Introduction
Breast cancer is an important public health threat
to women worldwide. Triple-negative breast cancer (TNBC) accounts
for 20% of total breast cancer cases, and is characterized as
estrogen receptor (ER)-negative, progesterone receptor
(PR)-negative, and HER2-negative (1). As a result of its aggressive biological
characteristics and the lack of effective treatment options, TNBC
has been associated with poor prognosis, when compared with other
subtypes of breast cancer (2).
Therefore, the development of novel TNBC therapies is urgently
required. Recently, several studies have shown that other types of
hormone receptors are expressed in TNBC, such as androgen receptor
(AR) (3). AR was proved to have a
high level of structural similarity to that of ER and PR (4). Several studies have demonstrated that
different types of breast cancer exhibited diverse function of AR.
AR-associated signaling pathway was speculated to act as either
tumorigenic or antitumor effect in multiple studies in breast
cancer. A previous study identified AR as a commonly expressed
nuclear receptor in breast cancer, which was crucial in promoting
tumorigenesis (5). In ER-positive
breast cancer, AR could predict promising outcome, while in TNBC,
the prognostic value of AR expression is still controversial. In
TNBC, multiple researches have illustrated different results on the
function of AR (5–8). Furthermore, it has been found that
bicalutamide, an AR inhibitor, could reduce TNBC proliferation and
activity (1). Nevertheless, the
precise molecular mechanisms of AR-induced cell proliferation,
migration and invasion in TNBC cells remains unclear. The present
study identified BCL11A, a novel oncogene, and its potential role
in the AR-associated regulation of TNBC.
BCL11A, also known as Evi9 and CTIP1, was firstly
discovered in leukemia with chromosome translocation t
(2;14)(p13;q32.3). The BCL11A gene is located on human chromosome
2p13 and is ~102 kb in length (9).
BCL11A is primarily expressed in brain and hematopoietic tissues
(10), though accumulating evidence
suggests that it also plays an essential role in the formation of
other tumors, including prostate cancer, lung cancer, laryngeal
squamous cell carcinoma and acute leukemia (11–16). In
lung cancer, Jiang et al identified BCL11A could induced by
miR-30a and act as a potential prognostic factor in NSCLC (17). Additionally, in lung squamous cell
carcinoma (LUSC), Lazarus et al found that BCL11A was
integral to LUSC pathology and could interact with SOX2 to regulate
several downstream transcription factors (18). In prostate cancer and colorectal
cancer, it has been demonstrated that BCL11A overexpression
strongly reversed the influence of tumor progression induced by
FOXQ1 inhibition (19,20). Notably, BCL11A may also promote
stemness in breast cancer cells by inducing Wnt/β-catenin signaling
(21). BCL11A activation is
associated with the induction of numerous carcinogenic signaling
pathways, and there is growing evidence to suggest that BCL11A
downregulation inhibits tumorigenesis (22,23).
However, the specific functions of BCL11A and the underlying
mechanism of its involvement in TNBC have not been fully clarified.
The results of the present study demonstrated the importance of
BCL11A in the development and progression of TNBC, and for the
first time, identified the potential mechanism of BCL11A-associated
AR regulation in TNBC.
Materials and methods
Clinical samples
Tissue microarray HBre-Ducl40Sur-01 was obtained
from Shanghai Outdo Biotech Co., Ltd, and contained tissue samples
collected between January 2001 and August 2004. The corresponding
patients had not received radiotherapy or chemotherapy prior to
surgery. Experiments with tumor tissues were performed according to
the Declaration of Helsinki and were approved by the Ethics
Committee of Jinling Hospital. Construction of the tissue
microarray has previously been demonstrated (24). The surgical time was between
2001.07–2004.08. The follow-up time was up to 2013.07. The median
survival time was 76 months.
TCGA database
Breast cancer tissue samples with RNA-seq data and
clinicopathological information were downloaded from the cancer
genome atlas (TCGA) database released on October 31, 2018. Samples
were divided into four breast cancer subtypes according to their
ER, PR and HER2 status. At the time of analysis, 466 luminal A
samples, 103 luminal B samples, 39 HER2-positive samples and 140
TNBC samples were included in our study. Overall survival (OS) was
measured from the date of diagnosis to the date of death due to all
causes or the last follow-up.
Immunohistochemistry
Immunohistochemical staining was performed as
previously described (24). Staining
was scored as: i) ++++ if >80% tumor cells were immuno-positive;
ii) +++ for 51–80%; iii) ++ for 11–50%; iv) + for 1–10% cells; and
v) -if <1% of the tumor cells were positively stained. The
staining intensity was categorized as negative (−), weak (+),
moderate (++) or strong (+++). The H-score method was utilized via
multiplying the percentage score by the staining intensity score
and produce a summed score between 0 and 12. The standard of
classification was shown as follows: Intensity score (0=no
staining, 1=weak, 2=moderate and 3=strong); stained cell proportion
score (0=0%, 1=1-10, 2=11-50, 3=51–80 and 4=81–100%). BCL11A high
expression was defined as H score ≥6. AR positive was defined as
≥10% expression. Immunostained sections were observed using a
microscope (Carl Zeiss Inc.).
Cell lines and culture conditions
TNBC cell lines (MDA-MB-231 and Hs578T) were
provided by Chinese Academy of Science Committee Type Culture
Collection Cell Bank (Shanghai, China). Cell culture experiment was
conducted as recently described (25). MDA-MB-231 and Hs578T cells were
cultured in RPMI 1640 and DMEM medium, respectively, both
supplemented with 10% fetal bovine serum (FBS) and 100 units/ml
penicillin/streptomycin (all Gibco; Thermo Fisher Scientific,
Inc.). The cells were maintained at 37°C with 5%
CO2.
Plasmids and transient
transfection
Short hairpin (sh)RNA sh-BCL11A and the negative
control sh-NC were cloned into expression plasmids to generate
recombinant lentiviral vectors (LV-shRNA-BCL11A and LV-shRNA-NC).
Cells were transfected with recombinant lentivirus plasmid
LV-shRNA-BCL11A or LV-shRNA-NC in the presence of 5 µg/µl polyene
(Sigma-Aldrich; Merck KGaA). Lentivirus plasmids were provided by
Suzhou GenePharma Co., Ltd. and si-BCL11A was provided by Guangzhou
RiboBio Co., Ltd. The sequences were: sh-BCL11A:
5′-GCAGATAAACTTCTGCACTGG-3′; sh-NC: 5′-TTCTCCGAACGTGTCACGT-3′;
si-BCL11A: 5′-GAACACTCATGGATTAAGA-3′.
Western blotting
Samples were prepared using a RIPA lysis buffer
containing a protease inhibitor cocktail. The protein
concentrations were quantified with the BCA kit (All kits from
KeyGen Biotech). Subsequent experiment was conducted as recently
described (26). The antibodies used
for western blotting were as follows: Rabbit anti-BCL11A (ab191401,
1:10,000, Abcam), rabbit anti-AR (ab74272, 1:300, Abcam), rabbit
anti-GAPDH (ab181603, 1:10,000, Abcam) and rabbit IgG (#7074,
1:10,000, Cell Signaling Technologies, Inc.).
Colony formation assay
Transfected cells were seeded into a 6-well plate
(600 cells per well) and incubated in fresh medium at 37°C for 14
days. The resulting colonies were fixed with 100% methanol. And
then colonies were stained with 0.5% crystal violet. Colonies
containing >50 cells were manually numbered.
Wound-healing assay
Following transfection, cells were seeded into
6-well plates and incubated at 37°C until 90% confluence. A sterile
pipette tips was used to create a wound across each cell monolayer,
and the cells were incubated in serum-free medium for a further 12
h. Images of the wound area were captured at 0 and 12 h, and wound
closure was assessed using Image-Pro Plus 6.0 (Media Cybernetics,
Inc.).
Transwell assay
Following transfection, 2×104 cells (in
200 µl serum-free medium) were added to the upper chamber of a
Transwell insert; 800 µl of medium (supplemented with 10% FBS) was
added to the lower chamber as a chemoattractant. After culturing at
37°C for 24 h, cells that had migrated to the lower matrix membrane
were stained with crystal violet; stained cells were counted using
a light microscope.
Cell cycle analysis
Cells transfected with LV-shRNA-BCL11A, LV-shRNA-NC
or the mock were cultured in 6-well plates at 2×105 per
well. Cells were harvested and centrifuged at 1500 rpm for 5 min.
Subsequent steps were performed as recently described (27). The cells were washed with ice-cold
PBS and fixed with 70% ethanol overnight at −20°C. The fixed cells
were washed with PBS for 10 min and treated with RNAase A for 30
min followed by incubation with propidium iodide for 30 min at room
temperature. The cell cycle in each specific sample were evaluated
by the flow cytometer (FACSCalibur) following the manufacture's
instruction. The ModFit software was used to analyze the cell cycle
result.
Statistical analysis
Data analysis was performed with SPSS v.20 (IBM
Corp.) and GraphPad Prism software v.7 (GraphPad Software, Inc.).
All experiments were independently conducted ≥3 times. Comparisons
between two groups were analyzed using Student's t-test.
Comparisons between multiple groups were analyzed using one-way
ANOVA, followed by Tukey's post hoc test. For the IHC results,
P-values were calculated using the χ2 test to compare
the distribution of demographic variables. Survival analysis were
performed according to Kaplan-Meier method. Prognostic factor was
calculated using Cox proportional hazards regression model.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Elevated expression of BCL11A
correlates with an unfavorable outcome of breast cancer
patients
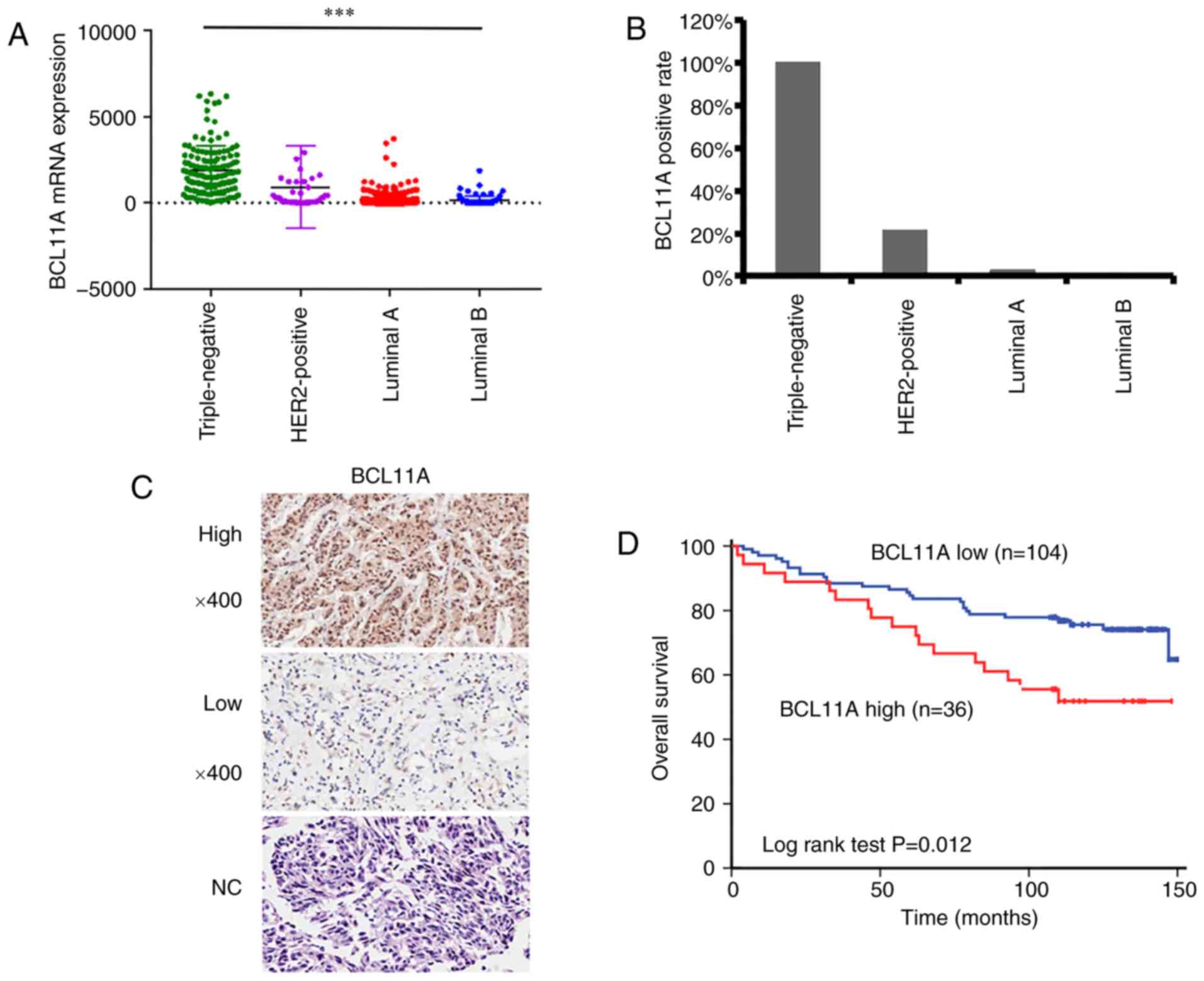
To evaluate the expression level of BCL11A in breast
cancer, BCL11A mRNA expression data was extracted from TCGA
database, which showed that TNBC tumors expressed significantly
increased levels of BCL11A compared with those of other types of
breast cancer (P<0.001) (Fig.
1A). Subsequently, IHC staining was performed in a breast
cancer microarray (140 tissue samples) to detect the expression of
BCL11A (Fig. 1B-C). The positive
rates of BCL11A in TNBC, HER-2 breast cancer and luminal A breast
cancer were 100% (31/31), 21.4% (2/14) and 2.9% (2/70),
respectively. Demographic, pathological and clinical variables were
also analyzed. Furthermore, the association between BCL11A
expression and clinical characteristics of 140 cases was reviewed
and analyzed (Table I). Results
showed that 36 cases (25.7%) exhibited high BCL11A levels, while
104 cases (74.3%) exhibited low BCL11A levels. In addition, these
data revealed that BCL11A expression was not significantly
correlated with age, histological or TNM stage. Notably, a high
BCL11A level was strongly associated with decreased level of ER, PR
and HER-2, and an increased level of AR. Then, correlation between
BCL11A level with patient clinical outcomes was investigated,
revealing that high BCL11A expression was correlated with poor OS
(Fig. 1D, P=0.012). In order to
elucidate whether OS was correlated with any of the
clinicopathological variables, multivariate analysis with Cox
proportional hazard model was applied. As displayed in Table II, several factors were
significantly associated. Therefore, BCL11A was speculated to act
as an independent unfavorable prognostic biomarker in breast cancer
[(HR)=2.099; 95% (CI)=1.123–3.925; P=0.020]. In summary, these
results suggest that in breast cancer patients, BCL11A expression
is correlated with a poor outcome.
 | Table I.Relationship between expression of
BCL11A and clinicopathologic features of patients with breast
cancer. |
Table I.
Relationship between expression of
BCL11A and clinicopathologic features of patients with breast
cancer.
|
| BCL11A high
(n=36) | BCL11A low
(n=104) |
|
|
|---|
|
|
|
|
|
|
|---|
| Variables | n | % | n | % | χ2 | P-value |
|---|
| Age |
|
|
|
| 0.946 | 0.331 |
| ≤51
years | 16 | 44.4 | 56 | 53.8 |
|
|
| >51
years | 20 | 55.6 | 48 | 46.2 |
|
|
|
Histologicala |
|
|
|
| 3.278 | 0.166 |
| I | 1 | 2.8 | 11 | 10.9 |
|
|
|
II–III | 35 | 97.2 | 90 | 89.1 |
|
|
| TNM
stageb |
|
|
|
| 0.714 | 0.700 |
|
I–II | 24 | 66.7 | 67 | 65.7 |
|
|
|
III | 12 | 33.3 | 35 | 34.3 |
|
|
| ER
statusc |
|
|
|
| 80.545 |
<0.001 |
|
Negative | 34 | 94.4 | 13 | 13.5 |
|
|
|
Positive | 2 | 5.6 | 83 | 86.5 |
|
|
| PR
statusd |
|
|
|
| 62.177 |
<0.001 |
|
Negative | 35 | 97.2 | 23 | 24.0 |
|
|
|
Positive | 1 | 2.8 | 73 | 76.0 |
|
|
| HER-2
statuse |
|
|
|
| 9.503 | 0.009 |
|
Negative | 33 | 91.7 | 68 | 70.8 |
|
|
|
Positive | 3 | 8.3 | 28 | 29.2 |
|
|
| AR
statusf |
|
|
|
| 4.223 | 0.040 |
|
Negative | 8 | 22.2 | 43 | 41.3 |
|
|
|
Positive | 28 | 77.8 | 61 | 58.7 |
|
|
 | Table II.Univariate Cox proportional hazard
regression model analysis of breast cancer for overall
survival. |
Table II.
Univariate Cox proportional hazard
regression model analysis of breast cancer for overall
survival.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age | 1.397
(0.769–2.538) | 0.272 |
|
|
| TNM stage | 2.361
(1.307–4.25) | 0.004 | 2.543
(1.398–4.625) | 0.002 |
| Tumor size |
|
|
|
|
| ≤2
cm | 1.000 |
| 1.000 |
|
| 2–5
cm | 1.403
(0.617–3.10) | 0.419 | 1.237
(0.541–2.828) | 0.614 |
| >5
cm | 2.355
(0.786–7.01) | 0.126 | 1.584
(0.478–5.251) | 0.452 |
| Node status |
|
|
|
|
| 0 | 1.000 |
| 1.000 |
|
|
1–3 | 1.003
(0.439–2.29) | 0.994 | 0.897
(0.381–2.113) | 0.840 |
|
4–9 | 2.454
(1.190–5.01) | 0.015 | 2.464
(0.231–26.27) | 0.455 |
|
≥10 | 2.177
(0.709–6.64) | 0.174 | 2.474
(0.212–28.93) | 0.470 |
| Histological | 0.953
(0.341–2.67) | 0.928 |
|
|
| ER status | 0.532
(0.288–0.984) | 0.044 | 0.597
(0.314–1.134) | 0.115 |
| PR status | 0.528
(0.284–0.99) | 0.043 | 0.599
(0.313–1.146) | 0.121 |
| HER-2 status | 0.829
(0.382–1.76) | 0.634 | 0.718
(0.324–1.592) | 0.415 |
| BCL11A status | 2.150
(1.169–3.96) | 0.014 | 2.099
(1.123–3.925) | 0.020 |
BCL11A-knockdown reduces TNBC cell
proliferation
Multiple researches have demonstrated that BCL11A is
critically associated with tumor development and progression.
However, the function of BCL11A in breast cancer, particularly in
TNBC, requires additional in-depth exploration. The present study
demonstrated that TNBC tissues exhibited significantly higher
BCL11A expression compared with other types of breast cancer. To
further gain evidence of its crucial role in TNBC, the effects of
BCL11A inhibition on the tumorigenic capacity of TNBC cells were
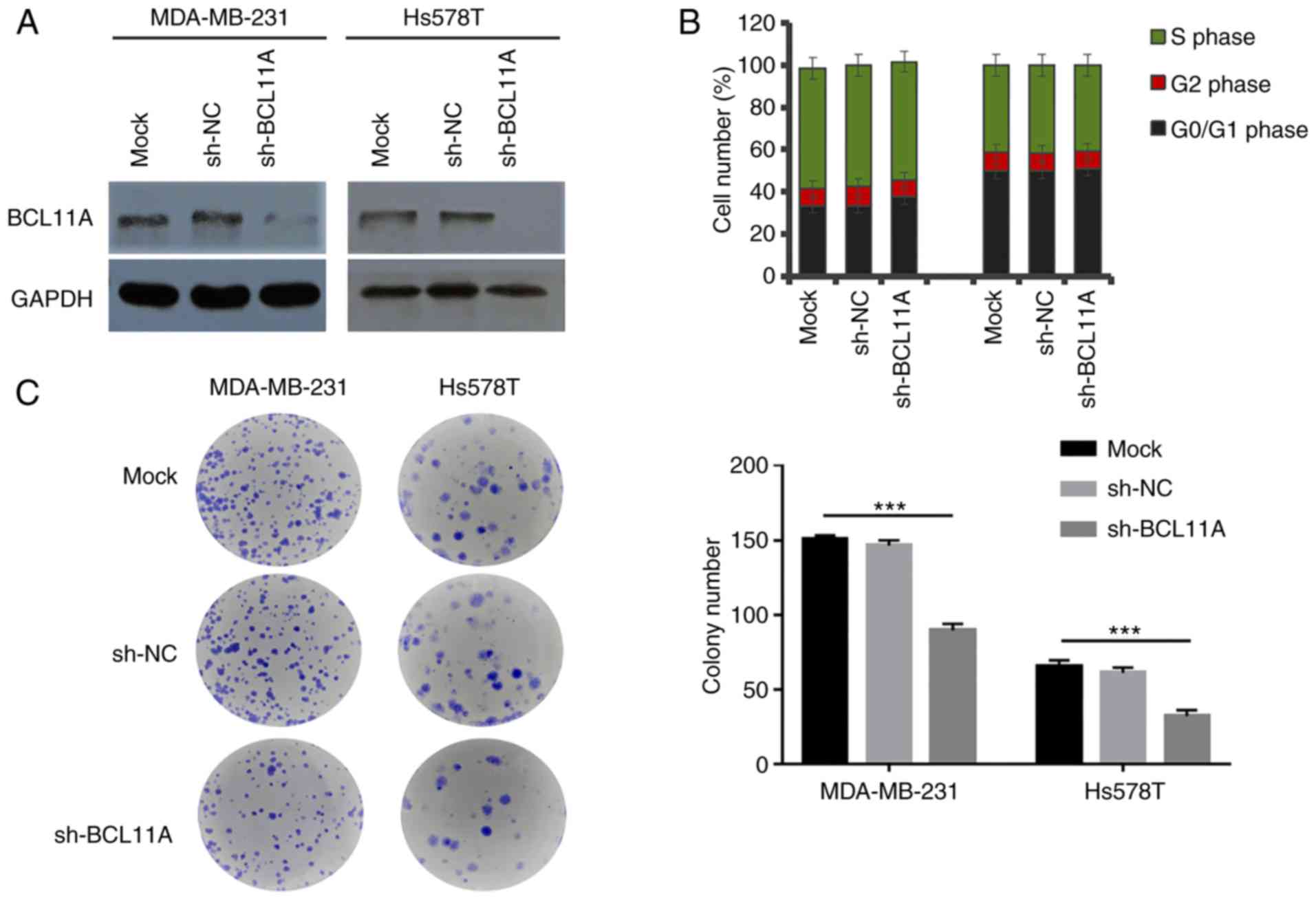
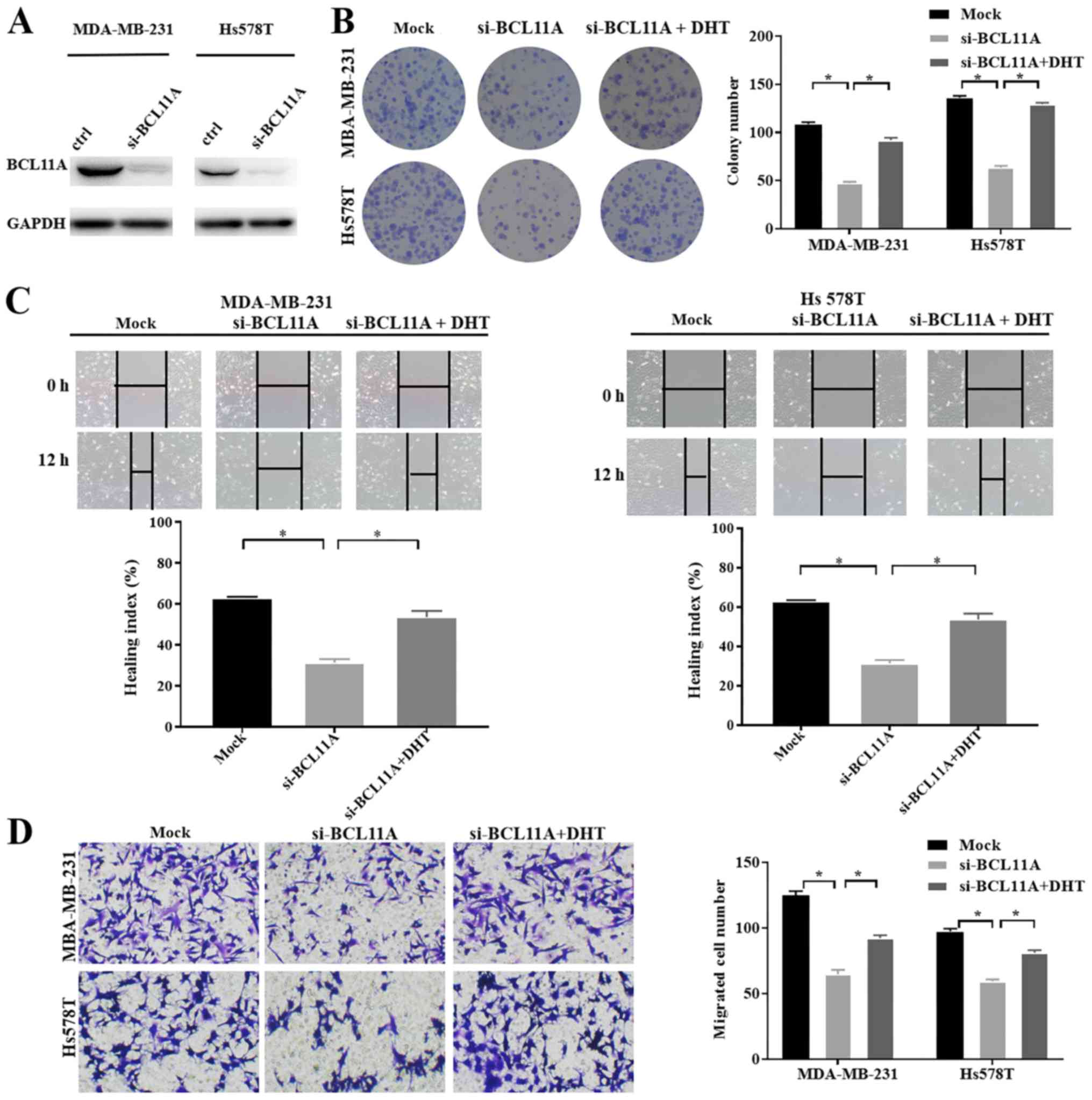
investigated, using an shRNA-knockdown assay (Fig. 2A). BCL11A expression was not
significantly related with cell cycle (Fig. 2B, Fig.
S1A-F). However, the results of the colony formation assay
showed that BCL11A-knockdown significantly reduced the cell
proliferative ability in both cell lines (Fig. 2C). These data thus indicated that
BCL11A could induce TNBC cell proliferation and promote cancer
development and progression.
BCL11A induces TNBC cell migration and
invasion
Few studies have elucidated the correlation between
BCL11A expression and cell proliferation in TNBC. The present study
indicated that BCL11A promoted TNBC proliferation. To further
evaluate its biological function, the influence of BCL11A protein
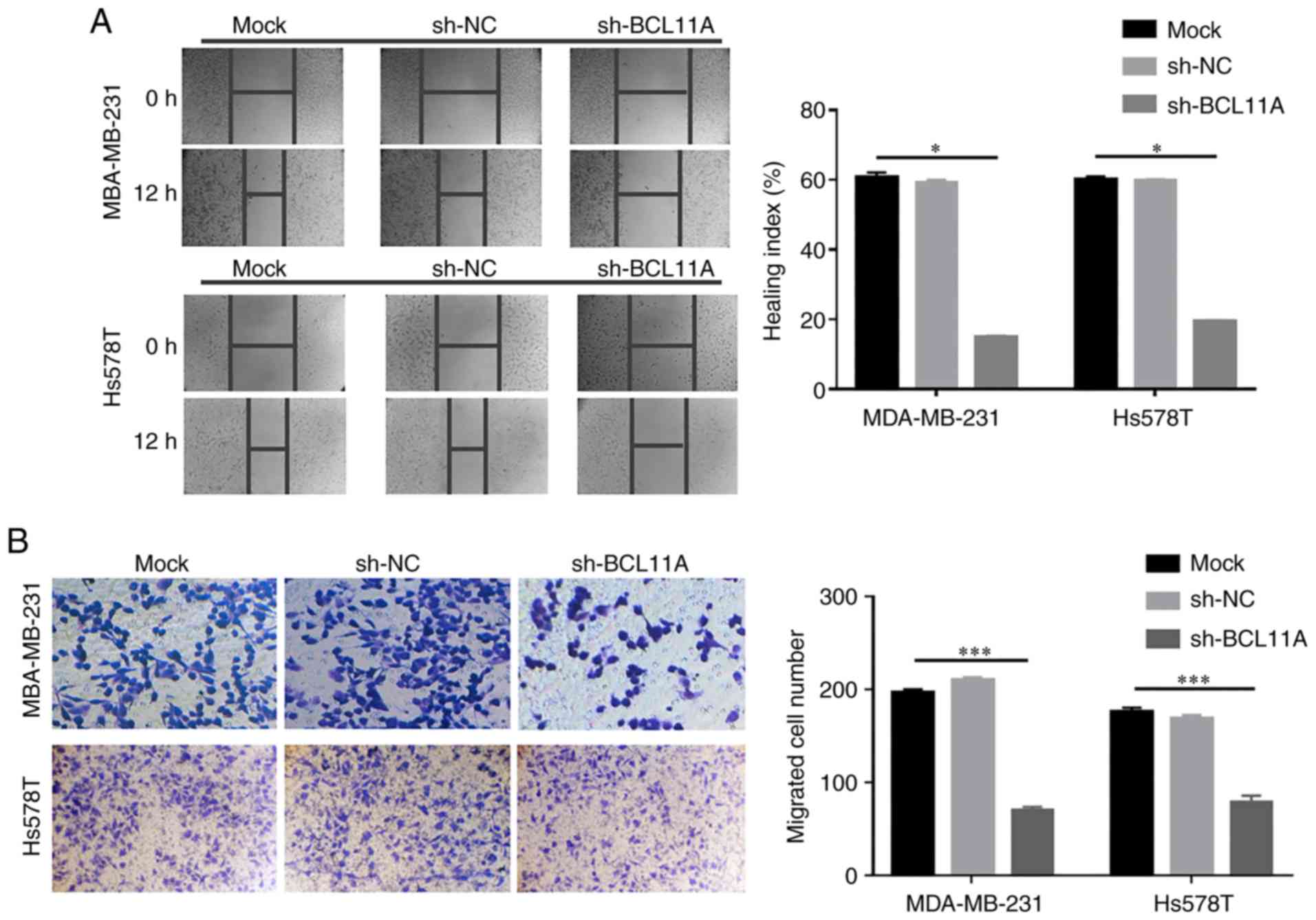
on TNBC migration and invasiveness ability was determined. Using a
wound-healing assay, BCL11A knockdown noticeably inhibited cell
migration in both cell lines (Fig.
3A). Likewise, a Transwell assay revealed that BCL11A-knockdown
significantly decreased the metastatic potential of TNBC cells
(Fig. 3B). These data thus revealed
that BCL11A could facilitate TNBC migration and invasion, which may
result in carcinogenesis and metastasis. Collectively, these
results confirmed that exogenous BCL11A knockdown inhibited tumor
progression in TNBC, and support the tumorigenic role of BCL11A in
TNBC cell function.
BCL11A regulates the expression of AR
in TNBC cells
The aforementioned data showed that BCL11A was
upregulated and served as a carcinogenic factor in TNBC. Although
TNBC is characterized as lacking of ER, PR and HER-2, it expresses
other receptors like AR. Yet the correlation of BCL11A and AR in
TNBC cells remains unknown. To address it, the present study
evaluated if inhibiting BCL11A could influence AR expression level
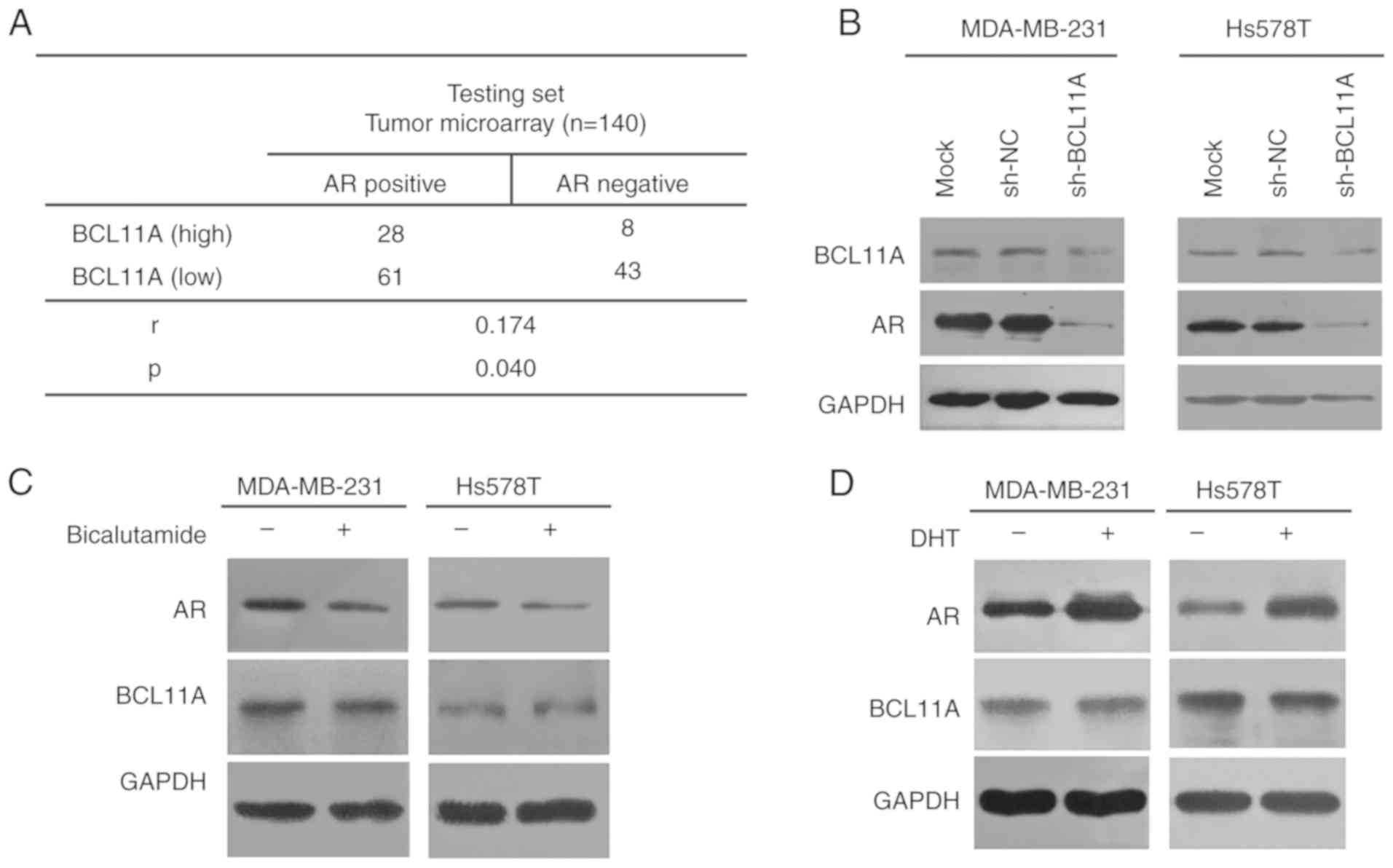
in TNBC cell lines. Using a tissue microarray, a significant
positive association was illustrated between BCL11A level and AR
(Fig. 4A). Furthermore, knockdown of
BCL11A expression significantly down-regulated the expression level
of AR in TNBC cells (Fig. 4B). To
determine whether the AR reversely regulated BCL11A expression,
TNBC cells were treated for 48 h with dihydrotestosterone (DHT), an
AR agonist, or bicalutamide, an AR antagonist. BCL11A protein
expression was then analyzed by western blotting. The results show
that the activation or inhibition of AR signaling had no
significant impact on BCL11A protein expression level (Fig. 4C and D). Moreover, in the functional
assay, the AR agonist, DHT, significantly reversed the antitumor
effect of si-BCL11A in both TNBC cell lines (Fig. 5A-D). Collectively, these results
provided solid evidence that BCL11A could induce the expression of
AR and further had an influence on proliferation, migration and
invasion in TNBC cell lines.
Discussion
In the current study, BCL11A was shown to be
upregulated, and to be a predictor of poor clinical outcome in
patients with TNBC. BCL11A was determined to serve as an
independent indicator of unfavorable outcome in breast cancer.
Experimentally, down-regulating BCL11A expression in TNBC cells
significantly inhibited tumor progression. Statistical analysis
demonstrated that high BCL11A level was associated with higher
expression of AR. Subsequent experimentation also revealed that
knockdown of BCL11A expression significantly down-regulated the
expression level of AR in TNBC cell lines and further had an
influence on cell function in TNBC.
It has previously been disclosed that as well as
normal lymphoid development and globin switching, BCL11A plays an
active role in mediating tumorigenesis (28,29). In
bone metastasis of breast cancer, the expression of BCL11A is
significantly increased compared with control group (30). In the present study, BCL11A was shown
to be upregulated in patients with TNBC, and high BCL11A protein
level was associated with poor OS, suggesting that there is a
correlation of BCL11A expression with breast cancer. Regarding
tumorigenesis in prostate cancer, it was previously shown that
BCL11A overexpression strongly reversed the influence of tumor
progression induced by FOXQ1 inhibition (19). On the basis of previous results, in
TNBC cells, disrupting BCL11A expression significantly reduced cell
proliferation and invasion. Additionally, gene expression profiling
of BCL11A siRNA-treated SUDHL6 cells proved that BCL11A could be
associated with the signaling network of cell cycle (22). However, the present data did not
indicate a relationship between BCL11A and cell cycle in TNBC. The
possible reason may be that BCL11A cannot increase transcriptional
rate in TNBC, which would lead to the decreased expression of
cyclin genes. Tsang et al investigated the cell cycle
phenotype in the BCL11A-deficiency hematopoietic stem cells (HSCs),
suggesting the reduction of quiescence in the
Bcl11a−/− HSC compartment (31). However, changes in the transcriptome
are one of the earliest events in entry to the cell cycle.
Therefore, the cell cycle progression requires further
translational and post-translational regulation, which might accout
for our cell cycle result. During neocortical development, BCL11A
is necessary for the cell polarity switch and upper layer neuron
migration (32). In current study,
disrupting BCL11A expression significantly reduced cell migration
in TNBC. Finally, our data provided compelling evidence that BCL11A
is critical in the tumor progression of TNBC.
In addition, it is important to elucidate the
potential underlying mechanism in TNBC and the downstream targets
of BCL11A in TNBC. As we all known, TNBC is characterized as
lacking of ER, PR and HER-2. However, it expresses other hormone
receptors like AR. AR has been recognized to act as a very
important factor in recent years. Our present data indicated that
high BCL11A level correlated with higher level of AR in breast
cancer patients. Although correlation coefficient r is only 0.174,
we evaluated to focus on the significance of P-value, which was
0.040 and statistically significant. Previous study has reported
that AR was expressed in a particular group of TNBC patients and
was strongly correlated with clinical prognosis. Increasing
evidence has indicated that AR signaling pathway played a critical
role in modulating oncogenesis and tumor metastasis. To gain more
solid evidence, we designed the following experiment in TNBC cell
lines to further investigate if inhibiting BCL11A could influence
AR expression level and subsequently influence tumor cell function.
The present study showed that BCL11A knockdown significantly
down-regulated the expression level of AR in TNBC cells and further
had an influence on cell function, which suggests that AR may be a
downstream target of BCL11A. Also, the results of DHT or
bicalutamide treatment in TNBC cells demonstrated that activating
or inhibiting AR signaling had no significant impact on BCL11A
protein level. To the best of our knowledge, the present study is
the first to reveal a unique molecular mechanism by which BCL11A
regulates the level of AR in TNBC, and further research in the
future is required to determine the precise mechanism of BCL11A and
AR in TNBC tumorigenesis and potential targeted therapies.
In summary, the present study suggests that BCL11A
as a novel biomarker of tumorigenesis, which acts through the
upregulation of the AR in breast cancer. Therefore, BCL11A may act
as both a new prognostic predictor and a feasible target for TNBC
therapy. Moreover, these data indicate that in-depth research of
BCL11A and AR as promising implications for TNBC-targeted
therapeutic options is warranted.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by The National
Natural Science Foundation of China (grant no. 81773102) and The
Key International Cooperation of the National Natural Science
Foundation of China (grant no. 81920108029 to XXG).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XW and YX performed the experiments, wrote the main
manuscript and analyzed the data. KX performed the experiments. YC
helped with the statistical analysis. XG and XX designed the
project and revised the manuscript. All authors contributed to the
revised manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by The Ethics
Committee of Jinling Hospital. Written informed consent was
obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
BCL11A
|
B-cell lymphoma/leukemia 11A
|
|
TNBC
|
triple-negative breast cancer
|
|
TCGA
|
The Cancer Genome Atlas
|
|
OS
|
overall survival
|
|
DHT
|
dihydrotestosterone
|
|
HR
|
hazard ratio
|
|
HC
|
immunohistochemistry
|
|
ER
|
estrogen receptor
|
|
PR
|
progesterone receptor
|
|
AR
|
androgen receptor
|
References
|
1
|
Lehmann BD, Bauer JA, Chen X, Sanders ME,
Chakravarthy AB, Shyr Y and Pietenpol JA: Identification of human
triple-negative breast cancer subtypes and preclinical models for
selection of targeted therapies. J Clin Invest. 121:2750–2767.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Millikan RC, Newman B, Tse CK, Moorman PG,
Conway K, Dressler LG, Smith LV, Labbok MH, Geradts J, Bensen JT,
et al: Epidemiology of basal-like breast cancer. Breast Cancer Res
Treat. 109:123–139. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Skor MN, Wonder EL, Kocherginsky M, Goyal
A, Hall BA, Cai Y and Conzen SD: Glucocorticoid receptor antagonism
as a novel therapy for triple-negative breast cancer. Clin Cancer
Res. 19:6163–6172. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lonergan PE and Tindall DJ: Androgen
receptor signaling in prostate cancer development and progression.
J Carcinogenesis. 10:202011. View Article : Google Scholar
|
|
5
|
Mrklic I, Pogorelic Z, Capkun V and Tomić
S: Expression of androgen receptors in triple negative breast
carcinomas. Acta Histochem. 115:344–348. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vera-Badillo FE, Templeton AJ, de Gouveia
P, Diaz-Padilla I, Bedard PL, Al-Mubarak M, Seruga B, Tannock IF,
Ocana A and Amir E: Androgen receptor expression and outcomes in
early breast cancer: A systematic review and meta-analysis. J Natl
Cancer Inst. 106:djt3192014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
He J, Peng R, Yuan Z, Wang S, Peng J, Lin
G, Jiang X and Qin T: Prognostic value of androgen receptor
expression in operable triple-negative breast cancer: A
retrospective analysis based on a tissue microarray. Med Oncol.
29:406–410. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gerratana L, Basile D, Buono G, De Placido
S, Giuliano M, Minichillo S, Coinu A, Martorana F, De Santo I, Del
Mastro L, et al: Androgen receptor in triple negative breast
cancer: A potential target for the targetless subtype. Cancer Treat
Rev. 68:102–110. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Satterwhite E, Sonoki T, Willis TG, Harder
L, Nowak R, Arriola EL, Liu H, Price HP, Gesk S, Steinemann D, et
al: The BCL11 gene family: Involvement of BCL11A in lymphoid
malignancies. Blood. 98:3413–3420. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Saiki Y, Yamazaki Y, Yoshida M, Katoh O
and Nakamura T: Human EVI9, a homologue of the mouse myeloid
leukemia gene, is expressed in the hematopoietic progenitors and
down-regulated during myeloid differentiation of HL60 cells.
Genomics. 70:387–391. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jin C, Yu W, Lou X, Zhou F, Han X, Zhao N
and Lin B: UCHL1 is a putative tumor suppressor in ovarian cancer
cells and contributes to cisplatin resistance. J Cancer. 4:662–670.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Boelens MC, Kok K, van der Vlies P, van
der Vries G, Sietsma H, Timens W, Postma DS, Groen HJ and van den
Berg A: Genomic aberrations in squamous cell lung carcinoma related
to lymph node or distant metastasis. Lung Cancer. 66:372–378. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Podgornik H, Pretnar J, Skopec B,
Andoljšek D and Černelč P: Concurrent rearrangements of BCL2, BCL3,
and BCL11A genes in atypical chronic lymphocytic leukemia.
Hematology. 19:45–48. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kapatai G and Murray P: Contribution of
the Epstein Barr virus to the molecular pathogenesis of Hodgkin
lymphoma. J Clin Pathol. 60:1342–1349. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chetaille B, Bertucci F, Finetti P,
Esterni B, Stamatoullas A, Picquenot JM, Copin MC, Morschhauser F,
Casasnovas O, Petrella T, et al: Molecular profiling of classical
Hodgkin lymphoma tissues uncovers variations in the tumor
microenvironment and correlations with EBV infection and outcome.
Blood. 113:2765–3775. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Agueli C, Cammarata G, Salemi D, Dagnino
L, Nicoletti R, La Rosa M, Messana F, Marfia A, Bica MG, Coniglio
ML, et al: 14q32/miRNA clusters loss of heterozygosity in acute
lymphoblastic leukemia is associated with up-regulation of BCL11a.
Am J Hematol. 85:575–578. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jiang BY, Zhang XC, Su J, Meng W, Yang XN,
Yang JJ, Zhou Q, Chen ZY, Chen ZH, Xie Z, et al: BCL11A
overexpression predicts survival and relapse in non-small cell lung
cancer and is modulated by microRNA-30a and gene amplification. Mol
Cancer. 12:612013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lazarus KA, Hadi F, Zambon E, Bach K,
Santolla MF, Watson JK, Correia LL, Das M, Ugur R, Pensa S, et al:
BCL11A interacts with SOX2 to control the expression of epigenetic
regulators in lung squamous carcinoma. Nat Commun. 9:33272018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang X, Wang L, Wang Y, Shi S, Zhu H,
Xiao F, Yang J, Yang A and Hao X: Inhibition of FOXQ1 induces
apoptosis and suppresses proliferation in prostate cancer cells by
controlling BCL11A/MDM2 expression. Oncol Rep. 36:2349–2356. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kaneda H, Arao T, Tanaka K, Tamura D,
Aomatsu K, Kudo K, Sakai K, De Velasco MA, Matsumoto K, Fujita Y,
et al: FOXQ1 is overexpressed in colorectal cancer and enhances
tumorigenicity and tumor growth. Cancer Res. 70:2053–2063. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu L, Pan R, Zhou D, Ye G and Tan W:
BCL11A enhances stemness and promotes progression by activating
Wnt/β-catenin signaling in breast cancer. Cancer Manag Res.
11:2997–3007. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He D, Wu H, Ding L and Li Y: Combination
of BCL11A siRNA with vincristine increases the apoptosis of SUDHL6
cells. Eur J Med Res. 19:342014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gao Y, Wu H, He D, Hu X and Li Y:
Downregulation of BCL11A by siRNA induces apoptosis in B lymphoma
cell lines. Biomed Rep. 1:47–52. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang F, Shen Y, Zhang W, Jin J, Huang D,
Fang H, Ji W, Shi Y, Tang L, Chen W, et al: An androgen receptor
negatively induced long non-coding RNA ARNILA binding to miR-204
promotes the invasion and metastasis of triple-negative breast
cancer. Cell Death Differ. 25:2209–2220. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu A, Li Y, Song W, Xu Y, Yang F, Zhang
W, Yin Y and Guan X: Antiproliferative effect of androgen receptor
inhibition in mesenchymal Stem-like triple-negative breast cancer.
Cell Physiol Biochem. 38:1003–1014. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang W, Luo J, Yang F, Wang Y, Yin Y,
Strom A, Gustafsson JÅ and Guan X: BRCA1 inhibits AR-mediated
proliferation of breast cancer cells through the activation of
SIRT1. Sci Rep. 6:220342016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ji W, Shi Y, Wang X, He W, Tang L, Tian S,
Jiang H, Shu Y and Guan X: Combined androgen receptor blockade
overcomes the resistance of breast cancer cells to palbociclib. Int
J Biol Sci. 15:522–532. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sankaran VG, Xu J, Ragoczy T, Ippolito GC,
Walkley CR, Maika SD, Fujiwara Y, Ito M, Groudine M, Bender MA, et
al: Developmental and species-divergent globin switching are driven
by BCL11A. Nature. 460:1093–1097. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin Y, Zhang Q, Zhang HM, Liu W, Liu CJ,
Li Q and Guo AY: Transcription factor and miRNA co-regulatory
network reveals shared and specific regulators in the development
of B cell and T cell. Sci Rep. 5:152152015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hayashi N, Manyam GC, Gonzalez-Angulo AM,
Niikura N, Yamauchi H, Nakamura S, Hortobágyi GN, Baggerly KA and
Ueno NT: Reverse-phase protein array for prediction of patients at
low risk of developing bone metastasis from breast cancer.
Oncologist. 19:909–914. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tsang JC, Yu Y, Burke S, Buettner F, Wang
C, Kolodziejczyk AA, Teichmann SA, Lu L and Liu P: Single-cell
transcriptomic reconstruction reveals cell cycle and multi-lineage
differentiation defects in Bcl11a-deficient hematopoietic stem
cells. Genome Biol. 16:1782015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wiegreffe C, Simon R, Peschkes K, Kling C,
Strehle M, Cheng J, Srivatsa S, Liu P, Jenkins NA, Copeland NG, et
al: Bcl11a (Ctip1) controls migration of cortical projection
neurons through regulation of Sema3c. Neuron. 87:311–325. 2015.
View Article : Google Scholar : PubMed/NCBI
|