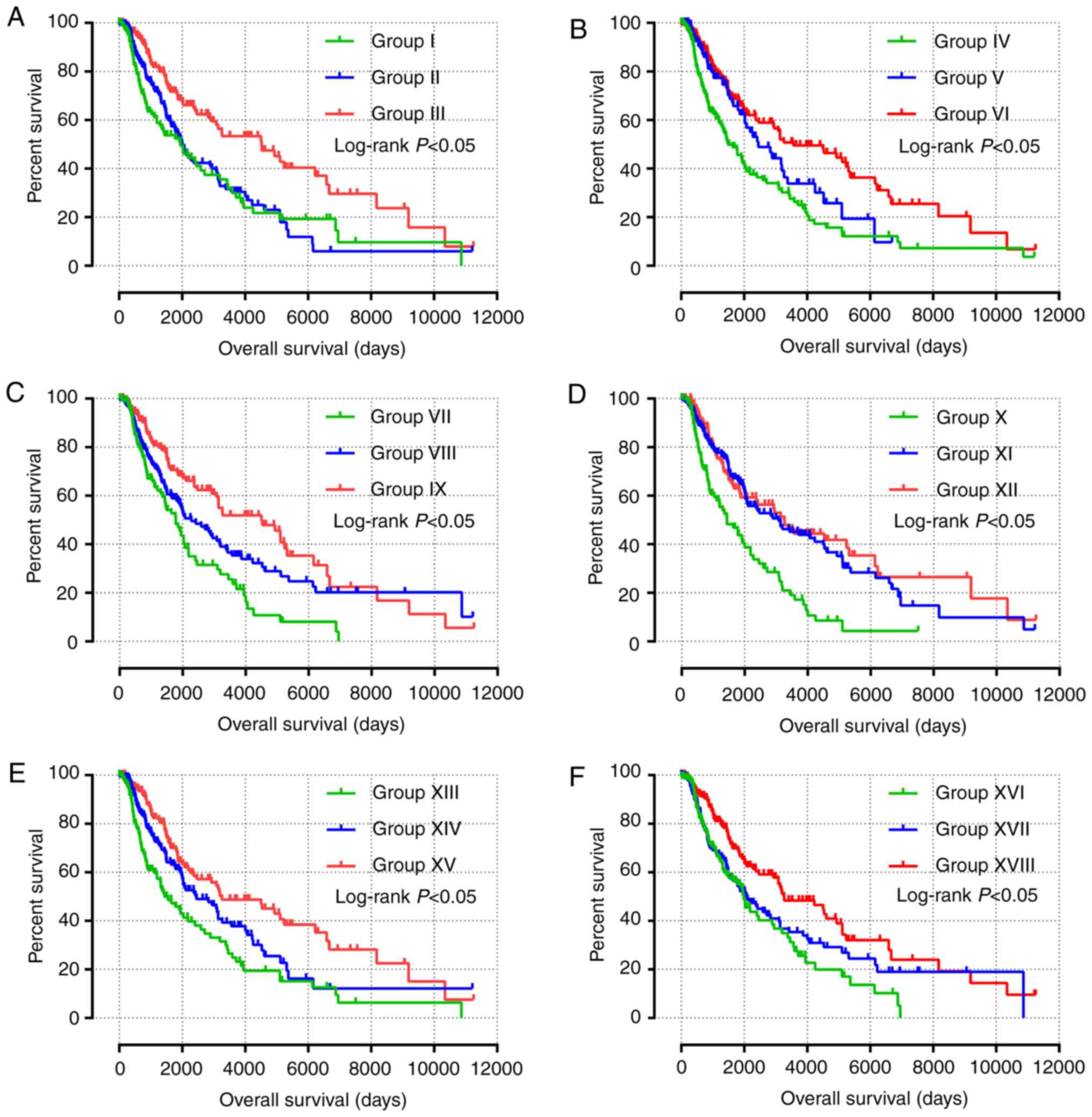
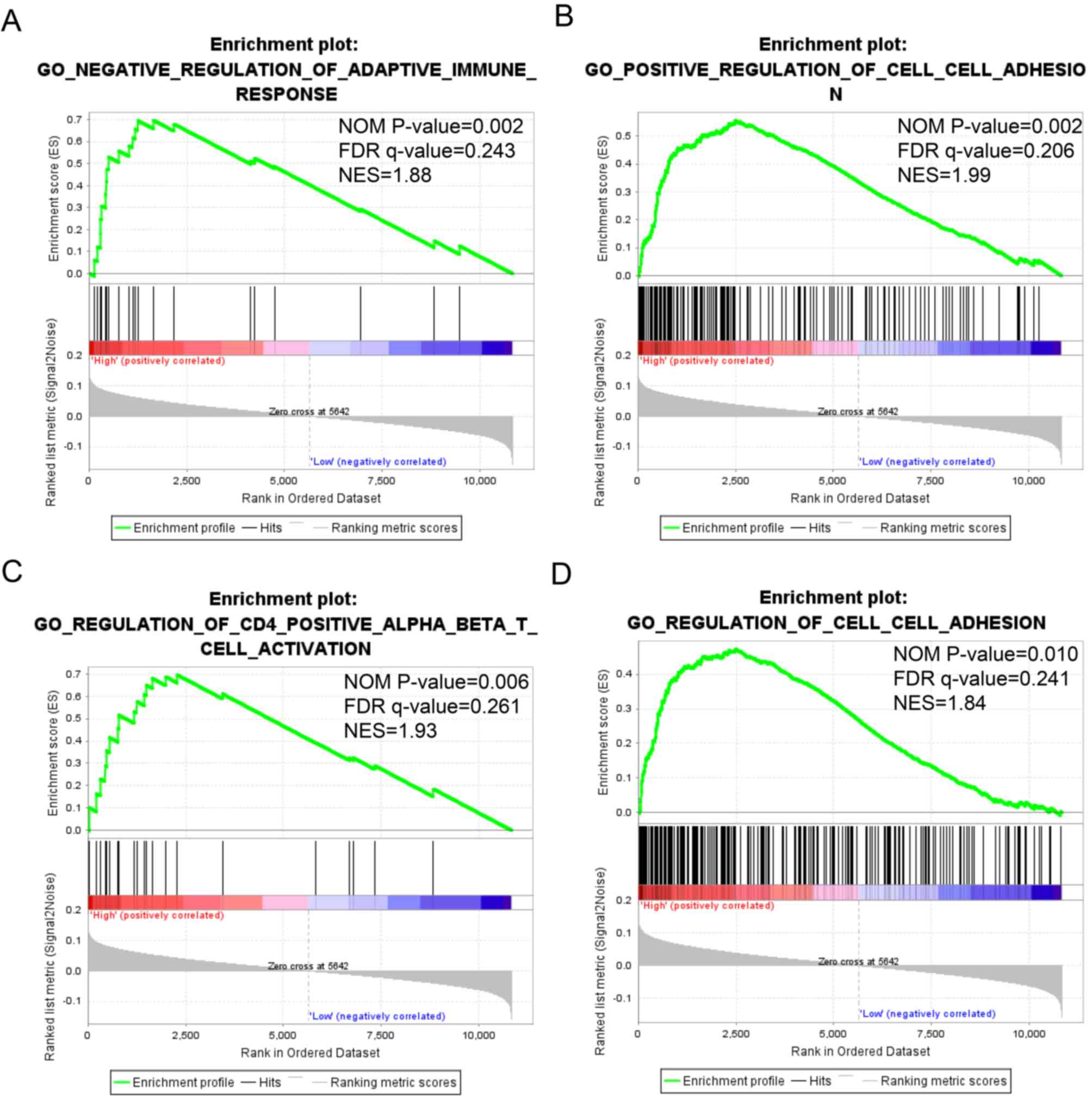
|
1
|
Goldstein BG and Goldstein AO: Diagnosis
and management of malignant melanoma. Am Fam Physician.
63:1359–1368. 2001.PubMed/NCBI
|
|
2
|
Situm M, Buljan M, Kolić M and Vučić M:
Melanoma-clinical, dermatoscopical, and histopathological
morphological characteristics. Acta Dermatovenerol Croat. 22:1–12.
2014.PubMed/NCBI
|
|
3
|
Grimaldi AM, Simeone E and Ascierto PA:
The role of MEK inhibitors in the treatment of metastatic melanoma.
Curr Opin Oncol. 26:196–203. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
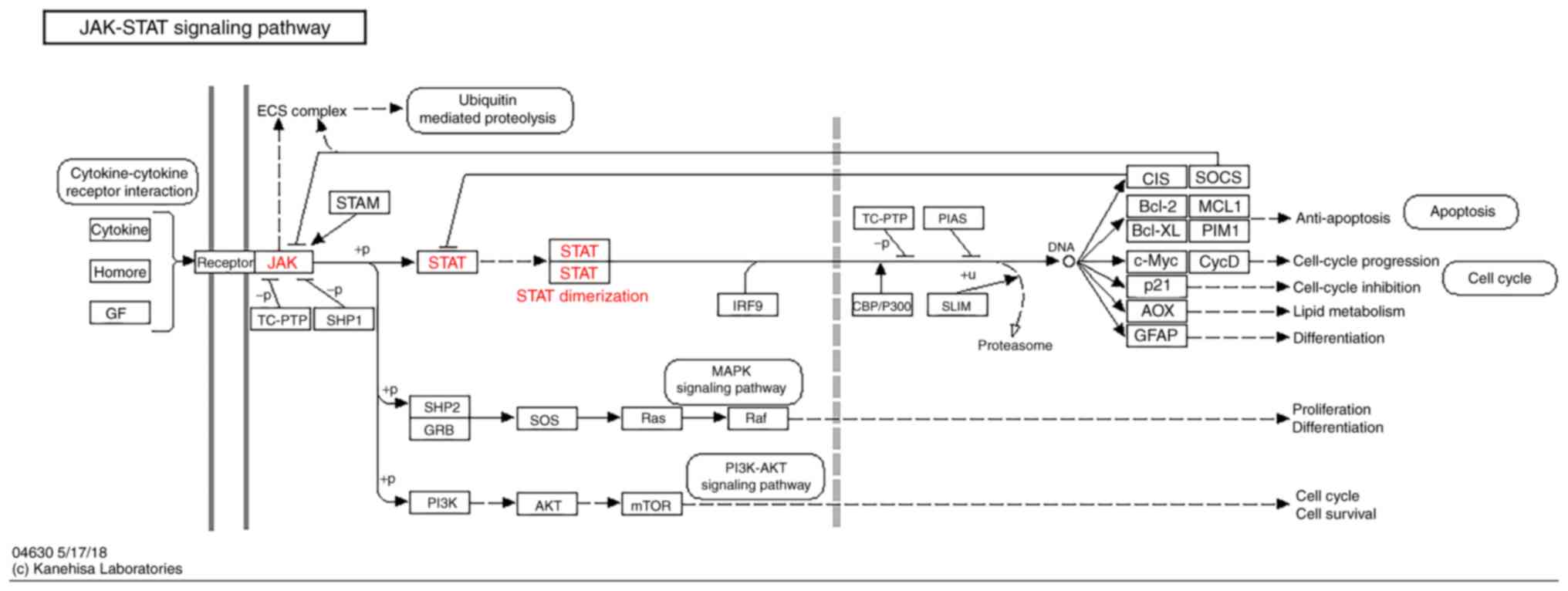
Aaronson DS and Horvath CM: A road map for
those who don't know JAK-STAT. Science. 296:1653–1655. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tymoszuk P, Charoentong P, Hackl H, Spilka
R, Müller-Holzner E, Trajanoski Z, Obrist P, Revillion F, Peyrat
JP, Fiegl H and Doppler W: High STAT1 mRNA levels but not its
tyrosine phosphorylation are associated with macrophage
infiltration and bad prognosis in breast cancer. BMC Cancer.
14:2572014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu Y, Huang J, Li W, Chen Y, Liu X and
Wang J: Meta-analysis of STAT3 and phospho-STAT3 expression and
survival of patients with breast cancer. Oncotarget. 9:13060–13067.
2018.PubMed/NCBI
|
|
9
|
Cui X, Jing X, Yi Q, Long C, Tan B, Li X,
Chen X, Huang Y, Xiang Z, Tian J and Zhu J: Systematic analysis of
gene expression alterations and clinical outcomes of STAT3 in
cancer. Oncotarget. 9:3198–3213. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Han C, Sun B, Zhao X, Zhang Y, Gu Q, Liu
F, Zhao N and Wu L: Phosphorylation of STAT3 promotes vasculogenic
mimicry by inducing epithelial-to-mesenchymal transition in
colorectal cancer. Technol Cancer Res Treat. 16:1209–1219. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kanehisa M, Sato Y, Furumichi M, Morishima
K and Tanabe M: New approach for understanding genome variations in
KEGG. Nucleic Acids Res. 47((D1)): D590–D595. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45(D1):D353–D361. 2017.
View Article : Google Scholar
|
|
13
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
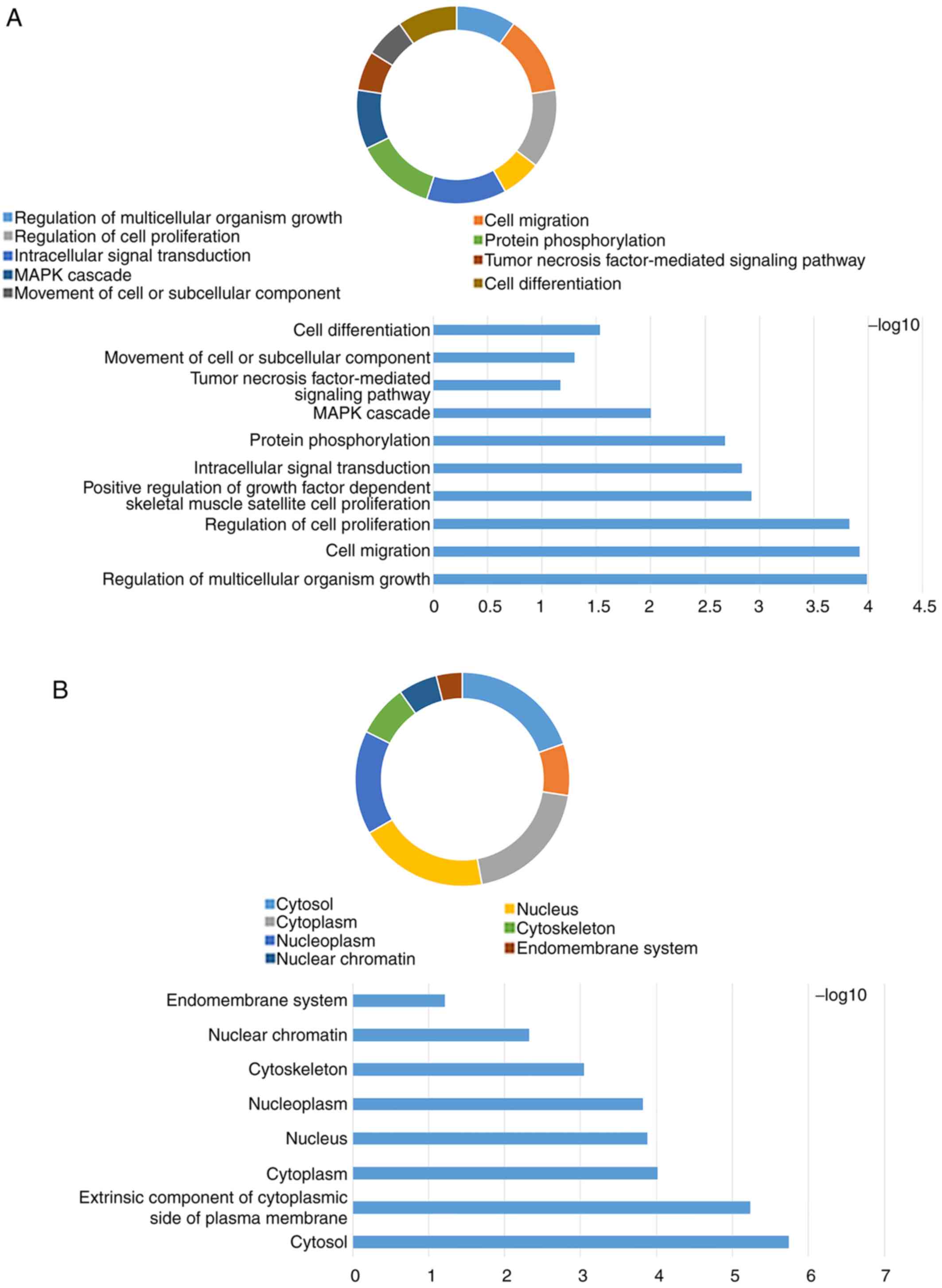
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
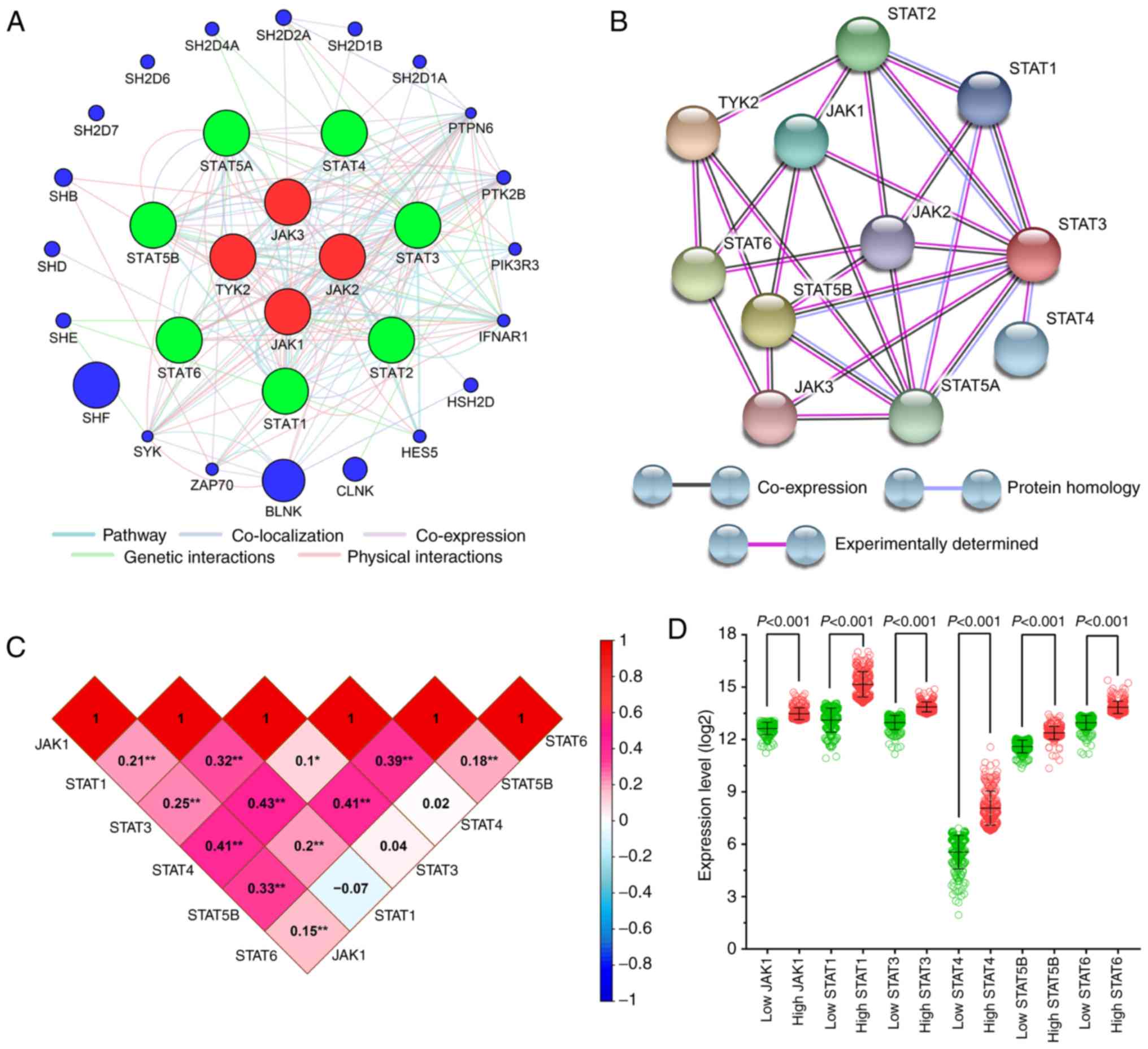
Mostafavi S, Ray D, Warde-Farley D,
Grouios C and Morris Q: GeneMANIA: A real-time multiple association
network integration algorithm for predicting gene function. Genome
Biol. 9 (Suppl 1):S42008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38:W214–W220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45((D1)): D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liberzon A, Birger C, Thorvaldsdóttir H,
Ghandi M, Mesirov JP and Tamayo P: The molecular signatures
database (MSigDB) hallmark gene set collection. Cell Syst.
1:417–425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Balch CM, Gershenwald JE, Soong SJ,
Thompson JF, Atkins MB, Byrd DR, Buzaid AC, Cochran AJ, Coit DG,
Ding S, et al: Final version of 2009 AJCC melanoma staging and
classification. J Clin Oncol. 27:6199–6206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Benjamini Y, Drai D, Elmer G, Kafkafi N
and Golani I: Controlling the false discovery rate in behavior
genetics research. Behav Brain Res. 125:279–284. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Reiner A, Yekutieli D and Benjamini Y:
Identifying differentially expressed genes using false discovery
rate controlling procedures. Bioinformatics. 19:368–375. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gerami P, Cook RW, Wilkinson J, Russell
MC, Dhillon N, Amaria RN, Gonzalez R, Lyle S, Johnson CE,
Oelschlager KM, et al: Development of a prognostic genetic
signature to predict the metastatic risk associated with cutaneous
melanoma. Clin Cancer Res. 21:175–183. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ghoreschi K, Laurence A and O'Shea JJ:
Janus kinases in immune cell signaling. Immunol Rev. 228:273–287.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Harrison DA: The Jak/STAT pathway. Cold
Spring Harb Perspect Biol. 4:a0112052012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zoranovic T, Grmai L and Bach EA:
Regulation of proliferation, cell competition, and cellular growth
by the drosophila JAK-STAT pathway. JAKSTAT.
2:e254082013.PubMed/NCBI
|
|
29
|
Rane SG and Reddy EP: Janus kinases:
Components of multiple signaling pathways. Oncogene. 19:5662–5679.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Messina JL, Yu H, Riker AI, Munster PN,
Jove RL and Daud AI: Activated stat-3 in melanoma. Cancer Control.
15:196–201. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen X, Du J, Jiang R and Li L:
MicroRNA-214 inhibits the proliferation and invasion of lung
carcinoma cells by targeting JAK1. Am J Transl Res. 10:1164–1171.
2018.PubMed/NCBI
|
|
32
|
Bi CL, Zhang YQ, Li B, Guo M and Fu YL:
MicroRNA-520a-3p suppresses epithelial-mesenchymal transition,
invasion, and migration of papillary thyroid carcinoma cells via
the JAK1-mediated JAK/STAT signaling pathway. J Cell Physiol.
234:4054–467. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jain N, Zhang T, Fong SL, Lim CP and Cao
X: Repression of Stat3 activity by activation of mitogen-activated
protein kinase (MAPK). Oncogene. 17:3157–3167. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rawlings JS, Rosler KM and Harrison DA:
The JAK/STAT signaling pathway. J Cell Sci. 117:1281–1283. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nissan MH, Pratilas CA, Jones AM, Ramirez
R, Won H, Liu C, Tiwari S, Kong L, Hanrahan AJ, Yao Z, et al: Loss
of NF1 in cutaneous melanoma is associated with RAS activation and
MEK dependence. Cancer Res. 74:2340–2350. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Davies MA and Samuels Y: Analysis of the
genome to personalize therapy for melanoma. Oncogene. 29:5545–5555.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Koti M, Siu A, Clément I, Bidarimath M,
Turashvili G, Edwards A, Rahimi K, Mes-Masson AM and Squire JA: A
distinct pre-existing inflammatory tumour microenvironment is
associated with chemotherapy resistance in high-grade serous
epithelial ovarian cancer. Br J Cancer. 112:1215–1222. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Simpson JA, Al-Attar A, Watson NF,
Scholefield JH, Ilyas M and Durrant LG: Intratumoral T cell
infiltration, MHC class I and STAT1 as biomarkers of good prognosis
in colorectal cancer. Gut. 59:926–933. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang Y, Molavi O, Su M and Lai R: The
clinical and biological significance of STAT1 in esophageal
squamous cell carcinoma. BMC Cancer. 14:7912014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Au KK, Le Page C, Ren R, Meunier L,
Clément I, Tyrishkin K, Peterson N, Kendall-Dupont J, Childs T,
Francis JA, et al: STAT1-associated intratumoural TH1
immunity predicts chemotherapy resistance in high-grade serous
ovarian cancer. J Pathol Clin Res. 2:259–270. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chan SR, Rickert CG, Vermi W, Sheehan KC,
Arthur C, Allen JA, White JM, Archambault J, Lonardi S, McDevitt
TM, et al: Dysregulated STAT1-SOCS1 control of JAK2 promotes
mammary luminal progenitor cell survival and drives ERalpha(+)
tumorigenesis. Cell Death Differ. 21:234–246. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Thota B, Arimappamagan A, Kandavel T,
Shastry AH, Pandey P, Chandramouli BA, Hegde AS, Kondaiah P and
Santosh V: STAT-1 expression is regulated by IGFBP-3 in malignant
glioma cells and is a strong predictor of poor survival in patients
with glioblastoma. J Neurosurg. 121:374–383. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
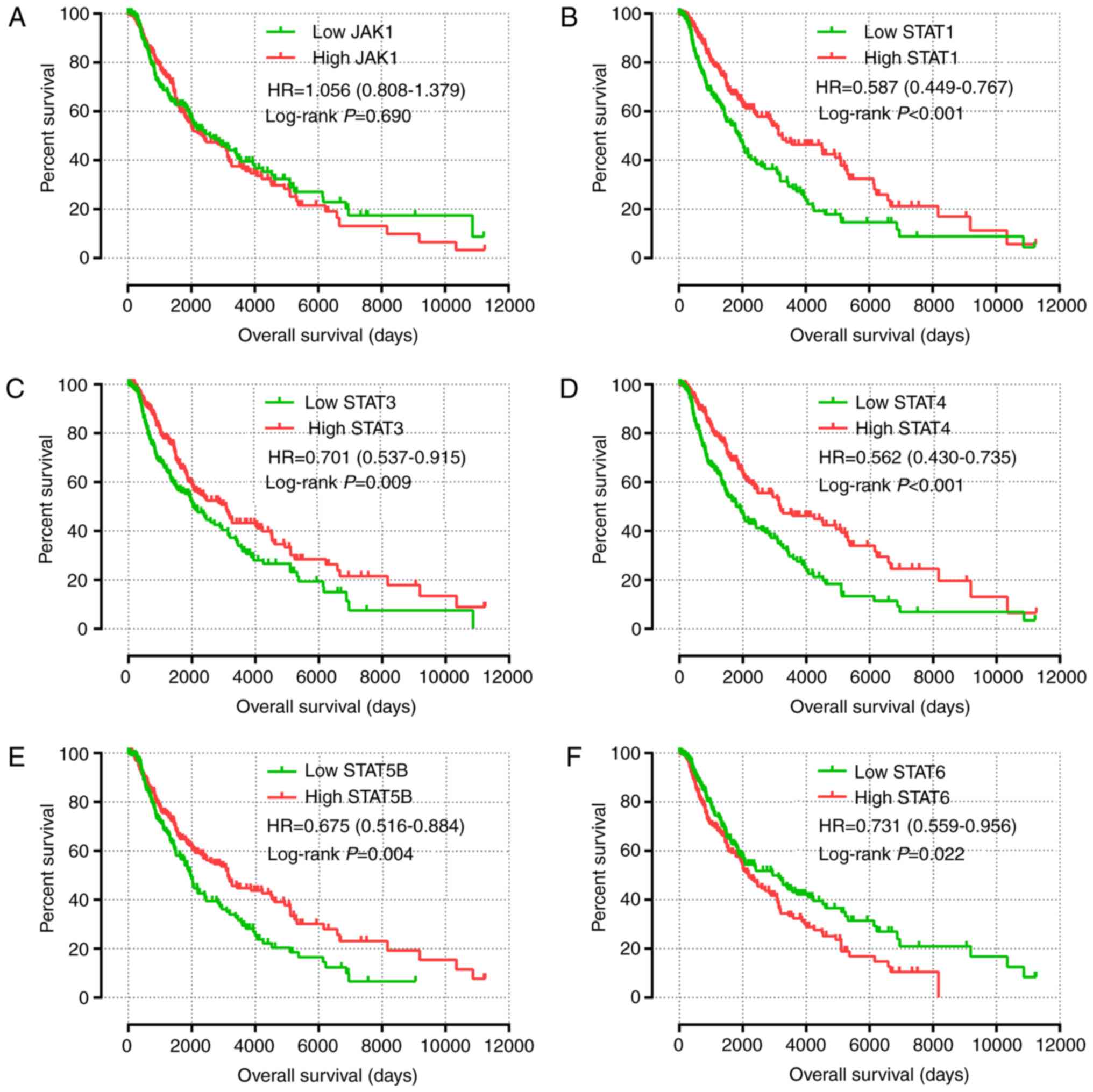
Arzt L, Kothmaier H, Halbwedl I,
Quehenberger F and Popper HH: Signal transducer and activator of
transcription 1 (STAT1) acts like an oncogene in malignant pleural
mesothelioma. Virchows Arch. 465:79–88. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jung SH, Ahn SY, Choi HW, Shin MG, Lee SS,
Yang DH, Ahn JS, Kim YK, Kim HJ and Lee JJ: STAT3 expression is
associated with poor survival in non-elderly adult patients with
newly diagnosed multiple myeloma. Blood Res. 52:293–299. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li WM, Huang CN, Lee YC, Chen SH, Lin HH,
Wu WJ, Li CC, Yeh HC, Chang LL, Hsu WC and Ke HL: Over-expression
of activated signal transducer and activator of transcription 3
predicts poor prognosis in upper tract urothelial carcinoma. Int J
Med Sci. 14:1360–1367. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ni Z, Lou W, Lee SO, Dhir R, DeMiguel F,
Grandis JR and Gao AC: Selective activation of members of the
signal transducers and activators of transcription family in
prostate carcinoma. J Urol. 167:1859–1862. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu S, Li L, Zhang Y, Zhang Y, Zhao Y, You
X, Lin Z, Zhang X and Ye L: The oncoprotein HBXIP uses two pathways
to up-regulate S100A4 in promotion of growth and migration of
breast cancer cells. J Biol Chem. 287:30228–30239. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhou X, Xia Y, Su J and Zhang G:
Down-regulation of miR-141 induced by helicobacter pylori promotes
the invasion of gastric cancer by targeting STAT4. Cell Physiol
Biochem. 33:1003–1012. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Silver DL, Naora H, Liu J, Cheng W and
Montell DJ: Activated signal transducer and activator of
transcription (STAT) 3: Localization in focal adhesions and
function in ovarian cancer cell motility. Cancer Res. 64:3550–3558.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang G, Chen JH, Qiang Y, Wang DZ and Chen
Z: Decreased STAT4 indicates poor prognosis and enhanced cell
proliferation in hepatocellular carcinoma. World J Gastroenterol.
21:3983–3993. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kuo YH, Chen YT, Tsai HP, Chai CY and Kwan
AL: Nucleophosmin overexpression is associated with poor survival
in astrocytoma. APMIS. 123:515–522. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang CG, Ye YJ, Yuan J, Liu FF, Zhang H
and Wang S: EZH2 and STAT6 expression profiles are correlated with
colorectal cancer stage and prognosis. World J Gastroenterol.
16:2421–2427. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Nivarthi H, Gordziel C, Themanns M, Kramer
N, Eberl M, Rabe B, Schlederer M, Rose-John S, Knösel T, Kenner L,
et al: The ratio of STAT1 to STAT3 expression is a determinant of
colorectal cancer growth. Oncotarget. 7:51096–51106. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sun Y, Han Y, Wang X, Wang W, Wang X, Wen
M, Xia J, Xing H, Li X and Zhang Z: Correlation of EGFR Del 19 with
Fn14/JAK/STAT signaling molecules in non-small cell lung cancer.
Oncol Rep. 36:1030–1040. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Jeon M, You D, Bae SY, Kim SW, Nam SJ, Kim
HH, Kim S and Lee JE: Dimerization of EGFR and HER2 induces breast
cancer cell motility through STAT1-dependent ACTA2 induction.
Oncotarget. 8:50570–50581. 2016.PubMed/NCBI
|
|
56
|
Chen G, Wang Y, Wu P, Zhou Y, Yu F, Zhu C,
Li Z, Hang Y, Wang K, Li J, et al: Reversibly stabilized polycation
nanoparticles for combination treatment of early- and late-stage
metastatic breast cancer. ACS Nano. 12:6620–6636. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Merk BC, Owens JL, Lopes MB, Silva CM and
Hussaini IM: STAT6 expression in glioblastoma promotes invasive
growth. BMC Cancer. 11:1842011. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Sumiyoshi H, Matsushita A, Nakamura Y,
Matsuda Y, Ishiwata T, Naito Z and Uchida E: Suppression of STAT5b
in pancreatic cancer cells leads to attenuated gemcitabine
chemoresistance, adhesion and invasion. Oncol Rep. 35:3216–3226.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Tian X, Guan W, Zhang L, Sun W, Zhou D,
Lin Q, Ren W, Nadeem L and Xu G: Physical interaction of STAT1
isoforms with TGF-β receptors leads to functional crosstalk between
two signaling pathways in epithelial ovarian cancer. J Exp Clin
Cancer Res. 37:1032018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wei M, Liu B, Gu Q, Su L, Yu Y and Zhu Z:
Stat6 cooperates with Sp1 in controlling breast cancer cell
proliferation by modulating the expression of p21(Cip1/WAF1) and
p27 (Kip1). Cell Oncol (Dordr). 36:79–93. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Morgan EL, Wasson CW, Hanson L, Kealy D,
Pentland I, McGuire V, Scarpini C, Coleman N, Arthur JSC, Parish
JL, et al: STAT3 activation by E6 is essential for the
differentiation-dependent HPV18 life cycle. PLoS Pathog.
14:e10069752018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hosui A, Klover P, Tatsumi T, Uemura A,
Nagano H, Doki Y, Mori M, Hiramatsu N, Kanto T, Hennighausen L, et
al: Suppression of signal transducers and activators of
transcription 1 in hepatocellular carcinoma is associated with
tumor progression. Int J Cancer. 131:2774–2784. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhang Y, Cheng X, Liang H and Jin Z: Long
non-coding RNA HOTAIR and STAT3 synergistically regulate the
cervical cancer cell migration and invasion. Chem Biol Interact.
286:106–110. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Khan MA, El-Gamal MI and Oh CH: A
progressive review of V600E-B-RAF-dependent melanoma and drugs
inhibiting it. Mini Rev Med Chem. 17:351–365. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Liu LJ, Wang W, Huang SY, Hong Y, Li G,
Lin S, Tian J, Cai Z, Wang HD, Ma DL and Leung CH: Inhibition of
the Ras/Raf interaction and repression of renal cancer xenografts
in vivo by an enantiomeric iridium(iii) metal-based compound. Chem
Sci. 8:4756–4763. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Bosserhoff AK: Novel biomarkers in
malignant melanoma. Clin Chim Acta. 367:28–35. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Kugel CH III and Aplin AE: Adaptive
resistance to RAF inhibitors in melanoma. Pigment Cell Melanoma
Res. 27:1032–1038. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Harpio R and Einarsson R: S100 proteins as
cancer biomarkers with focus on S100B in malignant melanoma. Clin
Biochem. 37:512–518. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Xiong TF, Pan FQ and Li D: Expression and
clinical significance of S100 family genes in patients with
melanoma. Melanoma Res. 29:23–29. 2019. View Article : Google Scholar : PubMed/NCBI
|