Introduction
Ovarian cancer is the cancer that carries the worst
prognosis among gynecological malignancies (1), and the majority of cases are diagnosed
at an advanced stage with peritoneal metastases. The standard
therapy for advanced ovarian cancer is primary debulking surgery
(PDS) followed by chemotherapy, and complete/optimal primary
cytoreduction can improve the survival (2). However, in cases where incomplete
surgery is anticipated, neoadjuvant chemotherapy (NAC) followed by
interval debulking surgery (IDS) is recommended as a treatment
option (3–5). Although platinum/taxane-based
chemotherapy is usually chosen as a 1st line regimen for NAC, a
significant number of patients show resistance to NAC. Furthermore,
it is a disadvantage that sufficient tumor samples may not be
acquired by cytology or laparoscopic biopsy before the start of
NAC. Some studies have indicated that change in the serum levels of
cancer antigen 125 (CA125) could serve as a predictor of monitoring
the response to NAC (6–8), however its usefulness is often limited
because no significant elevation of any specific serum tumor marker
is observed in some histological types of ovarian cancer. Thus,
novel and reliable molecular biomarkers related more closely to the
intrinsic molecular/biological characteristics of independent
ovarian cancers are needed, rather than the conventional serum
tumor markers to monitor the treatment response and predict the
efficiency of NAC.
Liquid biopsies of circulating tumor DNA (ctDNA)
released into the plasma from necrotic and apoptotic cancer tissues
have recently been used as a non-invasive diagnostic tool for
detecting tumor-specific gene mutations. Liquid biopsy has
advantages over tumor biopsy in that it is minimally invasive and
the samples can be analyzed in real-time and repeatedly.
Furthermore, the molecular characteristics reflected by liquid
biopsy may more closely represent tumor heterogeneity compared to
those reflected by tumor biopsy (9).
Recently, digital PCR (10,11) and next-generation sequencing (NGS)
(12,13) have enabled molecular profiling of
plasma ctDNA. Digital PCR-based methods have very high analytical
sensitivity for minor alleles (~0.01%) with improved specificity,
but in general, can only interrogate one or a few genomic positions
simultaneously (12,13). As a tool that allows these
limitations to be overcome, Cancer Personalized Profiling by deep
sequencing (CAPP-Seq) has recently been developed as the first
NGS-based ctDNA analysis method that allows both an ultralow
detection limit and broad patient coverage, allowing for a lower
amount of input DNA and a lower sequencing cost (14,15).
CAPP-Seq shows similar sensitivity for hotspot alleles as digital
PCR, and can simultaneously interrogate thousands of additional
genomic positions without its sensitivity or specificity being
affected. This ultrasensitive technique can detect plasma ctDNA in
patients with early and advanced stages of various human
malignancies, including lung cancer, lymphoma, and leiomyosarcoma
(15–17). We first reported the feasibility of
CAPP-Seq-based liquid biopsy in gynecological cancers including
ovarian cancer, cervical cancer, and endometrial cancer (18,19).
Otsubo et al reported using liquid biopsy-based CAPP-Seq,
that the T790M mutation of EGFR is associated with
amplification of MET, ERBB2, or EGFR in non-small
cell lung cancer (NSCLC) patients resistant to EGFR-TKIs (20). However, there have been no reports
about gene mutation profiles using CAPP-Seq for plasma ctDNA
obtained from advanced ovarian cancer patients receiving NAC.
In addition, recent studies have shown that the
tumor mutation burden (TMB) as determined by targeted NGS might be
associated with the response to immunotherapy in patients with lung
cancer (21). Furthermore,
tissue-based TMB (tTMB) has been reported to be positively
correlated with blood-TMB (bTMB) (22), suggesting bTMB could be a surrogate
marker of TMB. Gandara et al demonstrated that the bTMB
might be associated with the response to immune checkpoint
inhibitor therapy in patients with non-small lung cancer (22). It has been reported that CAPP-Seq may
be potentially useful as a technique for measuring the bTMB in
early/-advanced cancer patients and for monitoring the ctDNA during
treatment (14,15). In the gynecologic oncology field, it
has been reported that in patients with high grade serous ovarian
cancer, the TMB is associated with the treatment response and
survival (23), although there have
been no reports on the usefulness of the bTMB in patients with
gynecologic cancer.
In the present study, we applied CAPP-seq, a
ctDNA-based form of targeted NGS, to compare the variant allele
frequency (VAF) of tumor-derived somatic mutations in ctDNA
measured pre- and post-NAC in plasma samples obtained from ovarian
cancer patients receiving NAC. We also examined the changes of the
bTMB during NAC treatment as a potential novel biomarker.
Materials and methods
Patients and samples
This study was conducted in 10 patients who were
diagnosed as having stage III or IV ovarian cancer and received NAC
between May 2017 and February 2019 at Wakayama Medical University
Hospital. The initial diagnosis was based on the clinical findings,
including the findings of imaging [computed tomography (CT),
magnetic resonance imaging (MRI), and positron emission
tomography/computed tomography (PET/CT)] and the cytology/histology
of ascitic and pleural fluids. The regimen used for NAC was
paclitaxel (175 mg/m2, day 1) and carboplatin (5 areas
under the curve, day 1) with or without of bevacizumab (15 mg/kg,
day 1), every 21 days. Patient no. 1 developed allergy to
paclitaxel during the first course, and docetaxel (60
mg/m2) with carboplatin was administered during the
2nd/3rd course. Patient no. 8 received weekly paclitaxel (80
mg/m2, day 1/8/15/22) and bevacizumab (10 mg/kg, day
1/15) because of renal dysfunction. Patient no. 9 showed resistance
to paclitaxel and carboplatin, and the regimen was changed to
cisplatin (60 mg/m2, day 1) plus irinotecan (60
mg/m2, day 1/8/15). The number of cycles of NAC was
determined based on the clinical treatment response including the
clinical findings and serum levels of tumor markers, at the
discretion of the treating physician. Nine patients received IDS
following NAC. Patient no. 9 did not receive IDS because of her
poor performance status. The postoperative pathological diagnosis
was determined based on the findings of the resected tumors at the
time of IDS.
Blood samples were obtained pre- and post-NAC. The
response to chemotherapy was assessed using Response Evaluation
Criteria in Solid Tumors (RECIST) version 1.1 (24). In this study, patients with complete
response (CR) and partial response (PR) were defined as
NAC-sensitive, while those with stable disease (SD) and progressive
disease (PD) were defined as NAC-resistant. This study was
conducted with approval of the Ethics Committee of Wakayama Medical
University (authorization no. 2025) and Kindai University Faculty
of Medicine (authorization no. 29-066). Written informed consent
was obtained from each of the patients.
Circulating tumor DNA extraction
Peripheral blood samples (8.5 ml) were collected
from the patients in cell-free DNA collection tubes (Roche
Diagnostics). Plasma ctDNA was purified using an AVENIO cfDNA
isolation kit (Roche Diagnostics), in accordance with the
manufacturer's instructions. The quality and quantity of the DNA
was verified using the PicoGreen dsDNA assay kit (Life
Technologies; Thermo Fisher Scientific, Inc.). The extracted ctDNA
was stored at −80°C until the analysis.
Circulating tumor DNA sequencing
The CAPP-Seq of ctDNA (10–50 ng) was performed using
the AVENIO ctDNA surveillance kit (Roche Diagnostics, 197 genes) as
recently described (18–20). The purified libraries were pooled and
sequenced on an Illumina NextSeq 500 (Illumina) using the 300-cycle
high output kit. Variants were called with the AVENIO ctDNA
Analysis Software (Roche Diagnostics), which includes
bioinformatics methods from CAPP-Seq (14) and integrated digital error
suppression (15). Genetic variants
previously cataloged by the Exome Aggregation Consortium at a
frequency of ≥1% were excluded, and only non-synonymous single
nucleotide variants (SNVs), insertions-deletions (Indels), copy
number variations (CNVs), and gene fusions involving 197
cancer-related genes were extracted. Twenty plasma samples obtained
from 10 patients treated with NAC were successfully analyzed by
CAPP-Seq. The bTMB in each sample was evaluated as the number of
non-synonymous mutations number per Mb.
Statistical analysis
Statistical analyses were performed using the JMP
Pro statistical software version 13.1.1 for Windows (SAS Institute
Inc.). Statistical comparisons between the groups were performed
using the Mann-Whitney U test. Differences were considered to be
significant at P-values of <0.05.
Results
Patient characteristics
The clinicopathological characteristics and response
to NAC of the 10 patients are summarized in Table I. The median age of the patients was
63.5 years (44–74 years). Of the 10 patients, six (patient no. 1-6)
were NAC-sensitive, and four (patient no. 7-10) were NAC-resistant.
The postoperative histological diagnosis in all the NAC-sensitive
patients was high grade serous carcinoma. In the NAC-resistant
patient group, 2 cases had high-grade serous carcinoma, and one
case was mucinous carcinoma. We could not diagnose the histological
type in patient no. 9 because she did not receive IDS on account of
her poor performance status.
 | Table I.Clinico-pathological characteristics
of 10 NAC-treated patients with ovarian cancer. |
Table I.
Clinico-pathological characteristics
of 10 NAC-treated patients with ovarian cancer.
| Case no. | Age | Stage | Postoperative
histological diagnosis | Chemotherapy (no.
of cycles) | Clinical response
to NAC |
|---|
| 1 | 44 | III | HGSC | TC (1), DC (2) | PR |
| 2 | 71 | III | HGSC | TC (4) | PR |
| 3 | 72 | III | HGSC | TC (4) | PR |
| 4 | 70 | III | HGSC | TC (9) | CR |
| 5 | 48 | IV | HGSC | TC (3) | PR |
| 6 | 55 | IV | HGSC | TC (1), TC+Bev
(2) | PR |
| 7 | 51 | III | Mucinous
carcinoma | TC (3) | PD |
| 8 | 57 | III | HGSC | weekly PTX (1),
weeklyPTX+Bev (2) | PD |
| 9 | 72 | III | Not diagnosed | TC (3), CDDP+CPT-11
(1) | PD |
| 10 | 74 | IV | HGSC | TC (6) | PD |
Association of the ctDNA concentration
with the response to chemotherapy
The concentrations of ctDNA in plasma samples
collected pre- and post-NAC were measured by fluorometry (Fig. S1). The median ctDNA concentrations
in the pre- and post-NAC samples of NAC-sensitive patients were
2,335 and 2,198 copies/ml, respectively. On the other hand, in the
NAC-resistant cases, the median ctDNA concentrations in the pre-
and post-NAC samples were 3,558 and 5,155 copies/ml, respectively.
There were no significant differences in the baseline ctDNA
concentration between the NAC-sensitive and NAC-resistant patients.
There were no significant changes in the ctDNA concentration
between the pre- and post-NAC samples in either NAC-sensitive or
NAC-resistant patients.
Mutation status in pre- and post-NAC
ctDNA samples
The non-synonymous somatic mutations detected by
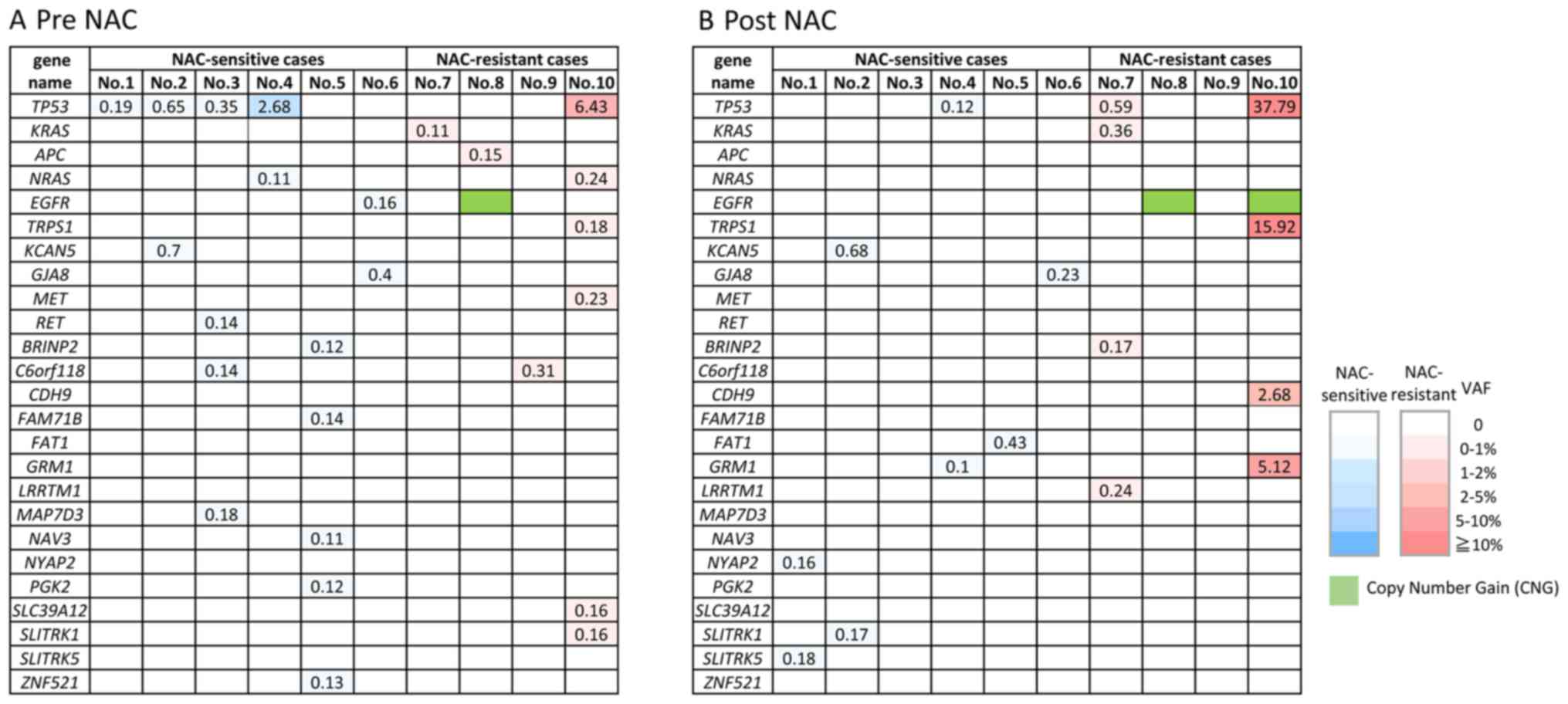
CAPP-Seq are shown in the NAC-sensitive and NAC-resistant patients
in ctDNA obtained pre-NAC (Fig. 1A)
and post-NAC (Fig. 1B). Various
types of non-synonymous mutations were detected in all the ctDNA
samples, however, TP53 mutations were the most frequently
detected (6/10, 60%). It is noteworthy that EGFR
amplification was detected in the post-NAC sample from the
NAC-resistant group (patient no. 8 and 10). Some of the mutations
such as NYAP2, SLITRK5, RET, GRM1, FAT1, LRRTM1, BRINP2,
CDH9 and GRM1 were detected only in the post NAC
samples, like EGFR amplification. These mutations,
especially LRRTM1, BRINP2, CDH9 and GRM1 might be related to
acquired resistance to NAC, however, it still remains unclear
whether these mutations are actionable or not.
On the other hand, mutations of several genes were
detected in the pre-NAC but not post-NAC samples: EGFR, APC,
NRAS, C6orf118, MAP7D3, BRINP2, NAV3, ZNF521, FAM71B, PGK2, ZFPM2,
NPAP1, SLC39A12 and MET. Interestingly, EGFR mutation
(p.L833F) detected in pre-NAC ctDNA in one case (patient no. 6),
but its mutation disappeared in post NAC ctDNA. EGFR
mutation is often detected in non-small cell lung cancer but is
rare in ovarian cancers. Therefore, the detection of the
EGFR mutations in ovarian cancer is a noteworthy finding,
although it remains unclear whether this mutation is pathogenic
(actionable) or not.
Change in the variant allele frequency
(VAF) during NAC
Next, we focused on the changes in the VAFs of
TP53 mutation and other somatic mutations, which could be
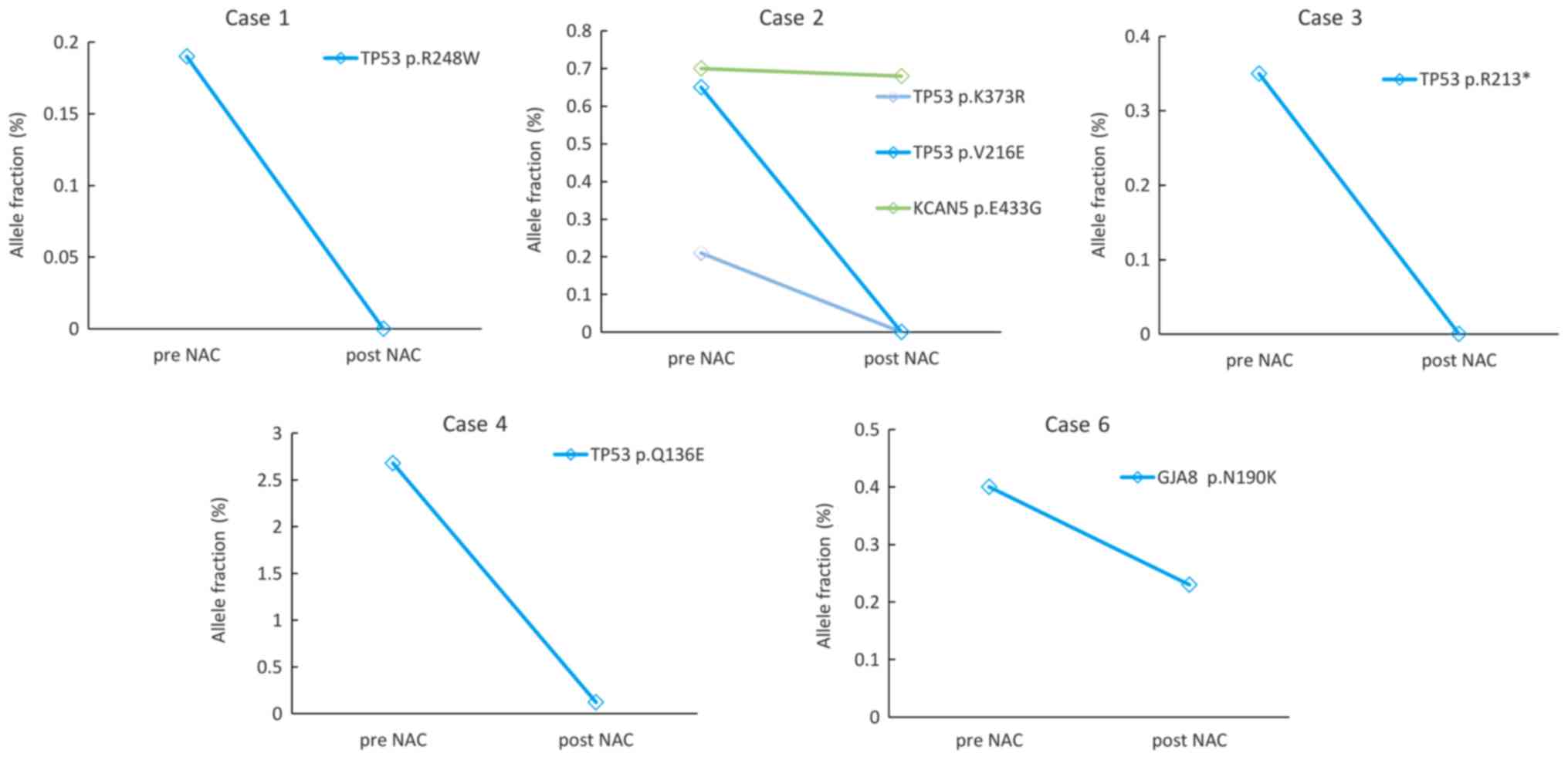
monitored in ctDNA both at pre- and post-NAC (Figs. 2 and 3). Five cases from the NAC-sensitive
patient group were monitored (Fig.
2). In patient no. 1, 2 and 3, TP53 mutations were
detected in the pre-NAC samples but not in the post-NAC samples. In
patient no. 2, the VAF of KCAN5 mutations decreased slightly
during the treatment. The VAF of TP53 in patient no. 4 and
the GJA8 mutation in patient no. 6 decreased during
treatment.
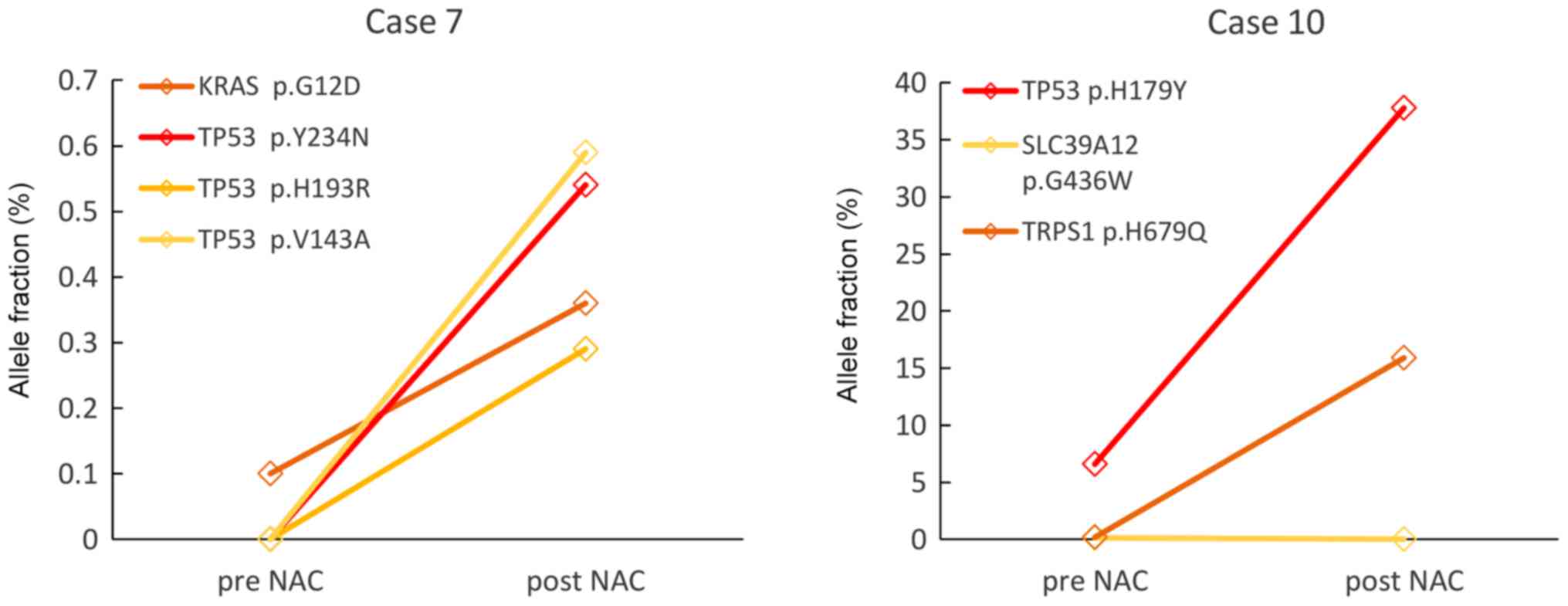
Two patients from the NAC-resistant group (patient
no. 7 and 10) could be monitored (Fig.
3). In Patient no. 7, KRAS mutation was detected in the
pre-NAC ctDNA but three other kinds of TP53 mutations as
well as KRAS mutations were detected in the post-NAC ctDNA.
In patient no. 10, VAFs of SLC39A12 mutation was not
detected, but VAFs of TP53 and TRPS1 mutations increased
after the NAC. These results suggest that minor clones present
before the NAC might expand under the selective pressure of
chemotherapy.
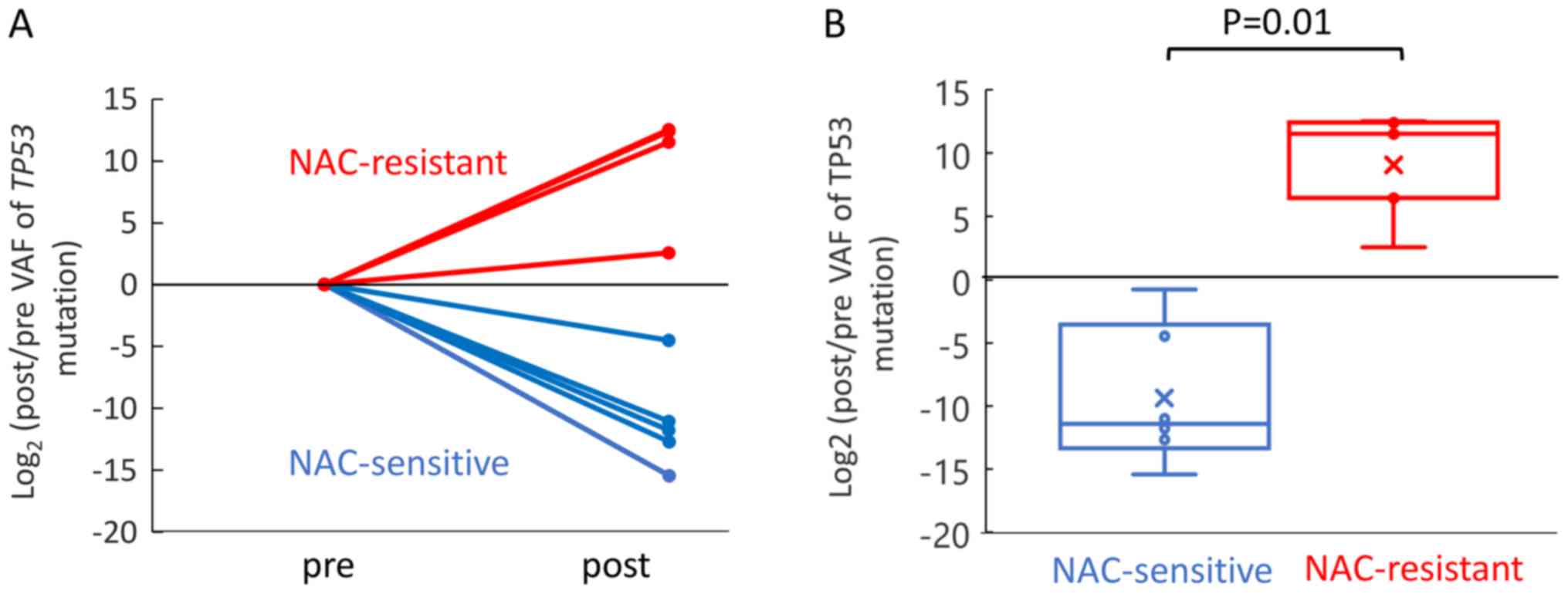
Association of TP53 VAF with the
response to NAC
According to The Cancer Genome Atlas (TCGA),
TP53 mutations are found in 94.6% of patients with
high-grade serous ovarian cancer (25). In this study, TP53 mutations
were detected at a high frequency. Thus, we compared the changes of
the TP53 VAF between the NAC-sensitive and NAC-resistant
patient groups. Five and four mutation sites of TP53 were
detected in the NAC-sensitive and NAC-resistant cases, respectively
(Fig. S2). Change of the
TP53 VAF was evaluated as a fold change in log based 2,
which was the ratio of the post TP53 to pre TP53 VAF.
The TP53 VAF increased significantly during NAC in the
NAC-resistant patient group as compared to the NAC-sensitive
patient groups (Fig. 4). These
results suggest that chemotherapy induces as increase of the mutant
allele frequency of TP53 in NAC-resistant cases.
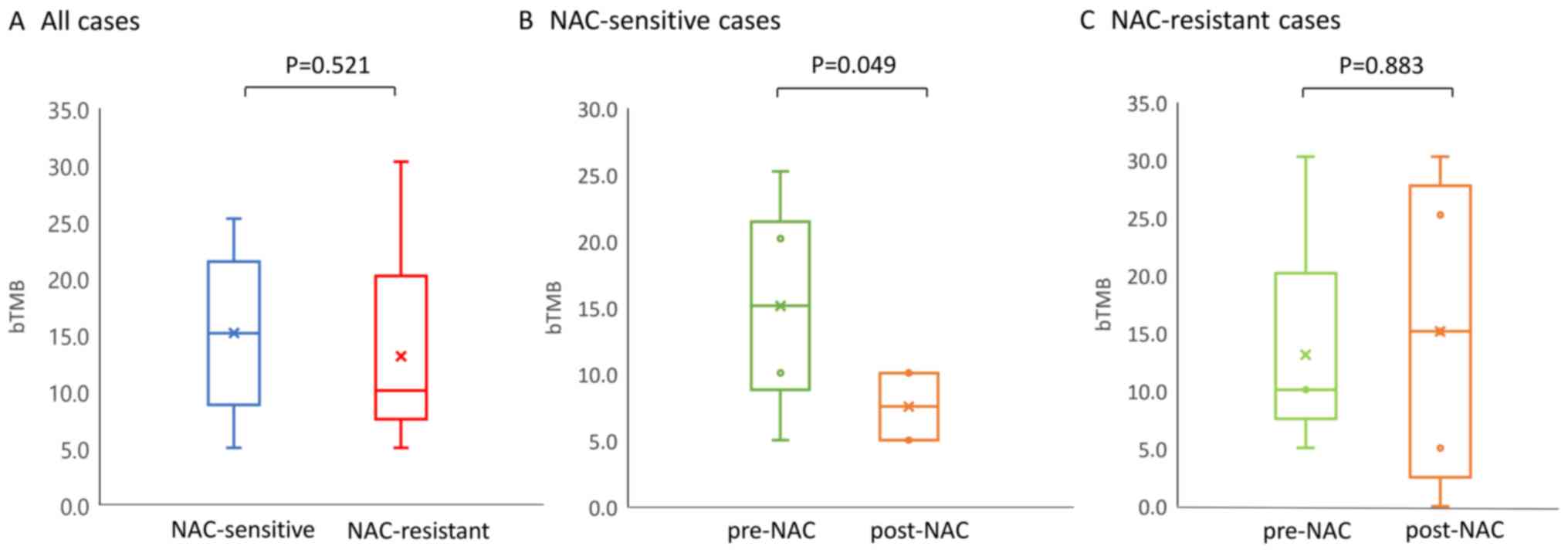
Blood tumor mutation burden (bTMB) and
response to chemotherapy
A recent study has shown a significant positive
correlation between the tissue-based TMB (tTMB) and bTMB (22), suggesting that the bTMB could serve
as a surrogate markers of the TMB. In this study, the bTMB was
evaluated as the number of non-synonymous mutation number per Mb,
and changes in the bTMB post-NAC relative the level at the baseline
were shown to be associated with the response to chemotherapy
(Fig. 5). There was no significant
difference in the pre-NAC baseline bTMB level between the
NAC-sensitive and -resistant patients (15.15/Mb and 10.10/Mb,
Fig. 5A). In the NAC-sensitive
group, the median values of the bTMB in the pre- and post- NAC
samples was 15.15/Mb and 7.58/Mb, respectively. Thus, the bTMB
decreased significantly during the NAC (P=0.049, Fig. 5B). On the other hand, in the
NAC-resistant patient group, a trend towards a light increase of
the bTMB value in the post-NAC ctDNA (15.15/Mb) as compared to the
pre-NAC ctDNA was observed (10.10/Mb), although this difference was
not significant (Fig. 5C). These
results suggest that the number of mutations detected in ctDNA by
CAPP-Seq may decrease as the tumor burden reduce during
treatment.
Discussion
To the best of our knowledge, the current study is
the first to demonstrate the usefulness of analyzing comprehensive
gene alterations in liquid biopsy-derived ctDNA using CAPP-Seq in
advanced ovarian cancer patients receiving NAC. Using CAPP-Seq, we
could detect changes in non-synonymous somatic mutations in the
plasma ctDNA between NAC-sensitive and NAC-resistant patients,
including changes of TP53 VAF and bTMB during treatment.
These findings suggest that CAPP-Seq based molecular profiling of
ctDNA may be useful for monitoring the treatment response in
advanced ovarian cancer patients receiving NAC.
Recently, some studies have demonstrated the
usefulness of detection of multiple somatic mutations by CAPP-seq.
The use of targeted hybrid capture with high-throughput sequencing
and a specialized bioinformatics workflow technique for plasma
ctDNA allows highly sensitive, non-invasive, and low-cost ctDNA
detection (14,15). Our previous studies have demonstrated
the feasibility of ctDNA gene mutation profiling using CAPP-Seq,
not only in cases of gynecological cancer, but also in metastasis
from colorectal cancer to the ovary, which exhibited the well-known
genetic signature of colorectal cancer (KRAS, APC and
TP53 mutations and MET copy-number gain) (18,19).
Previous reports showed the usefulness of the plasma
ctDNA concentration as a diagnostic, prognostic and predictive
biomarker in patients with ovarian cancer (26–30).
However, the clinical usefulness of this parameter is still
controversial because of the different assay methods, such as
real-time PCR and fluorescence staining, and different sample
sources such as plasma or serum. A recent meta-analysis reported
that quantitative analysis of ctDNA was associated with low
sensitivity, 0.70 (95% CI 0.65–0.74), but high specificity, 0.90
(95% CI 0.87–0.93) for the diagnosis of ovarian cancer (30). Capizzi et al showed that the
ctDNA concentration in patients with ovarian cancer measured before
chemotherapy decreased significantly after chemotherapy (26). In our study, there was no significant
difference in the overall ctDNA concentration during NAC treatment
between the NAC-sensitive and NAC-resistant cases, although the
sample size was limited. On the other hand, a recent review
reported that independent genetic alterations in the ctDNA showed
both higher sensitivity and specificity as compared to the overall
ctDNA concentration (31). Some
studies have focused on using TP53 mutations in the ctDNA as
the most common somatic mutation in ovarian cancer. Swisher et
al showed that the presence of TP53 mutation in the
ctDNA of ovarian cancer patients was an independent predictor of
decreased survival (32). Other
studies showed that undetectable TP53 mutations or reduction
in TP53 mutations after chemotherapy in ovarian cancer
patients was significantly associated with a better prognosis
(33,34). In our study, there were six sites of
TP53 mutations detected by CAPP-seq. We demonstrated a
significantly higher degree of change of TP53 mutations in
the NAC-resistant cases. Based on this result, we speculated that
chemotherapy may induce an increase in the mutant allele frequency
of TP53 in NAC-resistant cases.
In our study, CAPP-Seq enabled sensitive detection
of multiple classes of somatic mutations and deep sequencing.
Indeed, LRRTM1, BRINP2, CDH9 and GRM1 were detected only
after NAC in the NAC-resistant cases. It is suspected that minor
clones with these mutations before NAC might become major clones
during chemotherapy. Although there have been no previous reports
of these mutations in advanced ovarian cancer, these mutations
might be related to resistance to chemotherapy. We also found
EFGR amplification in post-NAC samples of NAC-resistant
cases (Patient no. 8 and 10). Some previous studies reported that
EGFR amplification was significantly associated with a
shorter overall survival in ovarian cancer patients (35,36).
Lassus et al reported that EGFR amplification and
overexpression in serous ovarian cancers were associated with a
shorter overall and disease-free survival (36). EGFR amplification detected in
the ctDNA might be a useful biomarker of the response to
chemotherapy in patients with ovarian cancer.
The detailed mechanism of changes in the gene
mutation profiles of ctDNA in each NAC-sensitive or NAC-resistant
patient shown in our study remains to be clarified. We speculated
that one reason for the phenomenon might be due to increasing or
decreasing tumor burden with response to chemotherapy. As another
mechanism, considering that ovarian cancer is likely to show tumor
heterogeneity, our data may reflect that NAC could induce clonal
evolution and might lead to appearance of chemotherapy-resistant
clones.
The present study demonstrated, for the first time,
changes in the bTMB in NAC -sensitive and NAC-resistant ovarian
cancer patients. The bTMB decreased significantly during treatment
in the NAC-sensitive cases, while no significant change of the bTMB
was observed in the NAC-resistant cases. However, the baseline bTMB
level measured before the NAC was not a predictor of the response
to chemotherapy within the small number of patients entered in this
study. In patients with non-small cell lung cancer, the high bTMB
(≥16 Mb) is
reported to be associated with better response to anti-PD-L1/PD-1
therapies (22). High-tissue TMB was
shown to be associated with longer survival in patients with
high-grade serous ovarian cancer carrying BRCA1 or BRCA2 mutations
(23). However, no reports have been
published between bTMB and chemosensitivity of at least ovarian
cancers, and the mechanism of the association of blood-based TMB
with efficacy of chemotherapy remains unclear in ovarian cancer.
Further study should be conducted whether bTMB, like tissue-TMB,
may be a predictive biomarker of response to chemotherapy or immune
checkpoint inhibitors for stratification of individual therapeutic
strategies in patients with advanced ovarian cancer.
There were several limitations of our study. First,
this was a retrospective study with a small sample size. Second, we
did not conduct the tumor tissue-derived DNA sequencing using
CAPP-Seq. CAPP-Seq is currently specialized for plasma ctDNA, and
therefore it remains a future challenge to expand its application
to formalin-fixed paraffin-embedded tumor tissue. Further
experiments using tumor DNA are needed to explore the
ability/sensitivity of detection of tumor mutations in ctDNA.
Third, blood samples were collected only at two points, pre- and
post-NAC in this study. If blood samples were collected at a
greater number of time points during chemotherapy, changes in the
allele frequency can also be detected in detail. Finally, approach
to analyze ctDNA remains technically challenge in order to
distinguish strictly ctDNA from cell-free DNA derived from normal
tissues and blood cells. CAPP-seq is a novel NGS-based
ultrasensitive method for ctDNA analysis and several studies showed
that the somatic mutations detected by CAPP-seq using the optimal
settings of cut-off values could reflect tumor-specific mutations
in ctDNA (14,15,18–20).
In conclusion, we demonstrated that molecular
profiling and monitoring of gene mutations could be successfully
performed by CAPP-Seq in advanced ovarian cancer patients receiving
NAC. Furthermore, we suggest that the TP53 mutation and bTMB
detected by liquid biopsy may be new biomarkers of the response to
NAC. These findings shown by liquid biopsy might enable detection
and assessment of post-treatment minimal residual disease (MRD).
Although extensive research has been conducted on the genetic
profiling of gynecological cancers so far conducted using tumor
tissue-derived DNA (25), tumor
biopsy or surgical resection for sufficient samples is often
difficult particularly in case of advanced/recurrent ovarian
cancer. Liquid biopsy is minimally invasive, allows easily serial
measurements and may also allow tumor heterogeneity to be
represented at real-time point. Genetic profiling of ctDNA in
gynecological cancers by CAPP-Seq may help in the establishment of
more efficient personalized therapeutic algorithms and real-time
therapy monitoring.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by Ministry of Education,
Culture, Sports, Science and Technology of Japan Grants-in-Aid for
Scientific Research Grant (grant no. JP19K18677).
Availability of data and materials
The datasets obtained and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Author contribution
TN and NI performed the experiments, interpreted the
data and wrote the manuscript. KS and TN analyzed the circulating
tumor DNA sequencing and interpreted the data. KM, HM, TY and ST
collected and prepared blood samples. KS contributed reagents,
materials and analytical tools and supervised the entire project.
KN and KI designed the research, interpreted the data and wrote the
paper. All authors reviewed the results and approved the final
version of the manuscript.
Ethics approval and consent to
participate
This study was approved by the ethics committee of
the Institutional Review Board of Wakayama Medical University
Faculty of Medicine (authorization no. 2025) and Kindai University
Faculty of Medicine (authorization no. 29-066). All patients in the
current study provided written informed consent for the use of
their plasma and tissue samples.
Patient consent for publication
Written informed consent for publication of the
present report was obtained from the patient.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chi DS, Eisenhauer EL, Zivanovic O, Sonoda
Y, Abu-Rustum NR, Levine DA, Guile MW, Bristow RE, Aghajanian C and
Barakat RR: Improved progression-free and overall survival in
advanced ovarian cancer as a result of a change in surgical
paradigm. Gynecol Oncol. 114:26–31. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fagotti A, Ferrandina G, Vizzielli G,
Fanfani F, Gallotta V, Chiantera V, Costantini B, Margariti PA,
Gueli Alletti S, Cosentino F, et al: Phase III randomised clinical
trial comparing primary surgery versus neoadjuvant chemotherapy in
advanced epithelial ovarian cancer with high tumour load (SCORPION
trial): Final analysis of peri-operative outcome. Eur J Cancer.
59:22–33. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kehoe S, Hook J, Nankivell M, Jayson GC,
Kitchener H, Lopes T, Luesley D, Perren T, Bannoo S, Mascarenhas M,
et al: Primary chemotherapy versus primary surgery for newly
diagnosed advanced ovarian cancer (CHORUS): An open-label,
randomised, controlled, non-inferiority trial. Lancet. 386:249–257.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vergote I, Tropé CG, Amant F, Kristensen
GB, Ehlen T, Johnson N, Verheijen RH, van der Burg ME, Lacave AJ,
Panici PB, et al: Neoadjuvant chemotherapy or primary surgery in
stage IIIC or IV ovarian cancer. N Engl J Med. 363:943–953. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pelissier A, Bonneau C, Chéreau E, de La
Motte Rouge T, Fourchotte V, Daraï E and Rouzier R: CA125 kinetic
parameters predict optimal cytoreduction in patients with advanced
epithelial ovarian cancer treated with neoadjuvant chemotherapy.
Gynecol Oncol. 135:542–546. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rodriguez N, Rauh-Hain JA, Shoni M,
Berkowitz RS, Muto MG, Feltmate C, Schorge JO, Del Carmen MG,
Matulonis UA and Horowitz NS: Changes in serum CA-125 can predict
optimal cytoreduction to no gross residual disease in patients with
advanced stage ovarian cancer treated with neoadjuvant
chemotherapy. Gynecol Oncol. 125:362–366. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zeng J, Yin J, Song X, Jin Y, Li Y and Pan
L: Reduction of CA125 levels during neoadjuvant chemotherapy can
predict cytoreduction to no visible residual disease in patients
with advanced epithelial ovarian cancer, primary carcinoma of
fallopian tube and peritoneal carcinoma. J Cancer. 7:2327–2332.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Diaz LA Jr and Bardelli A: Liquid
biopsies: Genotyping circulating tumor DNA. J Clin Oncol.
32:579–586. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Diehl F, Schmidt K, Choti MA, Romans K,
Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al:
Circulating mutant DNA to assess tumor dynamics. Nat Med.
14:985–990. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vogelstein B and Kinzler KW: Digital PCR.
Proc Natl Acad Sci USA. 96:9236–9241. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bettegowda C, Sausen M, Leary RJ, Kinde I,
Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al:
Detection of circulating tumor DNA in early- and late-stage human
malignancies. Sci Transl Med. 6:224ra242014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Narayan A, Carriero NJ, Gettinger SN,
Kluytenaar J, Kozak KR, Yock TI, Muscato NE, Ugarelli P, Decker RH
and Patel AA: Ultrasensitive measurement of hotspot mutations in
tumor DNA in blood using error-suppressed multiplexed deep
sequencing. Cancer Res. 72:3492–3498. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Newman AM, Bratman SV, To J, Wynne JF,
Eclov NC, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE, et
al: An ultrasensitive method for quantitating circulating tumor DNA
with broad patient coverage. Nat Med. 20:548–554. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Newman AM, Lovejoy AF, Klass DM, Kurtz DM,
Chabon JJ, Scherer F, Stehr H, Liu CL, Bratman SV, Say C, et al:
Integrated digital error suppression for improved detection of
circulating tumor DNA. Nat Biotechnol. 34:547–555. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Scherer F, Kurtz DM, Newman AM, Stehr H,
Craig AF, Esfahani MS, Lovejoy AF, Chabon JJ, Klass DM, Liu CL, et
al: Distinct biological subtypes and patterns of genome evolution
in lymphoma revealed by circulating tumor DNA. Sci Transl Med.
8:364ra1552016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Przybyl J, Chabon JJ, Spans L, Ganjoo KN,
Vennam S, Newman AM, Forgó E, Varma S, Zhu S, Debiec-Rychter M, et
al: Combination approach for detecting different types of
alterations in circulating tumor DNA in leiomyosarcoma. Clin Cancer
Res. 24:2688–2699. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Iwahashi N, Sakai K, Noguchi T, Yahata T,
Toujima S, Nishio K and Ino K: A comprehensive gene mutation
analysis of liquid biopsy samples from patients with metastatic
colorectal cancer to the ovary: A case report. Oncol Lett.
16:6431–6436. 2018.PubMed/NCBI
|
|
19
|
Iwahashi N, Sakai K, Noguchi T, Yahata T,
Matsukawa H, Toujima S, Nishio K and Ino K: Liquid biopsy-based
comprehensive gene mutation profiling for gynecological cancer
using cancer personalized profiling by deep sequencing. Sci Rep.
9:104262019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Otsubo K, Sakai K, Takeshita M, Harada D,
Azuma K, Ota K, Akamatsu H, Goto K, Horiike A, Kurata T, et al:
Genetic profiling of non-small cell lung cancer at development of
resistance to first- or second-generation EGFR-TKIs by CAPP-Seq
analysis of circulating tumor DNA. Oncologist. 24:1022–1026. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rizvi H, Sanchez-Vega F, La K, Chatila W,
Jonsson P, Halpenny D, Plodkowski A, Long N, Sauter JL, Rekhtman N,
et al: Molecular determinants of response to anti-programmed cell
death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in
patients with non-small-cell lung cancer profiled with targeted
next-generation sequencing. J Clin Oncol. 36:633–641. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gandara DR, Paul SM, Kowanetz M,
Schleifman E, Zou W, Li Y, Rittmeyer A, Fehrenbacher L, Otto G,
Malboeuf C, et al: Blood-based tumor mutational burden as a
predictor of clinical benefit in non-small-cell lung cancer
patients treated with atezolizumab. Nat Med. 24:1441–1448. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Birkbak NJ, Kochupurakkal B, Izarzugaza
JM, Eklund AC, Li Y, Liu J, Szallasi Z, Matulonis UA, Richardson
AL, Iglehart JD and Wang ZC: Tumor mutation burden forecasts
outcome in ovarian cancer with BRCA1 or BRCA2 mutations. PLoS One.
8:e800232013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Narahara M, Higasa K, Nakamura S, Tabara
Y, Kawaguchi T, Ishii M, Matsubara K, Matsuda F and Yamada R:
Large-scale East-Asian eQTL mapping reveals novel candidate genes
for LD mapping and the genomic landscape of transcriptional effects
of sequence variants. PLoS One. 9:e1009242014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cancer Genome Atlas Research Network, .
Integrated genomic analyses of ovarian carcinoma. Nature.
474:609–615. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Capizzi E, Gabusi E, Grigioni AD, De Iaco
P, Rosati M, Zamagni C and Fiorentino M: Quantification of free
plasma DNA before and after chemotherapy in patients with advanced
epithelial ovarian cancer. Diagn Mol Pathol. 17:34–38.
2008.PubMed/NCBI
|
|
27
|
Gu XH, Wu NW and Liu XS: Application of
serum DNA quantification in the diagnosis of epithelial ovarian
cancer. Matern Child Health Care China. 24:1413–1415. 2009.
|
|
28
|
Holdenrieder S, Stieber P, Bodenmuller H,
Busch M, Fertig G, Fürst H, Schalhorn A, Schmeller N, Untch M and
Seidel D: Nucleosomes in serum of patients with benign and
malignant diseases. Int J Cancer. 95:114–120. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kamat AA, Baldwin M, Urbauer D, Dang D,
Han LY, Godwin A, Karlan BY, Simpson JL, Gershenson DM, Coleman RL,
et al: Plasma cell-free DNA in ovarian cancer: An independent
prognostic biomarker. Cancer. 116:1918–1925. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhou Q, Li W, Leng B, Zheng W, He Z, Zuo M
and Chen A: Circulating cell Free DNA as the diagnostic marker for
ovarian cancer: A systematic review and meta-analysis. PLoS One.
11:e01554952016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Barbosa A, Peixoto A, Pinto P, Pinheiro M
and Teixeira MR: Potential clinical applications of circulating
cell-free DNA in ovarian cancer patients. Expert Rev Mol Med.
20:e62018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Swisher EM, Wollan M, Mahtani SM, Willner
JB, Garcia R, Goff BA and King MC: Tumor-specific p53 sequences in
blood and peritoneal fluid of women with epithelial ovarian cancer.
Am J Obstet Gynecol. 193:662–667. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim YM, Lee SW, Lee YJ, Lee HY, Lee JE and
Choi EK: Prospective study of the efficacy and utility of TP53
mutations in circulating tumor DNA as a non-invasive biomarker of
treatment response monitoring in patients with high-grade serous
ovarian carcinoma. J Gynecol Oncol. 30:e322019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pereira E, Camacho-Vanegas O, Anand S,
Sebra R, Catalina Camacho S, Garnar-Wortzel L, Nair N, Moshier E,
Wooten M, Uzilov A, et al: Personalized circulating tumor DNA
biomarkers dynamically predict treatment response and survival in
gynecologic cancers. PLoS One. 10:e01457542015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lafky JM, Wilken JA, Baron AT and Maihle
NJ: Clinical implications of the ErbB/epidermal growth factor (EGF)
receptor family and its ligands in ovarian cancer. Biochim Biophys
Acta. 1785:232–265. 2008.PubMed/NCBI
|
|
36
|
Lassus H, Sihto H, Leminen A, Joensuu H,
Isola J, Nupponen NN and Butzow R: Gene amplification, mutation,
and protein expression of EGFR and mutations of ERBB2 in serous
ovarian carcinoma. J Mol Med (Berl). 84:671–681. 2006. View Article : Google Scholar : PubMed/NCBI
|