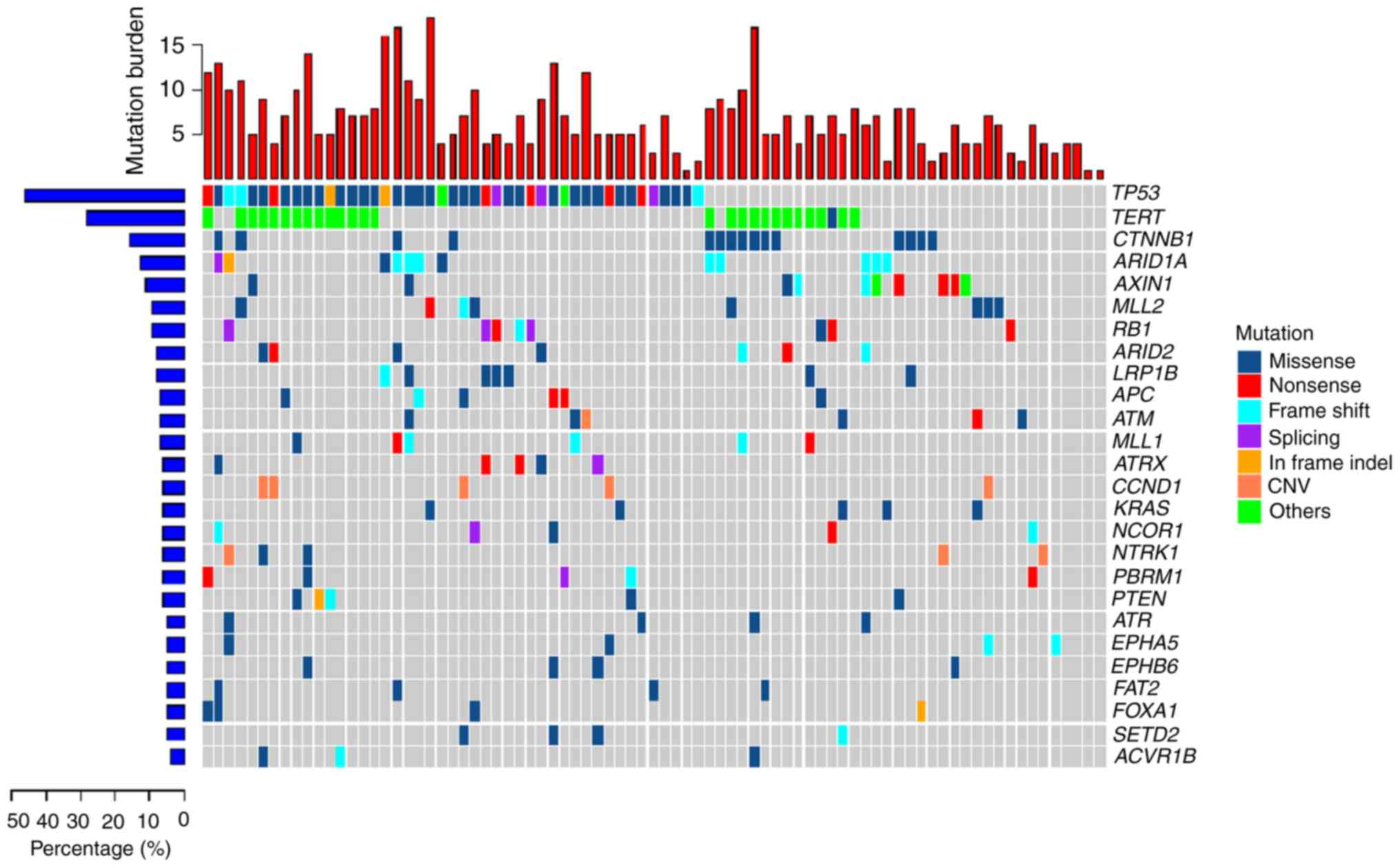
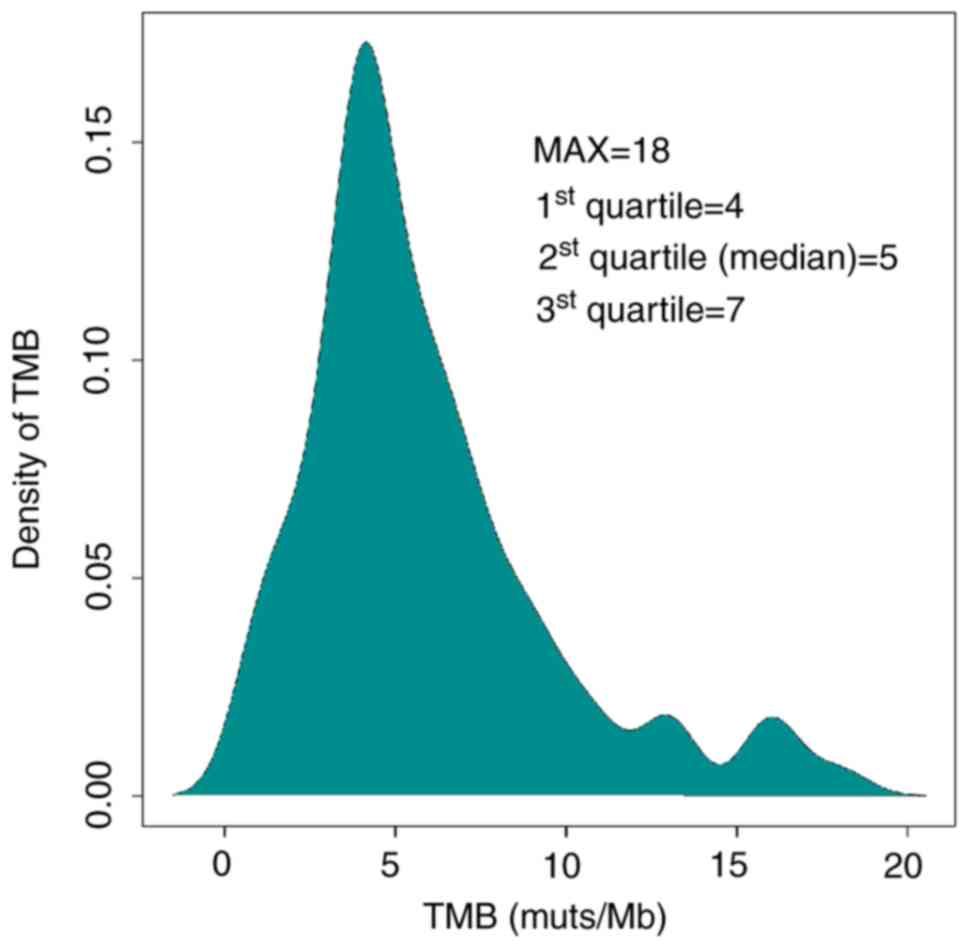
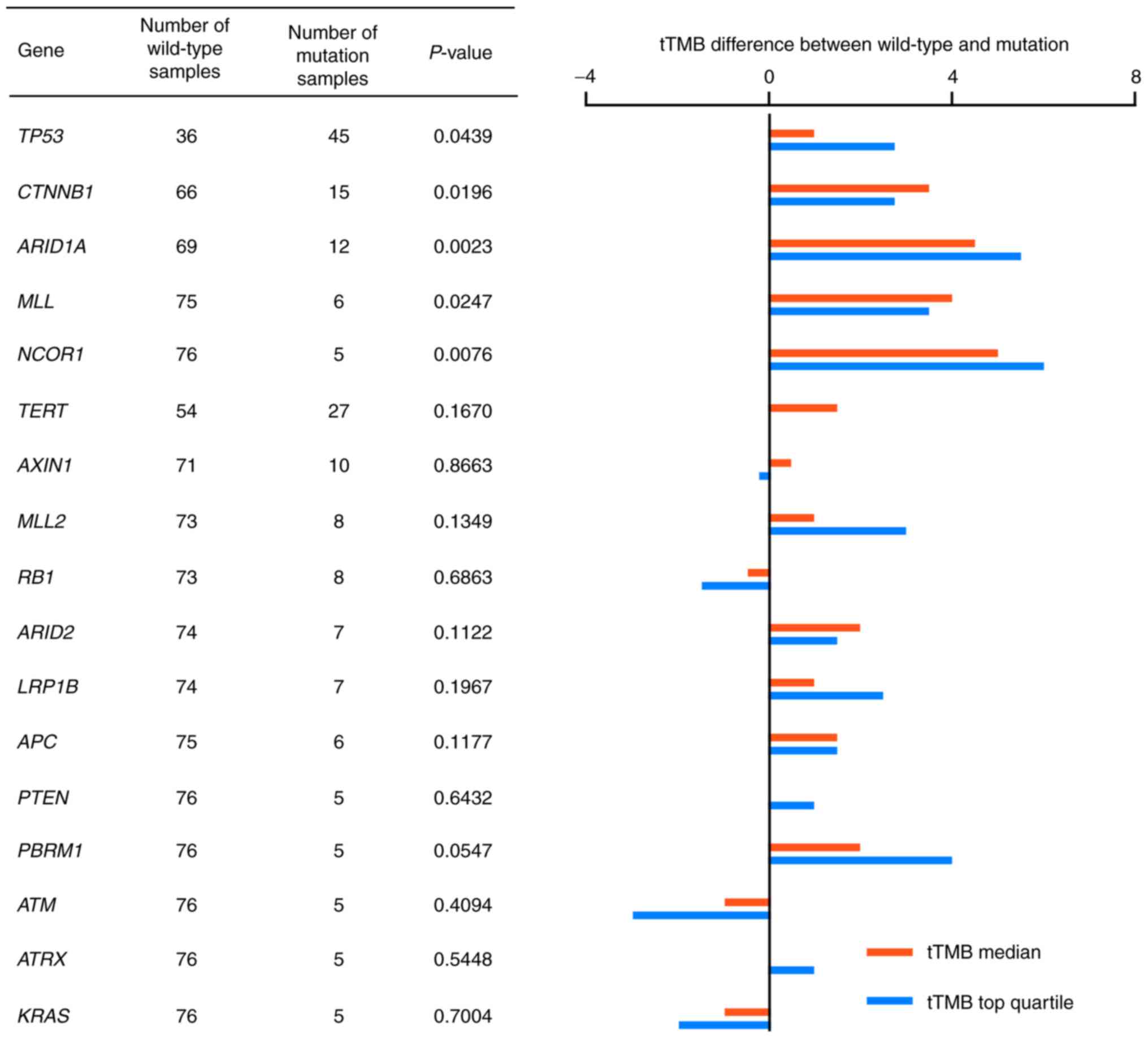
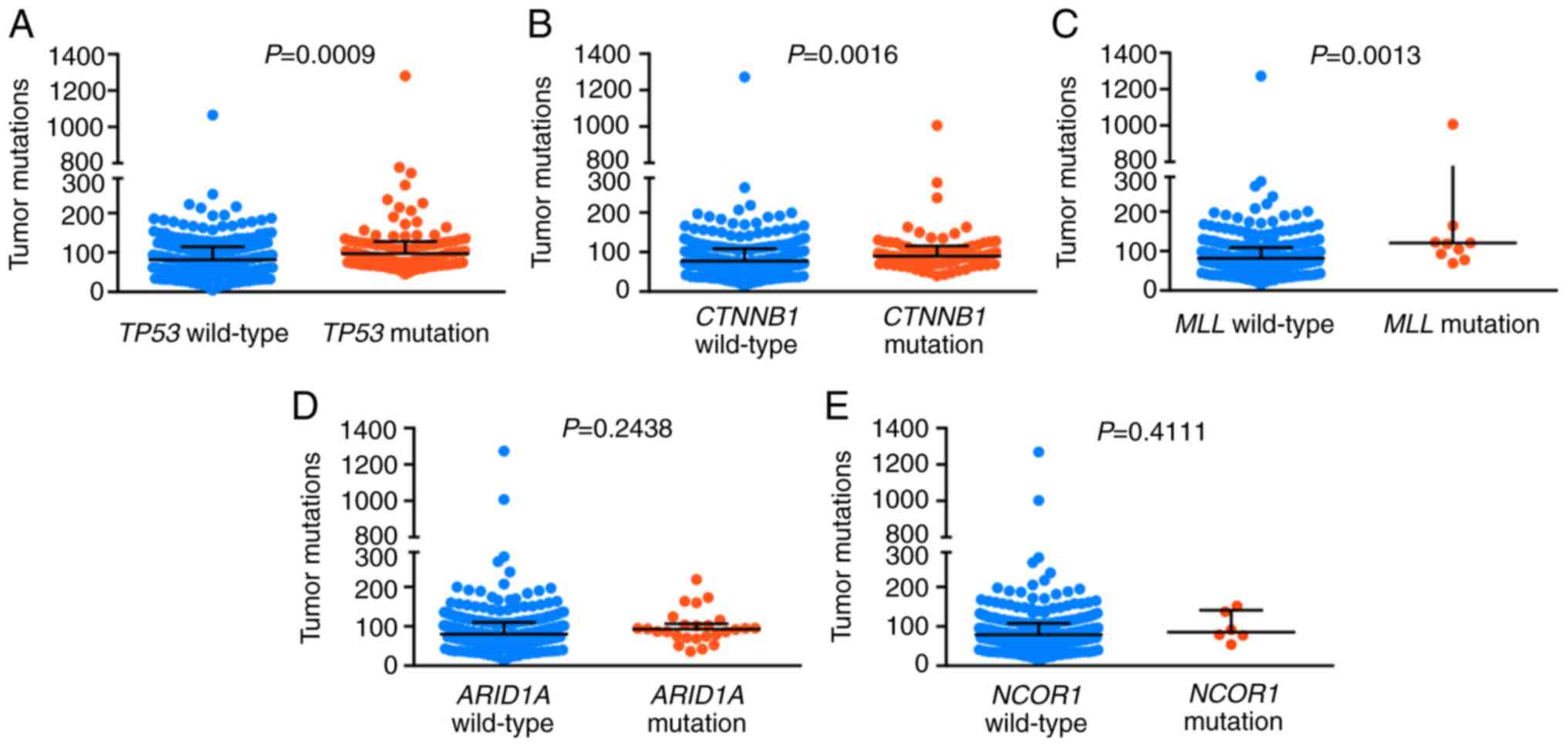
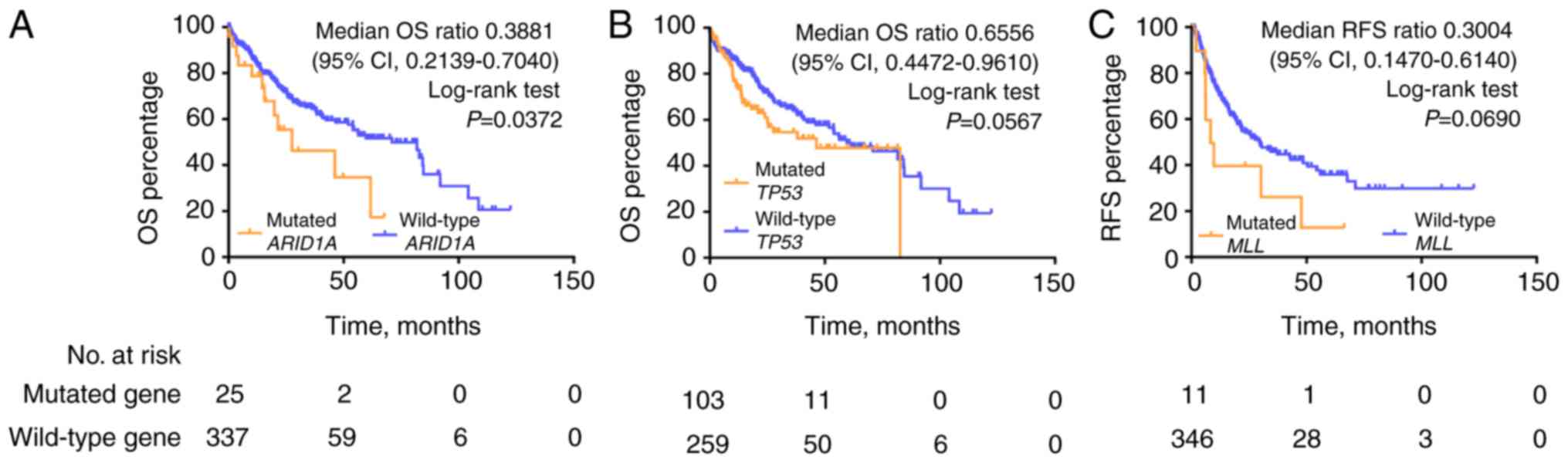
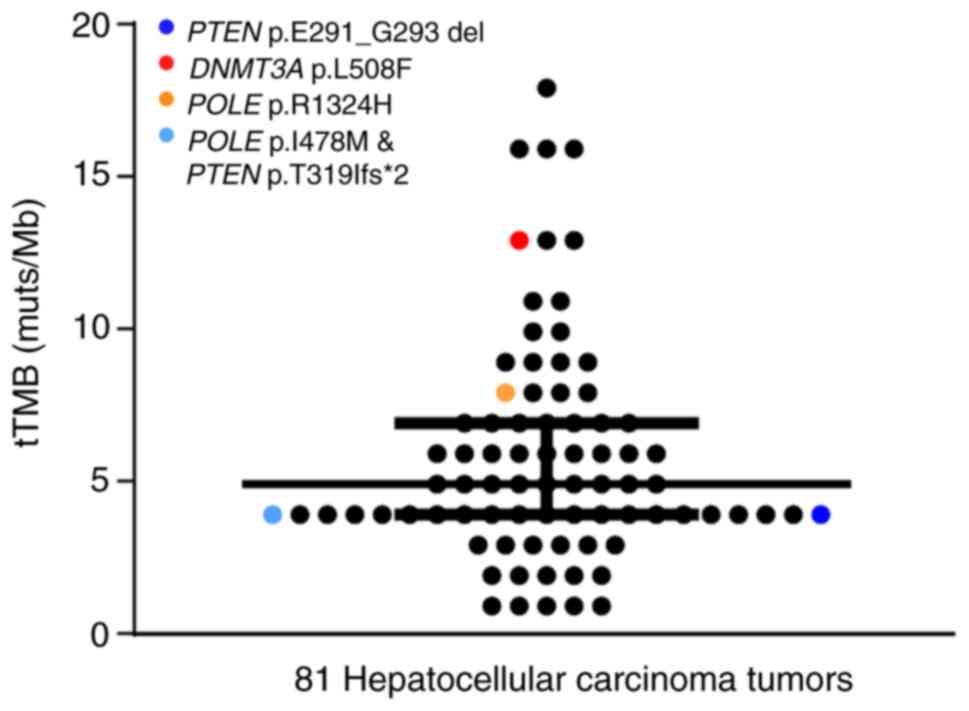
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yu SJ: A concise review of updated
guidelines regarding the management of hepatocellular carcinoma
around the world: 2010–2016. Clin Mol Hepatol. 22:7–17. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sprinzl MF and Galle PR: Current progress
in immunotherapy of hepatocellular carcinoma. J Hepatol.
66:482–484. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Topalian SL, Drake CG and Pardoll DM:
Immune checkpoint blockade: A common denominator approach to cancer
therapy. Cancer Cell. 27:450–461. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rotte A, Jin JY and Lemaire V: Mechanistic
overview of immune checkpoints to support the rational design of
their combinations in cancer immunotherapy. Ann Oncol. 29:71–83.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Topalian SL, Hodi FS, Brahmer JR,
Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD,
Sosman JA, Atkins MB, et al: Safety, activity, and immune
correlates of anti-PD-1 antibody in cancer. N Engl J Med.
366:2443–2454. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Q and Wu X: Primary and acquired
resistance to PD-1/PD-L1 blockade in cancer treatment. Int
Immunopharmacol. 46:210–219. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhu J, Armstrong AJ, Friedlander TW, Kim
W, Pal SK, George DJ and Zhang T: Biomarkers of immunotherapy in
urothelial and renal cell carcinoma: PD-L1, tumor mutational
burden, and beyond. J Immunother Cancer. 6:42018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Overman MJ, Lonardi S, Wong KYM, Lenz HJ,
Gelsomino F, Aglietta M, Morse MA, Van Cutsem E, McDermott R, Hill
A, et al: Durable clinical benefit with nivolumab plus ipilimumab
in DNA mismatch repair-deficient/microsatellite instability-high
metastatic colorectal cancer. J Clin Oncol. 36:773–779. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yarchoan M, Hopkins A and Jaffee EM: Tumor
mutational burden and response rate to PD-1 inhibition. N Engl J
Med. 377:2500–2501. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schumacher TN and Schreiber RD:
Neoantigens in cancer immunotherapy. Science. 348:69–74. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rizvi NA, Hellmann MD, Snyder A, Kvistborg
P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al: Cancer
immunology. Mutational landscape determines sensitivity to PD-1
blockade in non-small cell lung cancer. Science. 348:124–128. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu J, Zhang Y, Jia R, Yue C, Chang L, Liu
R, Zhang G, Zhao C, Zhang Y, Chen C, et al: Anti-PD-1 antibody
SHR-1210 combined with apatinib for advanced hepatocellular
carcinoma, gastric or esophagogastric junction cancer: An
open-label, dose escalation and expansion study. Clin Cancer Res.
25:515–523. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Alexandrov LB, Nik-Zainal S, Wedge DC,
Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A,
Børresen-Dale AL, et al: Signatures of mutational processes in
human cancer. Nature. 500:415–421. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Le DT, Uram JN, Wang H, Bartlett BR,
Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et
al: PD-1 blockade in tumors with mismatch-repair deficiency. N Engl
J Med. 372:2509–2520. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hugo W, Zaretsky JM, Sun L, Song C, Moreno
BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G,
et al: Genomic and transcriptomic features of response to Anti-PD-1
therapy in metastatic melanoma. Cell. 165:35–44. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pal SK, Agarwal N, Choueiri TK, Stephens
PJ, Ross JS, Miller VA, Ali SM, Chung J and Grivas P: Comparison of
tumor mutational burden (TMB) in relevant molecular subsets of
metastatic urothelial cancer (MUC). Ann Oncol. 28
(Suppl-5):v295–v329. 2017. View Article : Google Scholar
|
|
18
|
Lv X, Zhao M, Yi Y, Zhang L, Guan Y, Liu
T, Yang L, Chen R, Ma J and Yi X: Detection of rare mutations in
CtDNA using next generation sequencing. J Vis Exp. 2017.doi:
10.3791/56342. View
Article : Google Scholar
|
|
19
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cibulskis K, Lawrence MS, Carter SL,
Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES
and Getz G: Sensitive detection of somatic point mutations in
impure and heterogeneous cancer samples. Nat Biotechnol.
31:213–221. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li J, Lupat R, Amarasinghe KC, Thompson
ER, Doyle MA, Ryland GL, Tothill RW, Halgamuge SK, Campbell IG and
Gorringe KL: CONTRA: Copy number analysis for targeted
resequencing. Bioinformatics. 28:1307–1313. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nong J, Gong Y, Guan Y, Yi X, Yi Y, Chang
L, Yang L, Lv J, Guo Z, Jia H, et al: Circulating tumor DNA
analysis depicts subclonal architecture and genomic evolution of
small cell lung cancer. Nat Commun. 9:31142018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Totoki Y, Tatsuno K, Covington KR, Ueda H,
Creighton CJ, Kato M, Tsuji S, Donehower LA, Slagle BL, Nakamura H,
et al: Trans-ancestry mutational landscape of hepatocellular
carcinoma genomes. Nat Genet. 46:1267–1273. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Podolskiy DI, Lobanov AV, Kryukov GV and
Gladyshev VN: Analysis of cancer genomes reveals basic features of
human aging and its role in cancer development. Nat Commun.
7:121572016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tao Y, Ruan J, Yeh SH, Lu X, Wang Y, Zhai
W, Cai J, Ling S, Gong Q, Chong Z, et al: Rapid growth of a
hepatocellular carcinoma and the driving mutations revealed by
cell-population genetic analysis of whole-genome data. Proc Natl
Acad Sci USA. 108:12042–12047. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Breuhahn K, Gores G and Schirmacher P:
Strategies for hepatocellular carcinoma therapy and diagnostics:
Lessons learned from high throughput and profiling approaches.
Hepatology. 53:2112–2121. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zender L, Villanueva A, Tovar V, Sia D,
Chiang DY and Llovet JM: Cancer gene discovery in hepatocellular
carcinoma. J Hepatol. 52:921–929. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Guichard C, Amaddeo G, Imbeaud S, Ladeiro
Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M,
Degos F, et al: Integrated analysis of somatic mutations and focal
copy-number changes identifies key genes and pathways in
hepatocellular carcinoma. Nat Genet. 44:694–648. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fujimoto A, Totoki Y, Abe T, Boroevich KA,
Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, et al:
Whole-genome sequencing of liver cancers identifies etiological
influences on mutation patterns and recurrent mutations in
chromatin regulators. Nat Genet. 44:760–764. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
da Motta Girardi D, Correa TS, Crosara
Teixeira M and Dos Santos Fernandes G: Hepatocellular carcinoma:
Review of targeted and immune therapies. J Gastrointest Cancer.
49:227–236. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gomaa AI, Khan SA, Toledano MB, Waked I
and Taylor-Robinson SD: Hepatocellular carcinoma: Epidemiology,
risk factors and pathogenesis. World J Gastroenterol. 14:4300–4308.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Elsegood CL, Tirnitz-Parker JE, Olynyk JK
and Yeoh GC: Immune checkpoint inhibition: Prospects for prevention
and therapy of hepatocellular carcinoma. Clin Transl Immunology.
6:e1612017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wieder T, Eigentler T, Brenner E and
Röcken M: Immune checkpoint blockade therapy. J Allergy Clin
Immunol. 142:1403–1414. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
El-Khoueiry AB, Sangro B, Yau T, Crocenzi
TS, Kudo M, Hsu C, Kim TY, Choo SP, Trojan J, Welling TH Rd, et al:
Nivolumab in patients with advanced hepatocellular carcinoma
(CheckMate 040): An open-label, non-comparative, phase 1/2 dose
escalation and expansion trial. Lancet. 389:2492–2502. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zaretsky JM, Garcia-Diaz A, Shin DS,
Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY,
Abril-Rodriguez G, Sandoval S, Barthly L, et al: Mutations
associated with acquired resistance to PD-1 blockade in melanoma. N
Engl J Med. 375:819–829. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kato S, Goodman A, Walavalkar V,
Barkauskas DA, Sharabi A and Kurzrock R: Hyperprogressors after
Immunotherapy: Analysis of Genomic Alterations associated with
accelerated growth rate. Clin Cancer Res. 23:4242–4250. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Llosa NJ, Cruise M, Tam A, Wicks EC,
Hechenbleikner EM, Taube JM, Blosser RL, Fan H, Wang H, Luber BS,
et al: The vigorous immune microenvironment of microsatellite
instable colon cancer is balanced by multiple counter-inhibitory
checkpoints. Cancer Discov. 5:43–51. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lemery S, Keegan P and Pazdur R: First FDA
approval agnostic of cancer site-when a biomarker defines the
indication. N Engl J Med. 377:1409–1412. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Campbell BB, Light N, Fabrizio D, Zatzman
M, Fuligni F, de Borja R, Davidson S, Edwards M, Elvin JA, Hodel
KP, et al: Comprehensive analysis of hypermutation in human cancer.
Cell. 171:1042–1056.e10. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Peng W, Chen JQ, Liu C, Malu S, Creasy C,
Tetzlaff MT, Xu C, McKenzie JA, Zhang C, Liang X, et al: Loss of
PTEN promotes resistance to t cell-mediated immunotherapy. Cancer
Discov. 6:202–216. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cleary SP, Jeck WR, Zhao X, Chen K,
Selitsky SR, Savich GL, Tan TX, Wu MC, Getz G, Lawrence MS, et al:
Identification of driver genes in hepatocellular carcinoma by exome
sequencing. Hepatology. 58:1693–1702. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lee JS: The mutational landscape of
hepatocellular carcinoma. Clin Mol Hepatol. 21:220–229. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Braun DA, Burke KP and Van Allen EM:
Genomic approaches to understanding response and resistance to
immunotherapy. Clin Cancer Res. 22:5642–5650. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Teng F, Meng X, Kong L and Yu J: Progress
and challenges of predictive biomarkers of anti PD-1/PD-L1
immunotherapy: A systematic review. Cancer Lett. 414:166–173. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Giannakis M, Mu XJ, Shukla SA, Qian ZR,
Cohen O, Nishihara R, Bahl S, Cao Y, Amin-Mansour A, Yamauchi M, et
al: Genomic correlates of immune-cell infiltrates in colorectal
carcinoma. Cell Rep. 15:857–865. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Matsushita H, Vesely MD, Koboldt DC,
Rickert CG, Uppaluri R, Magrini VJ, Arthur CD, White JM, Chen YS,
Shea LK, et al: Cancer exome analysis reveals a T-cell-dependent
mechanism of cancer immunoediting. Nature. 482:400–404. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lee JK, Choi YL, Kwon M and Park PJ:
Mechanisms and consequences of cancer genome instability: Lessons
from genome sequencing studies. Annu Rev Pathol. 11:283–312. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang Z, Zhao J, Wang G, Zhang F, Zhang Z,
Zhang F, Zhang Y, Dong H, Zhao X, Duan J, et al: Co-mutations in
DNA damage response pathways serve as potential biomarkers for
immune checkpoint blockade. Cancer Res. 78:6486–6496.
2018.PubMed/NCBI
|
|
49
|
Scarbrough PM, Weber RP, Iversen ES,
Brhane Y, Amos CI, Kraft P, Hung RJ, Sellers TA, Witte JS, Pharoah
P, et al: A cross-cancer genetic association analysis of the DNA
repair and DNA damage signaling pathways for lung, ovary, prostate,
breast, and colorectal cancer. Cancer Epidemiol Biomarkers Prev.
25:193–200. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Muller PA and Vousden KH: Mutant p53 in
cancer: New functions and therapeutic opportunities. Cancer Cell.
25:304–317. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bieging KT, Mello SS and Attardi LD:
Unravelling mechanisms of p53-mediated tumor suppression. Nat Rev
Cancer. 14:359–370. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wilson BG and Roberts CW: SWI/SNF
nucleosome remodelers and cancer. Nat Rev Cancer. 11:481–492. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Shen J, Ju Z, Zhao W, Wang L, Peng Y, Ge
Z, Nagel ZD, Zou J, Wang C, Kapoor P, et al: ARID1A deficiency
promotes mutability and potentiates therapeutic antitumor immunity
unleashed by immune checkpoint blockade. Nat Med. 24:556–562. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Battaglia S, Maguire O and Campbell MJ:
Transcription factor co-repressors in cancer biology: Roles and
targeting. Int J Cancer. 126:2511–2519. 2010.PubMed/NCBI
|
|
55
|
Clevers H and Nusse R: Wnt/beta-catenin
signaling and disease. Cell. 149:1192–1205. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Schulze K, Imbeaud S, Letouzé E,
Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C,
Shinde J, Soysouvanh F, et al: Exome sequencing of hepatocellular
carcinomas identifies new mutational signatures and potential
therapeutic targets. Nat Genet. 47:505–511. 2015. View Article : Google Scholar : PubMed/NCBI
|