Introduction
According to the World Health Organization
statistics in 2018, lung cancer is the sixth leading cause of
cancer-associated mortality worldwide and NSCLC accounts for ~85%
of all lung cancer cases (1,2). Although a number of patients with lung
cancer may benefit from chemotherapy, radiotherapy or molecular
targeted therapy, more effective immunotherapies need to be
developed to aid our understanding of the molecular characteristics
of lung tumor tissues.
The body is able to recognize and destroy cancer
cells through immune surveillance mechanisms (3,4).
However, certain characteristics of cancer cells may lead to immune
tolerance and can be induced by multiple mechanisms in the tumor
microenvironment (TME), including a reduction in the expression of
co-stimulatory molecules and cytokines and through the expression
of negative immunoregulatory molecules (5,6).
Cytokines serve an important role in the antitumor immune response
(7,8); therefore, investigation of cytokine
expression levels in the TME may provide valuable novel insight
into the underlying molecular mechanisms of tumor behavior for
cancer immunotherapy.
Interleukin (IL)-36 is a member of the IL-1 family
and has several subtypes, including IL-36α, IL-36β, IL-36γ and
IL-36 receptor antagonist (9).
IL-36γ interacts with the IL-36 receptor/IL-1RAcP, activating the
NF-κB and mitogen-activated protein kinase signaling pathways.
These pathways result in the production of inflammatory mediators,
such as cytokines and chemokines, and regulate autoimmune diseases,
inflammatory responses and antitumor immune responses. IL-36γ is
primarily expressed in peripheral blood lymphocytes, keratinocytes
and bronchial epithelial cells (9).
In addition, human macrophages and murine dendritic cells (DCs)
express IL-36γ following stimulation by the toll-like receptor or
lipopolysaccharides (10,11). Previous studies have demonstrated
that IL-36γ induces autoimmune diseases such as psoriasis, allergic
rhinitis (11) and allergic asthma
(12), and is associated with type-1
immune responses (13–15). High IL-36γ expression levels can
stimulate immune differentiation of Th1-type cells, contributing to
a positive immune response to infectious diseases (16,17).
IL-36γ-transfected DCs can upregulate the expression levels of
T-bet, a T-box transcription factor, transforming the TME and
promoting the development of lymphoid organs and inhibiting tumor
growth (10,18). IL-36γ is a novel antitumor cytokine
that can promote proliferation of CD4+ T lymphocytes,
CD8+ T lymphocytes, NK cells and γδT cells in
vitro and in vivo, promoting tumor eradication in the
TME (7).
A previous study demonstrated that a low expression
level of IL-33, another member of the IL-1 family, was associated
with poor prognosis in patients with lung adenocarcinoma (19). Therefore, the present study aimed to
determine if IL-36γ had a similar association with the prognosis of
patients with non-small cell lung carcinoma (NSCLC). By reviewing
the The National Center for Biotechnology Information Gene
Expression Database (NCBI GEO) database (ncbi.nlm.nih.gov/geo), it was identified that IL-36γ
was expressed in lung cancer, especially in lung squamous cell
carcinoma. A previous study demonstrated that IL-36γ greatly
promoted the proliferation and activation of CD8+ cells
and enhanced the antitumor immune response using animal models
(7). Therefore, the present study
retrospectively analyzed clinical tissue specimens to investigate
the value of IL-36γ expression levels in the treatment and
diagnosis of patients with NSCLC. Immunohistochemistry and
quantitative (q)PCR was used to investigate IL-36γ mRNA and protein
expression levels during the progression of NSCLC, and to establish
the association between IL-36γ and the clinical and pathological
parameters of patients with NSCLC.
Materials and methods
Specimens
IL-36γ tissue microarrays of lung adenocarcinoma and
squamous cell lung cancer were purchased from the Shanghai Xinchao
Biological Technology Co., Ltd. Each chip contained 150 tissues,
including 75 tumor tissues and 75 corresponding adjacent normal
tissues. Among the 75 lung squamous cell carcinoma tissues, one was
classified as large cell carcinoma and was excluded from the
follow-up analysis. Immunohistochemistry was performed on the
tissue chips by Shanghai Xinchao Biological Technology Co., Ltd.,
in accordance with standard procedures.
Tumor tissues and adjacent normal tissues were also
collected from patients with NSCLC (age range, 38–76 years; median
age, 63 years; 40 men, 30 women) following surgery at The Third
Affiliated Hospital of Soochow University between March and
December 2009, and between January 2014 and February 2015. The
samples of lung cancer tissue were confirmed as NSCLC by senior
pathologists based on tissue histopathology and morphology, and
there were 57 cases of lung adenocarcinoma and 42 cases of lung
squamous cell carcinoma. According to the Tumor-Node-Metastasis
(TNM) stage criteria for lung cancer by the International
Association for the Study of Lung Cancer (20), stages I and IIa were classified as
early cases, whereas stages IIb, III and IV were classified as
advanced cases (19). The tissues
(100 mg) were frozen and stored in nitrogen immediately (−196°C).
The present study was approved by The Ethics Committee of Soochow
University (Suzhou, China) and all patients provided informed
written consent.
Total RNA extraction and qPCR
Total RNA was extracted from patient tumor tissues
and adjacent tissues using TRIzol® reagent (Ambion;
Thermo Fisher Scientific, Inc.). RNA quality was assessed using 1%
agarose gel electrophoresis and absorbance was measured at 260/280
nm using a NanoDrop™ 2000 UV spectrophotometer (Thermo Fisher
Scientific, Inc.). RNA was then reverse transcribed into cDNA using
a Reverse Transcription kit (Applied Biosystems, Thermo Fisher
Scientific, Inc.) on a Bio-Rad T100™ Thermal Cycler (Bio-Rad
Laboratories, Inc.), and the reaction conditions were as follows:
25°C for 10 min, 37°C for 120 min and 85°C for 5 min, followed by
maintaining at 4°C. The qPCR assay was performed using a QuantiNova
SYBR PCR kit (Qiagen China Co., Ltd.) using a CFX96™ Real-Time
system (Bio-Rad Laboratories, Inc.). The qPCR cycling conditions
were as follows: Preheating at 95°C for 2 min, denaturation at 95°C
for 5 sec, annealing at 60°C for 10 sec and a final extension at
60°C for 10 sec, for 40 amplification cycles. The results were
quantified using the 2−ΔΔCq method (21). The primers were designed and
synthesized by Nanjing GenScript Biotech Corp. GAPDH was used as
the internal reference and all primer sequences are shown in
Table I.
 | Table I.IL-36γ and GAPDH primer
sequences. |
Table I.
IL-36γ and GAPDH primer
sequences.
| Gene | Primer sequence
(5′-3′) | Fragment length,
bp |
|---|
| IL-36γ |
| 112 |
|
Forward |
AGGTTGGAGAACAGCCCACATT |
|
|
Reverse |
GTCCTACCAGTCTTGGCACGG |
|
| GAPDH |
| 189 |
|
Forward |
GGAAGGTGAAGGTCGGAGTC |
|
|
Reverse |
CGTTCTCAGCCTTGACGGT |
|
Pathological scoring criteria
All tissue chip staining scores of IL-36γ protein
expression levels were independently assessed by two pathologists
under a light microscope at ×200 magnification. A positive signal
was identified when the cytoplasm or nucleus showed a dark brown
color. A total of 10 fields were randomly selected, and the protein
positive ratio and color intensity were scored. The staining
positive ratio was scored on a 5-point scale based on the
percentage of positive staining as follows: 0 points, <5%; 1
point, 6–25%; 2 points, 26–50%; 3 points, 51–75%; and 4 points,
>75%. Color intensity was scored on 4 levels as follows: 0
points, no color; 1 point, light yellow; 2 points, brown; and 3
points, dark brown. The final score was calculated by multiplying
the positive ratio and color intensity, with four levels as
follows: -, 0 points; +, 1–4 points; ++, 5–8 points; and +++, 9–12
points. Low expression levels were denoted as −/+ and high
expression levels were denoted as ++/+++ for statistical analysis
(22,23).
Bioinformatics
The GEO (https://www.ncbi.nlm.nih.gov/) (dataset no.
GDS3966/220322_at/IL-36γ) and Oncomine databases (https://www.oncomine.org/) were used to retrieve
IL-36γ expression data from human tumors.
Statistical analysis
Statistical analyses and graphing were performed
using GraphPad Prism 5.0 (GraphPad Software, Inc.). The data are
presented as the mean ± standard deviation. IL-36γ mRNA results
were obtained using the Mann-Whitney U test. Fisher's exact test
was used to analyze protein expression levels of IL-36γ or the
association between IL-36γ protein expression levels and clinical
parameters. A χ2 test was used to analyze the
association between IL-36γ protein expression levels and tumor
pathological grade. Patient survival was analyzed using the
Kaplan-Meier survival analysis and log-rank test, and the Cox
hazard ratio model. P<0.05 was considered to indicate a
statistically significant difference.
Results
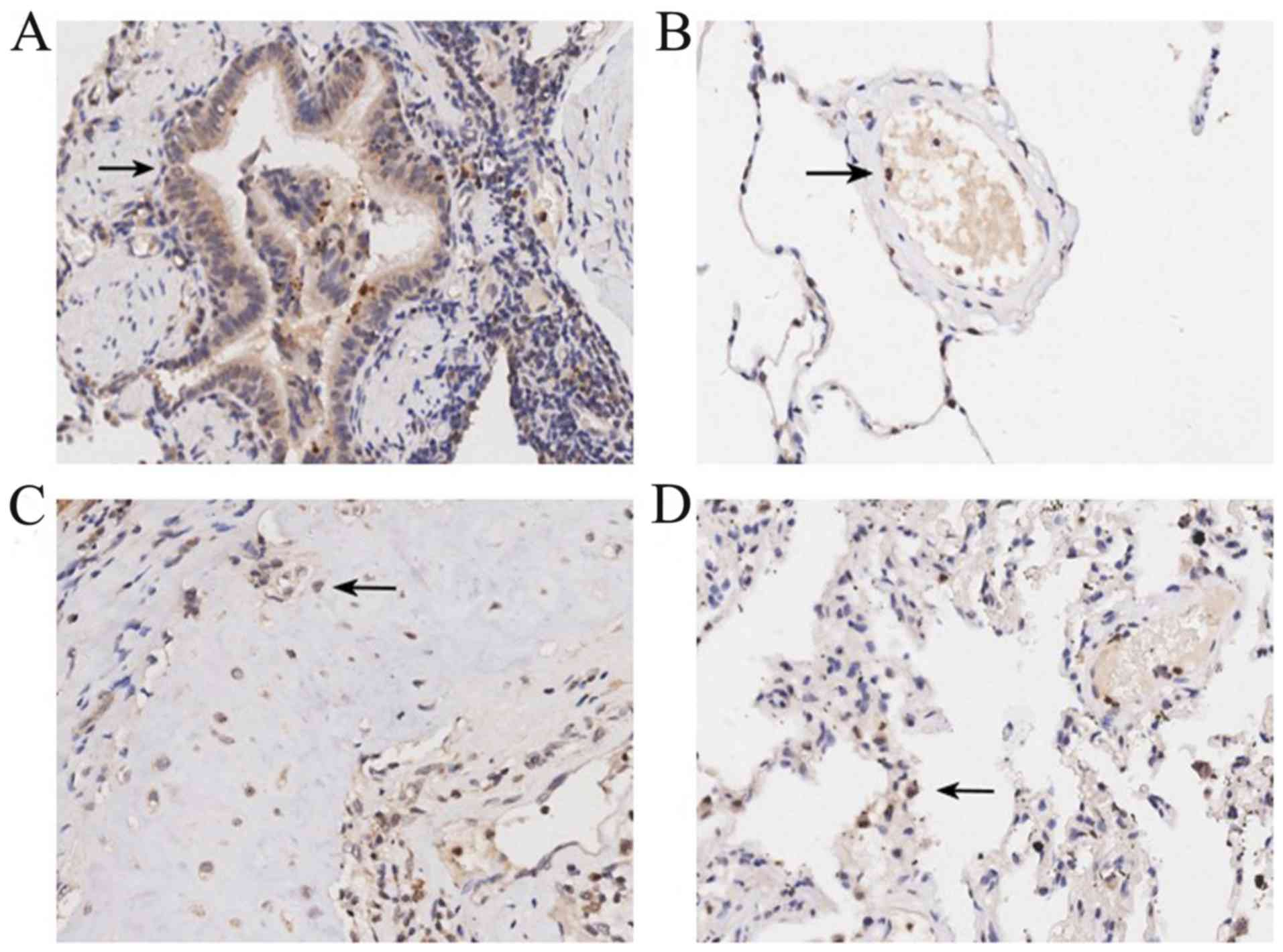
IL-36γ protein is expressed in normal
tissues
Immunohistochemical analysis of the tissue
microarrays was used to explore the expression patterns of IL-36γ
protein in tumor-adjacent normal tissues compared with NSCLC
tissues. Positive expression signals of IL-36γ were primarily
located in the cytoplasm, with weaker staining identified in the
nucleus, shown as brown particles. In tumor-adjacent normal
tissues, IL-36γ was expressed in various cell types, including
bronchial epithelial cells (Fig.
1A), vascular endothelial cells (Fig. 1B), chondrocytes (Fig. 1C) and alveolar epithelial cells
(Fig. 1D).
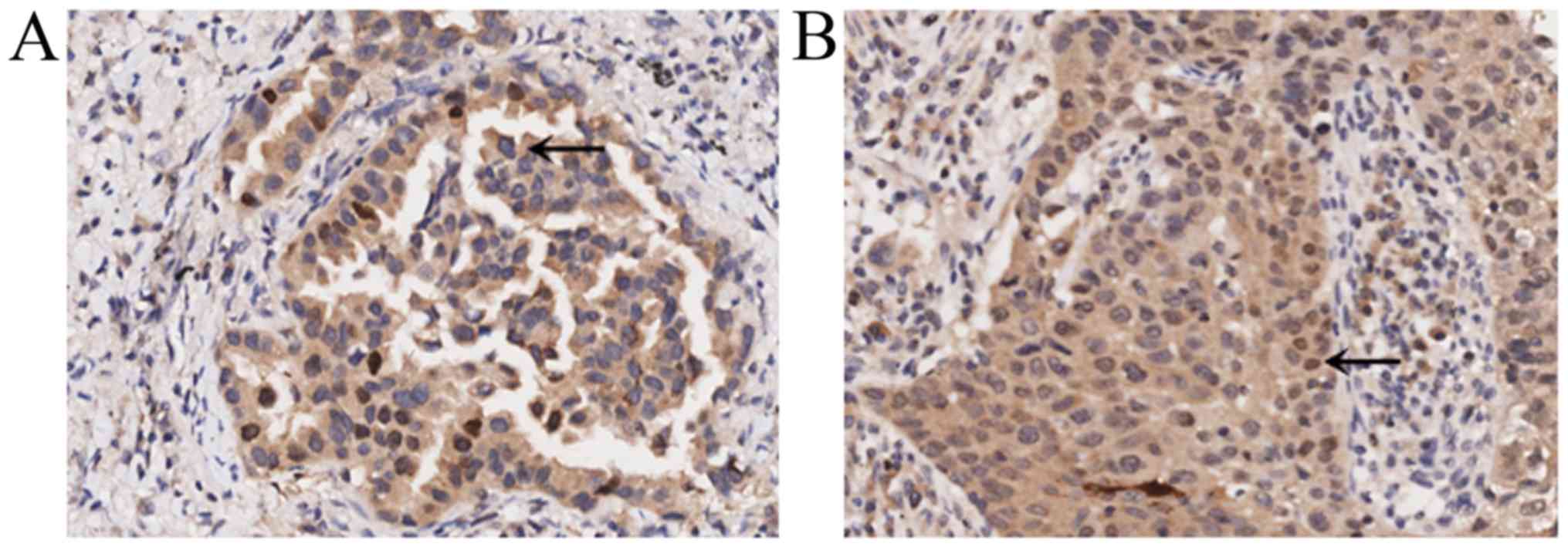
IL-36γ protein is expressed in tumor
cells
Based on the results of the immunohistochemical
staining, it was revealed that IL-36γ was expressed in lung cancer
cells, including lung adenocarcinoma (Fig. 2A) and lung squamous cell carcinoma
(Fig. 2B). Positive expression
signals (brown particles) of IL-36γ were also primarily located in
the cytoplasm, with weaker staining identified in the nucleus.
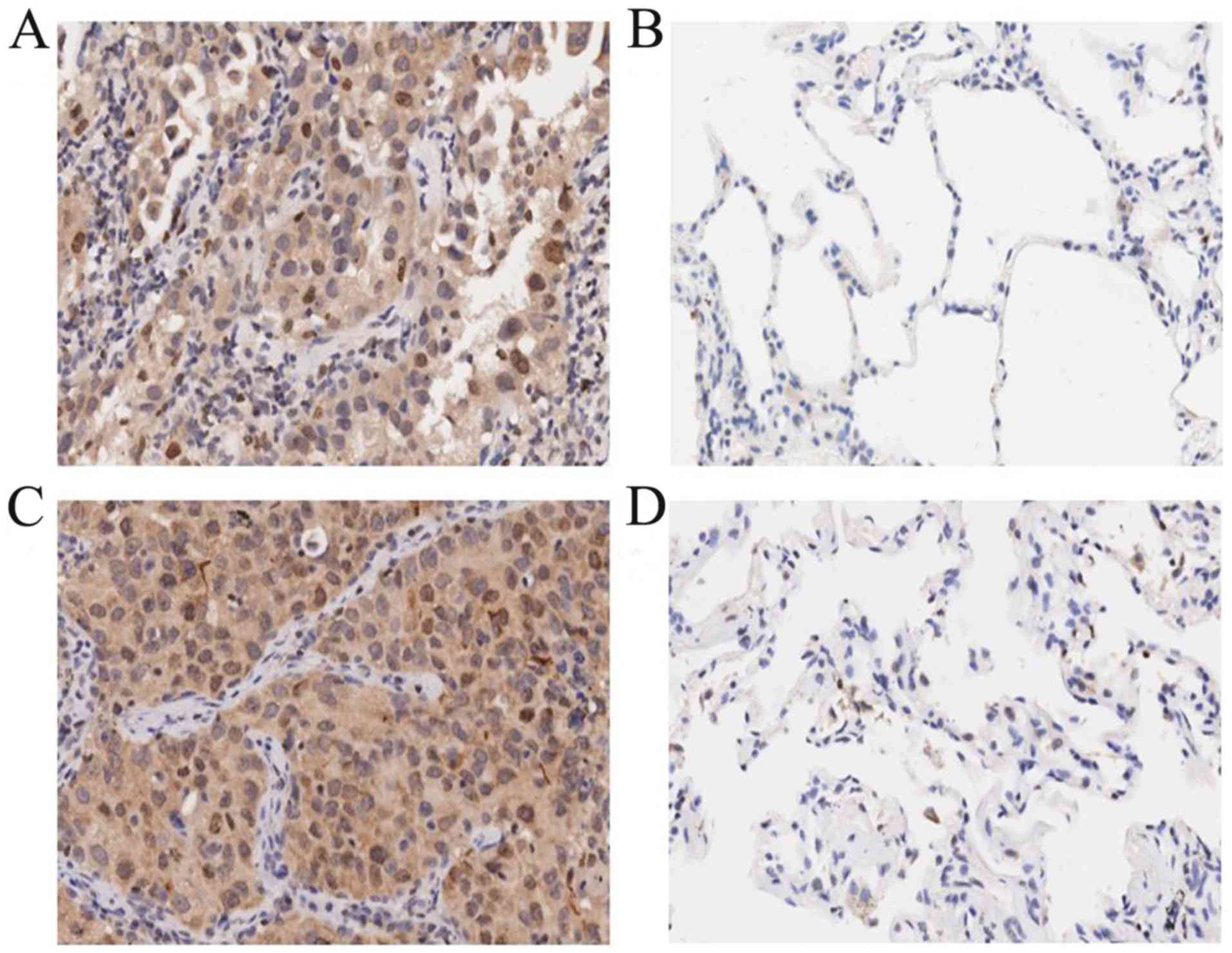
IL-36γ protein and mRNA expression
levels in tumor tissues of lung adenocarcinoma and squamous cell
carcinoma are significantly higher compared with those in adjacent
normal tissues
Based on the pathological scoring criteria, IL-36γ
protein expression levels were evaluated in 75 lung adenocarcinoma
tumor tissues, 74 squamous cell carcinoma tissues and the
corresponding adjacent normal tissues. IL-36γ protein expression
levels were higher in the cancer tissues compared with those in the
corresponding adjacent normal tissues (Fig. 3). Among the 75 patients with lung
adenocarcinoma, 39 (52%) exhibited higher IL-36γ expression levels
in tumor tissues, whereas only 2 (3%) exhibited higher IL-36γ
expression levels in adjacent normal tissues (P<0.0001; Table II). Among the 74 patients with
squamous cell carcinoma, 42 (57%) exhibited significantly higher
IL-36γ expression levels in tumor tissues, whereas only 1 (1%) of
the adjacent tissue samples exhibited higher IL-36γ expression
levels (P<0.0001; Table
III).
 | Table II.Interleukin-36γ expression levels in
lung adenocarcinoma and adjacent normal tissues (n=75). |
Table II.
Interleukin-36γ expression levels in
lung adenocarcinoma and adjacent normal tissues (n=75).
| Group | Low expression, n
(%) | High expression, n
(%) | P-value |
|---|
| Adenocarcinoma | 36 (48.0) | 39 (52.0) | <0.0001 |
| Adjacent
tissues | 73 (97.3) | 2 (2.7) | <0.0001 |
 | Table III.Interleukin-36γ expression levels in
lung squamous cell carcinoma and adjacent normal tissues
(n=74). |
Table III.
Interleukin-36γ expression levels in
lung squamous cell carcinoma and adjacent normal tissues
(n=74).
| Group | Low expression, n
(%) | High expression, n
(%) | P-value |
|---|
| Squamous cell
carcinoma | 32 (43.2) | 42 (56.8) | <0.0001 |
| Adjacent
tissues | 73 (98.6) | 1 (1.4) | <0.0001 |
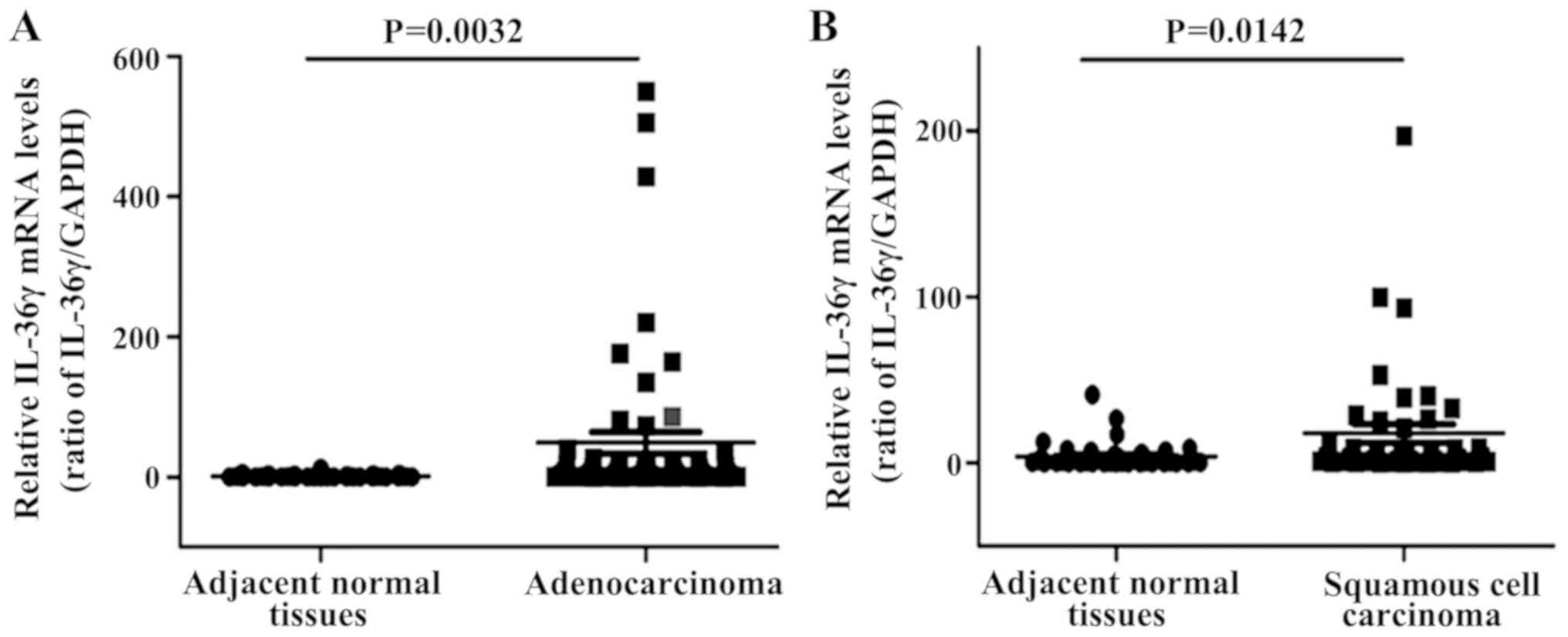
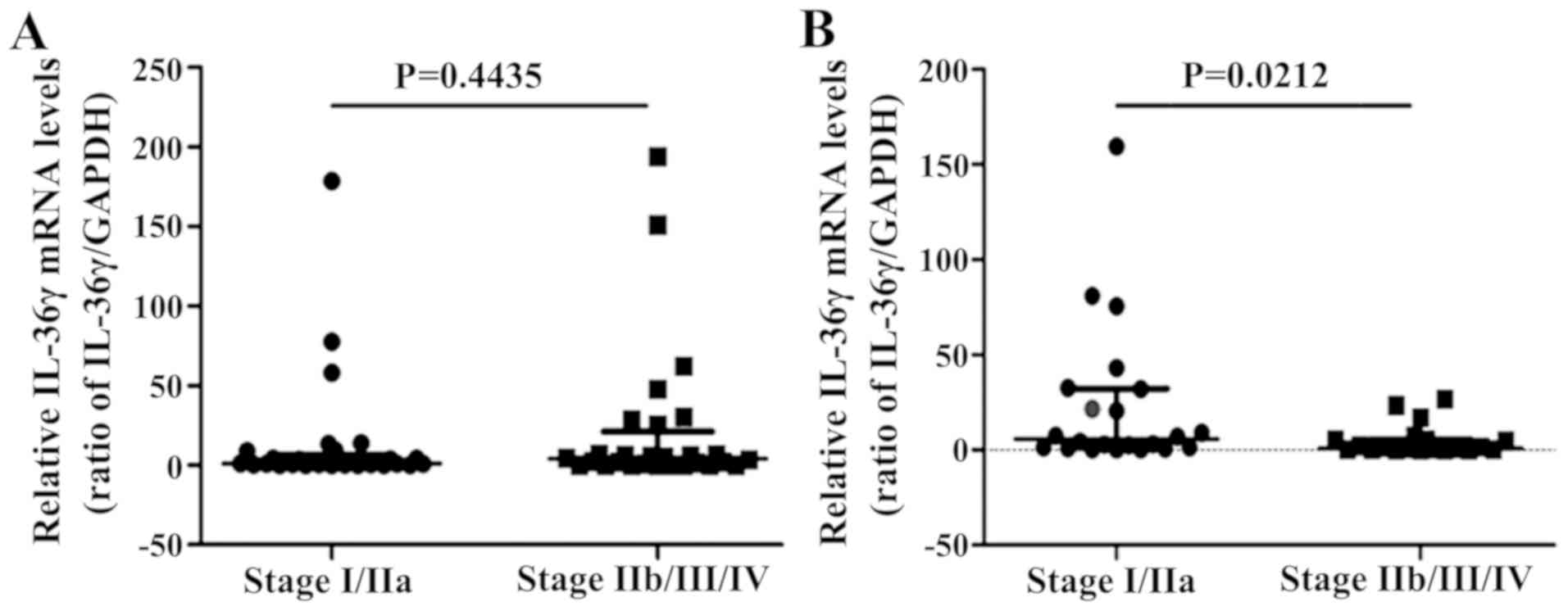
IL-36γ mRNA expression levels were also analyzed in
patients recruited from the Third Affiliated Hospital of Soochow
University, including 57 cases of lung adenocarcinoma (29 cases in
stage I/IIa and 28 cases in stage IIb/III/IV) and 42 cases of
squamous cell carcinoma (22 cases in stage I/IIa and 20 cases in
stage IIb/III/IV). IL-36γ mRNA expression levels were significantly
increased in both lung adenocarcinoma and squamous cell carcinoma
tumor tissues compared with those in normal tissues (P<0.01 and
P<0.05, respectively; Fig.
4).
High IL-36γ protein and mRNA
expression levels are associated with tumor pathological grade in
lung adenocarcinoma and clinical TNM stage in squamous cell
carcinoma
The association between IL-36γ protein expression
levels and the clinical pathological parameters of NSCLC were
investigated. Higher IL-36γ protein expression levels were
significantly associated with a higher tumor pathological grade of
lung adenocarcinoma (P<0.05; Table
IV). Meanwhile, there was no association between IL-36γ protein
expression level and all other assessed clinical pathological
parameters in the 74 cases of lung squamous cell carcinoma
(Table V). However, IL-36γ mRNA
expression level was inversely associated with the clinical TNM
stage of the patients with squamous cell carcinoma, which was lower
in the late stages (stage IIb/III/IV) than in the early stages
(stage I/IIa) (P<0.05; Fig.
5B).
 | Table IV.Association between IL-36γ protein
expression levels and clinicopathological features of patients with
adenocarcinoma. |
Table IV.
Association between IL-36γ protein
expression levels and clinicopathological features of patients with
adenocarcinoma.
|
|
| IL-36γ expression
levels |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
feature | n (%) | −/+ | ++/+++ | P-value |
|---|
| Sex |
|
|
| 0.6466 |
|
Men | 40/75 (53.3) | 18 | 22 |
|
|
Women | 35/75 (46.7) | 18 | 17 |
|
| Age, years |
|
|
| 0.8147 |
|
<60 | 31/73 (42.5) | 16 | 15 |
|
|
≥60 | 42/73 (57.5) | 20 | 22 |
|
| Pathological
grade |
|
|
| 0.0302a |
| I | 13/75 (17.3) | 8 | 5 |
|
| II | 49/75 (65.3) | 26 | 23 |
|
|
III | 13/75 (17.3) | 2 | 11 |
|
| Tumor size, cm |
|
|
| 0.2781 |
|
<5.5 | 58/75 (77.3) | 30 | 28 |
|
|
≥5.5 | 17/75 (22.7) | 6 | 11 |
|
| Lymph node
metastasis |
|
|
| 0.3668 |
|
N0-N1 | 43/57 (75.4) | 18 | 25 |
|
|
N2-N3 | 14/57 (24.6) | 8 | 6 |
|
| Clinical stage |
|
|
| >0.9999 |
|
I/IIa | 36/58 (62.1) | 17 | 19 |
|
|
IIb/III/IV | 22/58 (37.9) | 11 | 11 |
|
 | Table V.Association between IL-36γ protein
expression levels and clinicopathological features of patients with
squamous cell carcinoma. |
Table V.
Association between IL-36γ protein
expression levels and clinicopathological features of patients with
squamous cell carcinoma.
|
|
| IL-36γ expression
levels |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
feature | n (%) | −/+ | ++/+++ | P-value |
|---|
| Sex |
|
|
|
|
|
Men | 68/74 (91.9) | 31 | 37 | 0.2258 |
|
Women | 6/74 (8.1) | 1 | 5 |
|
| Age, years |
|
|
|
|
|
<60 | 26/73 (35.6) | 13 | 13 | 0.4682 |
|
≥60 | 47/73 (64.4) | 19 | 28 |
|
| Pathological
grade |
|
|
| 0.1475 |
| I | 5/74 (6.7) | 3 | 2 |
|
| II | 61/74 (82.4) | 28 | 33 |
|
|
III | 8/74 (10.8) | 1 | 7 |
|
| Tumor size, cm |
|
|
| 0.3312 |
|
<5.5 | 47/74 (63.5) | 18 | 29 |
|
|
≥5.5 | 27/74 (36.5) | 14 | 13 |
|
| Lymph node
metastasis |
|
|
|
|
|
N0-N1 | 56/62 (90.3) | 26 | 30 | >0.9999 |
|
N2-N3 | 6/62 (9.7) | 3 | 3 |
|
| Clinical stage |
|
|
|
|
|
I/IIa | 40/62 (64.5) | 18 | 22 | 0.7929 |
|
IIb/III/IV | 22/62 (35.5) | 11 | 11 |
|
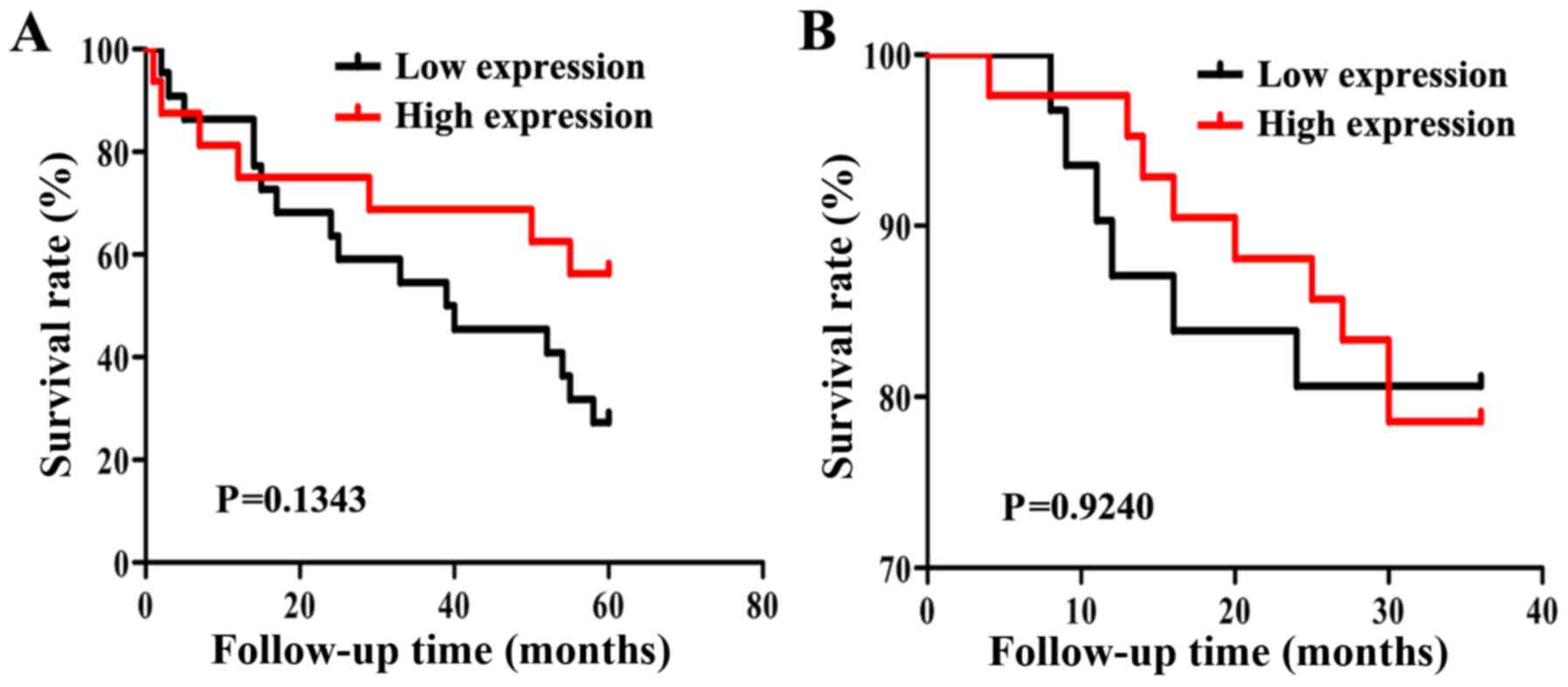
Association between IL-36γ protein
expression levels and prognosis in patients with NSCLC
After excluding patients with no clinical stage data
and a lack of follow-up data, 38 patients with lung adenocarcinoma
were followed for 5 years. The overall 5-year survival rate of
patients with NSCLC was 39% (15/38). In addition, 74 patients with
squamous cell carcinoma were followed up for 3 years and these
patients had a 3-year overall survival rate of 78% (58/74).
Kaplan-Meier survival analysis and a log-rank test demonstrated
that patients with lung adenocarcinoma and high IL-36γ protein
expression levels experienced a longer survival time; however, this
difference was not statistically significant (P=0.1343; Fig. 6A). In addition, IL-36γ protein
expression levels were not associated with survival in patients
with squamous cell carcinoma (P>0.05; Fig. 6B).
Association between clinical
parameters and survival of patients with NSCLC
A Cox hazard ratio model was also built. Univariate
and multivariate survival analyses were performed on IL-36γ
expression level, sex, age, pathological grade, tumor size and T
stage in patients with lung adenocarcinoma and squamous cell
carcinoma (Tables VI and VII). No correlation was discovered
between lung adenocarcinoma survival and any of the above variables
(Table VI). There was a significant
association between tumor size and survival in patients with lung
squamous cell carcinoma in the univariate and multivariate analyses
(Table VII). With HR<1 for
patients with squamous cell carcinoma, this suggests that the
smaller the tumor the longer the survival time (P<0.01; Table VII).
 | Table VI.Survival analysis using Cox's
regression model in patients with lung adenocarcinoma (n=75). |
Table VI.
Survival analysis using Cox's
regression model in patients with lung adenocarcinoma (n=75).
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Clinicopathological
feature | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| IL36γ expression
levels (low:high) | 1.276
(0.815–1.999) | 0.2860 | 1.402
(0.880–2.234) | 0.1547 |
| Sex
(men:women) | 0.610
(0.344–1.083) | 0.0912 | 0.563
(0.286–1.108) | 0.0965 |
| Age (<60:≥60
years) | 1.163
(0.663–2.041) | 0.5980 | 0.917
(0.499–1.684) | 0.7797 |
| Pathological grade
(I:II:III) | 0.729
(0.446–1.192) | 0.2080 | 0.737
(0.380–1.429) | 0.3667 |
| Tumor size
(<5.5:≥5.5 cm) | 0.414
(0.147–1.164) | 0.0944 | 0.385
(0.080–1.849) | 0.2329 |
| T stage
(T1:T2:T3) | 0.659
(0.319–1.362) | 0.2600 | 1.011
(0.346–2.953) | 0.9844 |
 | Table VII.Survival analysis using Cox's
regression model in patients with squamous cell carcinoma
(n=74). |
Table VII.
Survival analysis using Cox's
regression model in patients with squamous cell carcinoma
(n=74).
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Clinicopathological
feature | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| IL36γ protein
expression levels (low:high) | 0.920
(0.535–1.583) | 0.7630 | 1.439
(0.698–2.967) | 0.3237 |
| Sex
(men:women) | 0.882
(0.372–2.088) | 0.7750 | 1.072
(0.393–2.925) | 0.8922 |
| Age (<60:≥60
years) | 0.659
(0.343–1.265) | 0.2100 | 0.526
(0.231–1.195) | 0.1248 |
| Pathological grade
(I:II:III) | 1.014
(0.570–1.804) | 0.9610 | 0.807
(0.399–1.631) | 0.5504 |
| Tumor size
(<5.5:≥5.5 cm) | 0.355
(0.172–0.735) | 0.0053a | 0.258
(0.104–0.642) | 0.0036a |
| T stage
(T1:T2:T3) | 0.902
(0.490–1.663) | 0.7420 | 1.552
(0.778–3.093) | 0.2122 |
Discussion
In the present study, IL-36γ mRNA and protein
expression levels were upregulated in NSCLC. Elevated IL-36γ
protein expression levels were significantly associated with a
higher tumor grade of lung adenocarcinoma, and IL-36γ mRNA
expression levels were inversely associated with clinical TNM stage
in patients with squamous cell carcinoma. In addition, higher
IL-36γ expression in patients with adenocarcinoma tended to prolong
survival, although this was not statistically significant. These
data suggest that IL-36γ may have an antitumor role in NSCLC.
IL-33 and IL-36, both members of the cytokine IL-1
family, primarily function as an ‘alarmins’, which are released
following tissue injury (9) or
during infection (11,24) and are associated with the antitumor
immune response (7,25,26).
These cytokines can enhance the function of immune cells such as
CD8+ T lymphocytes and NK cells by promoting the
secretion and expression of effector cytokines, thereby functioning
in the antitumor immune response (7,25). In
our previous study, IL-33 had an antitumor effect in NSCLC
(19). IL-33 expression levels were
downregulated in tumor tissues and upregulated IL-33 expression
levels were associated with longer survival times in patients with
lung adenocarcinoma. Therefore, the present study aimed to
determine whether IL-36 had a similar effect.
As a subtype of IL-36, IL-36γ functions in a variety
of skin inflammatory reactions and immunopathological processes,
such as psoriasis, inflammatory megacolon and infectious diseases
(16,27–31).
Previous studies have demonstrated the involvement of IL-36γ in the
differentiation of Th cells and type-1 immune responses (7,16,17). Our
previous study showed that IL-36γ promoted cell activation and
expressed IFN-γ, granzyme-B and other type 1 effectors in
vitro by stimulating cultured human peripheral blood
CD4+ T lymphocytes and CD8+ T lymphocytes
(7). Therefore, due to these
characteristics of IL-36γ, melanoma tumor cells and breast cancer
cells have previously been transfected to overexpress full-length
IL-36γ in our laboratory (7). A
mouse model demonstrated that IL-36γ overexpression inhibited
tumorigenesis and metastasis by promoting the proliferation of
CD8+ T and NK cells, and the production of the effector
cytokines IFN-γ and TNF-α. Thus, the survival time of tumor-bearing
mice was prolonged. In addition, IL-36γ is also used as an adjuvant
for tumor vaccines to induce antigen-specific immune responses
(7). A recent breast cancer lung
metastasis model study indicated that IL-36γ has an important
effect in improving the antitumor immune response by enhancing the
type-1 immune response, inhibiting lung metastasis (32). Overall, these studies suggest that
IL-36γ, as an inflammatory cytokine, may serve an important role in
inflammatory diseases and antitumor immunotherapy.
The IL-36γ protein is expressed in keratinocytes,
bronchial epithelial cells and brain tissue, and IL-36γ expression
levels in macrophages and neutrophils are significantly increased
during infection (9,31,33).
IL-36γ may affect a variety of cells, including stromal cells, DCs,
macrophages and lymphocytes by inducing a series of related
inflammatory responses, including the promotion of synthesis and
activity of IL-12, IL-8 and IL-6, and the chemokines CXCL1 and
CCL20 (31,33–35). In
the present study, immunohistochemistry of tissue microarrays
showed that IL-36γ is expressed in various cell types, including
bronchial epithelial cells, vascular endothelial cells,
chondrocytes and alveolar epithelial cells. Positive signals were
primarily located in the cytoplasm, with weak staining in the
nucleus. According to previous reports and the NCBI GEO and
Oncomine databases, IL-36γ is expressed in several other tumor
tissues, including melanoma, colorectal cancer, head and neck
cancer and lung cancer. The results of the present study also
suggest that NSCLC cells express high levels of IL-36γ in a diffuse
pattern.
IL-36γ mRNA and protein expression levels were
significantly increased in NSCLC tissues compared with those in
adjacent normal tissues in the present study. Higher IL-36γ protein
expression levels in adenocarcinoma tissues were significantly
associated with higher tumor pathological grades, but there was no
association observed in squamous cell carcinoma. IL-36γ mRNA
expression levels in squamous cell carcinoma were inversely
associated with the clinical TNM stage, which is consistent with a
previous report investigating melanoma and lung cancer progression
that demonstrated that IL-36γ expression levels were higher in the
early stage compared with those in the advanced stage (7). It suggestes a potential antitumor
effect of IL-36γ in squamous cell carcinoma.
Furthermore, in the present study, survival analysis
showed that patients with adenocarcinoma with high IL-36γ protein
expression levels had longer survival times (P>0.05); however,
the lack of information on patient treatment is a limitation when
evaluating the survival time of patients. In addition, the Cox risk
model indicated that the survival of patients with squamous cell
carcinoma was associated with tumor size.
Prior to the present study, there have been a few
studies on the association between IL-36γ and tumors (7,32).
Although the underlying mechanism of IL-36γ in the antitumor immune
response has been studied in animal models, the role of IL-36γ in
human tumors is unclear (7). In the
present study, IL-36γ expression patterns in human tumor tissues
were investigated, aiming to determine the association between
IL-36γ mRNA and protein expression levels and clinical and
pathological parameters in NSCLC. The findings of the present study
have provided valuable information that may inform later studies of
potential mechanisms underlying the function of IL-36γ in NSCLC.
The present study may also provide novel insight into the value of
IL-36γ as an immunotherapy target for NSCLC treatment. Therefore,
further specimens should be collected and the sample size expanded
to further study the associations between IL-36γ mRNA and protein
expression levels and clinical parameters, and the mechanism
underlying IL-36γ function to better determine its value for
clinical application. Immunofluorescence co-localization of
cellular markers (such as CD4, CD8 and CD56) and IL-36γ in tumor
tissues may aid the identification of cell types that secret
IL-36γ, facilitating further investigation of the underlying
mechanisms of IL-36γ function.
Acknowledgements
Not applicable.
Funding
The present study was funded by The National Natural
Science Foundation of China (grant nos. 81273208 and 31607916), The
Major Basic Research Project of the Natural Science Foundation of
the Jiangsu Higher Education Institutions (grant no. 18KJA180011)
and The Suzhou Applied Basic Research-Medical Guidance Program
(grant no. SYSD2016106).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JW and YZ designed and directed the study. LL and HH
conducted the experiments and wrote the original manuscript. DX, HZ
and LS analyzed the data. YF and YG collected clinical specimens
and acquired the data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by The Ethics
Committee of Soochow University (Suzhou, China) and all patients
provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
DCs
|
dendritic cells
|
|
IL-36γ
|
interleukin-36γ
|
|
NSCLC
|
non-small cell lung cancer
|
|
qPCR
|
quantitative PCR
|
|
TME
|
tumor microenvironment
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Miura Y and Sunaga N: Role of
immunotherapy for oncogene-driven non-small cell lung cancer.
Cancers (Basel). 10:E2452018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Burnet M: Cancer; A biological approach.
I. The processes of control. Br Med J. 1:779–786. 1957. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schreiber RD, Old LJ and Smyth MJ: Cancer
immunoediting: Integrating immunity's roles in cancer suppression
and promotion. Science. 331:1565–1570. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sugie T: Immunotherapy for metastatic
breast cancer. Chin Clin Oncol. 7:282018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mellman I, Coukos G and Dranoff G: Cancer
immunotherapy comes of age. Nature. 480:480–489. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang X, Zhao X, Feng C, Weinstein A, Xia
R, Wen W, Lv Q, Zuo S, Tang P, Yang X, et al: IL-36γ transforms the
tumor microenvironment and promotes type 1 lymphocyte-mediated
antitumor immune responses. Cancer Cell. 28:296–306. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lippitz BE: Cytokine patterns in patients
with cancer: A systematic review. Lancet Oncol. 14:e218–228. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gresnigt MS and van de Veerdonk FL:
Biology of IL-36 cytokines and their role in disease. Semin
Immunol. 25:458–465. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bassoy EY, Towne JE and Gabay C:
Regulation and function of interleukin-36 cytokines. Immunol Rev.
281:169–178. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qin X, Liu M, Zhang S, Wang C and Zhang T:
The role of IL-36γ and its regulation in eosinophilic inflammation
in allergic rhinitis. Cytokine. 117:84–90. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ramadas RA, Li X, Shubitowski DM, Samineni
S, Wills-Karp M and Ewart SL: IL-1 receptor antagonist as a
positional candidate gene in a murine model of allergic asthma.
Immunogenetics. 58:851–855. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Blumberg H, Dinh H, Trueblood ES,
Pretorius J, Kugler D, Weng N, Kanaly ST, Towne JE, Willis CR,
Kuechle MK, et al: Opposing activities of two novel members of the
IL-1 ligand family regulate skin inflammation. J Exp Med.
204:2603–2614. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Johnston A, Xing X, Guzman AM, Riblett M,
Loyd CM, Ward NL, Wohn C, Prens EP, Wang F, Maier LE, et al:
IL-1F5, -F6, -F8, and -F9: A novel IL-1 family signaling system
that is active in psoriasis and promotes keratinocyte antimicrobial
peptide expression. J Immunol. 186:2613–2622. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chustz RT, Nagarkar DR, Poposki JA,
Favoreto S Jr, Avila PC, Schleimer RP and Kato A: Regulation and
function of the IL-1 family cytokine IL-1F9 in human bronchial
epithelial cells. Am J Respir Cell Mol Biol. 45:145–153. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gresnigt MS, Rosler B, Jacobs CW, Becker
KL, Joosten LA, van der Meer JW, Netea MG, Dinarello CA and van de
Veerdonk FL: The IL-36 receptor pathway regulates aspergillus
fumigatus-induced Th1 and Th17 responses. Eur J Immunol.
43:416–426. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vigne S, Palmer G, Martin P, Lamacchia C,
Strebel D, Rodriguez E, Olleros ML, Vesin D, Garcia I, Ronchi F, et
al: IL-36 signaling amplifies Th1 responses by enhancing
proliferation and Th1 polarization of naive CD4+ T
cells. Blood. 120:3478–3487. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Weinstein AM, Chen L, Brzana EA, Patil PR,
Taylor JL, Fabian KL, Wallace CT, Jones SD, Watkins SC, Lu B, et
al: Tbet and IL-36γ cooperate in therapeutic DC-mediated promotion
of ectopic lymphoid organogenesis in the tumor microenvironment.
Oncoimmunology. 6:e13222382017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang M, Feng Y, Yue C, Xu B, Chen L, Jiang
J, Lu B and Zhu Y: Lower expression level of IL-33 is associated
with poor prognosis of pulmonary adenocarcinoma. PLoS One.
13:e01934282018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Feng SH and Yang ST: The new 8th TNM
staging system of lung cancer and its potential imaging
interpretation pitfalls and limitations with CT image
demonstrations. Diagn Interv Radiol. 25:270–279. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen C, Qu QX, Xie F, Zhu WD, Zhu YH and
Huang JA: Analysis of B7-H4 expression in metastatic pleural
adenocarcinoma and therapeutic potential of its antagonists. BMC
Cancer. 17:6522017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun J, Chen LJ, Zhang GB, Jiang JT, Zhu M,
Tan Y, Wang HT, Lu BF and Zhang XG: Clinical significance and
regulation of the costimulatory molecule B7-H3 in human colorectal
carcinoma. Cancer Immunol Immunother. 59:1163–1171. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liew FY, Pitman NI and McInnes IB:
Disease-associated functions of IL-33: The new kid in the IL-1
family. Nat Rev Immunol. 10:103–110. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li X, Lv Q, Feng Y, Gu Y, Xia R, Ma J, He
H and Zhu Y: Interleukin-33, a potential cytokine expressed in
tumor microenvironment involves in antitumor immunotherapy through
facilitates CD8(+) T cells. J Interferon Cytokine Res. 38:491–499.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao X, Chen X, Shen X, Tang P, Chen C,
Zhu Q, Li M, Xia R, Yang X, Feng C, et al: IL-36β promotes CD8(+) T
cell activation and antitumor immune responses by activating
mTORC1. Front Immunol. 10:18032019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Boutet MA, Bart G, Penhoat M, Amiaud J,
Brulin B, Charrier C, Morel F, Lecron JC, Rolli-Derkinderen M,
Bourreille A, et al: Distinct expression of interleukin (IL)-36α, β
and γ, their antagonist IL-36Ra and IL-38 in psoriasis, rheumatoid
arthritis and Crohn's disease. Clin Exp Immunol. 184:159–173. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tomuschat C, O'Donnell AM, Coyle D and
Puri P: Altered expression of IL36γ and IL36 receptor (IL1RL2) in
the colon of patients with Hirschsprung's disease. Pediatr Surg
Int. 33:181–186. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gardner JK and Herbst-Kralovetz MM: IL-36γ
induces a transient HSV-2 resistant environment that protects
against genital disease and pathogenesis. Cytokine. 111:63–71.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xiaoling Y, Chao W, Wenming W, Feng L and
Hongzhong J: Interleukin (IL)-8 and IL-36γ but not IL-36Ra are
related to acrosyringia in pustule formation associated with
palmoplantar pustulosis. Clin Exp Dermatol. 44:52–57. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wein AN, Dunbar PR, McMaster SR, Li ZT,
Denning TL and Kohlmeier JE: IL-36γ protects against severe
influenza infection by promoting lung alveolar macrophage survival
and limiting viral replication. J Immunol. 201:573–582. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen Y, Sun J, Huang Y, Liu Y, Liang L,
Yang D, Lu B and Li S: Targeted codelivery of doxorubicin and
IL-36γ expression plasmid for an optimal chemo-gene combination
therapy against cancer lung metastasis. Nanomedicine. 15:129–141.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huynh J, Scholz GM, Aw J, Kwa MQ, Achuthan
A, Hamilton JA and Reynolds EC: IRF6 regulates the expression of
IL-36γ by human oral epithelial cells in response to porphyromonas
gingivalis. J Immunol. 196:2230–2238. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Foster AM, Baliwag J, Chen CS, Guzman AM,
Stoll SW, Gudjonsson JE, Ward NL and Johnston A: IL-36 promotes
myeloid cell infiltration, activation, and inflammatory activity in
skin. J Immunol. 192:6053–6061. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Vigne S, Palmer G, Lamacchia C, Martin P,
Talabot-Ayer D, Rodriguez E, Ronchi F, Sallusto F, Dinh H, Sims JE
and Gabay C: IL-36R ligands are potent regulators of dendritic and
T cells. Blood. 118:5813–5823. 2011. View Article : Google Scholar : PubMed/NCBI
|