Signal transducer and activator of transcription
(STAT) proteins are a class of transcription factor that are
activated by cytokines, growth factors and other peptide ligands.
STATs are activated by tyrosine phosphorylation in response to
diverse cytokine signals in the cytoplasm. Following activation,
the STAT proteins translocate to the nucleus, binding their
specific targets and serving as transcription factors (1–4). STATs
influence numerous physiological processes, including cell
proliferation, apoptosis, division and differentiation (5). In healthy cells, the activation of
STATs is tightly regulated to prevent uncontrolled gene expression;
however, prolonged activation of STATs in cancer cells may result
in significant adverse effects, such as drug resistance and poor
prognosis (5,6). In humans, the STAT family comprises
seven proteins, including STAT-1, −2, −3, −4, −5A, −5B and −6, and
the genes encoding the STAT family are located on chromosomes 2
(STAT-1 and −4), 12 (STAT-2 and −6) and 17 (STAT-3, −5A and −5B)
(7). Among the seven members, STAT-3
and −5 exhibit the strongest association with tumor progression.
Persistent activation of STAT-3 or STAT-5 (particularly STAT-3)
regulates a variety of functions, including proliferation, cell
cycle progression, apoptosis, angiogenesis and immune evasion
(8–10). Consequently, STAT-3 mainly
contributes to tumor proliferation and survival owing to its role
in stromal cells, including immune cells recruitment in the tumor
microenvironment to promote tumor growth, and is, therefore,
recognized as a promising target for cancer therapy (11–16).
Previous studies have demonstrated that DNA
methylation and chromatin modulation may also be regulated by
STAT-3 via epigenetic mechanisms (17,18).
Moreover, STAT-3 has been recognized as a potent immune checkpoint
regulator for multiple antitumor immune response pathways (12,13).
Despite the plethora of evidence implicating STAT-3 in the
progression of several types of cancer and indicating it as an
ideal target for cancer therapy, there is still no clinical drug
available that directly targets STAT-3 (19). Thus, a novel small molecule able to
directly target STAT-3 may represent a promising novel STAT-3
inhibitor. Notably, STAT-3 is highly complex in its diverse
biological functions, as well as its various activators. Therefore,
further investigations into STAT-3 biology and signaling pathways
are particularly important.
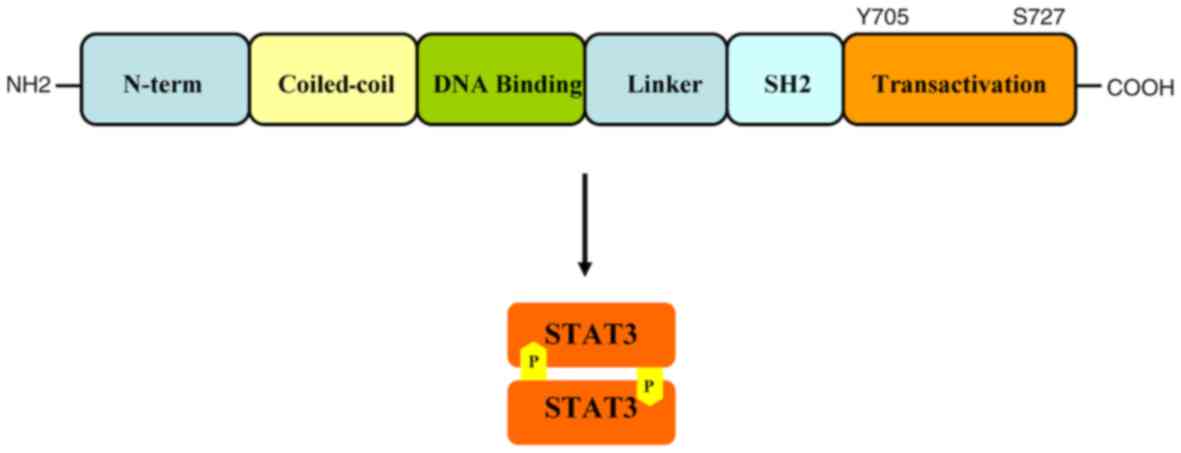
STAT-3 is comprised of ~800 amino acids, and its
relative mass is 92 kDa; it has several conserved functional
domains, including N-terminal, coiled-domain, DNA-binding domain
(DBD), Src-homology-2 (SH-2) domain and transactivation (TA) domain
(20–22). DBD has specificity with certain
regions of DNA, allowing STAT-3 to bind downstream of the target
gene-promoter element to induce the expression of target genes. The
SH-2 domain participates in phosphorylation of tyrosine residues,
facilitating protein-protein interactions with
tyrosine-phosphorylated proteins (23). Furthermore, the SH-2 domain is
critical in the formation of the STAT-3 dimer (the region that
STAT-3 binds to in order to activate receptors). In between the DBD
and SH-2 domain there is a linker protein that mediates the
stability of DNA binding and assembly of the transcriptional body
(24). The TA domain contains one
tyrosine phosphorylation site (Yyr705) and one serine
phosphorylation site (Ser727). During STAT-3 activation, tyrosine
and serine residues are phosphorylated by upstream kinases and
recognized by the SH-2 domain (25),
as indicated in Fig. 1.
However, it has been demonstrated that STAT-3
contains three different isomers: The full-length α form, and two
shorter isoforms, β and γ, which are the result of mRNA splicing
and proteolysis, respectively. STAT-3α primarily participates in
cell proliferation and transformation, while STAT-3β regulates cell
differentiation mediated by granulocyte colony-stimulating factor
(G-CSF) and lacks a serine phosphorylation site (26). Activated STAT-3γ has been identified
in differentiated neutrophils and does not contain a
transactivation domain (27,28). A previous study also suggested that
these isoforms may exert a dominant-negative effect on their
full-length counterparts (29).
Transient STAT3 activation is a key determinant of
tissue integrity restoration, wound healing and immune response
resolution, given its critical biological functions (30). STAT-3 is heavily regulated to ensure
transient activation under standard conditions. To mediate this
process, three classes of proteins located upstream of STAT-3
influence negative regulation of its activation, including tyrosine
phosphatases, protein inhibitors of activated STATs (PIAS) and
suppressors of cytokine signaling (SOCS) (31). Tyrosine phosphatases downregulate
Janus kinase (JAK)/STAT signaling via dephosphorylation of STAT and
its upstream kinases (32). It has
been reported that the protein tyrosine phosphatase receptor T
specifically dephosphorylates the Tyr705 residue in the TA domain
of STAT-3 (32). The PIAS family
comprises four members: PIAS-1, −3, -x and -y (33). Of these, PIAS-3 is associated with
STAT-3 inhibition via blocking its binding to DNA, recruiting
co-repressors or serving as SUMOylation E3 ligases (34–36). The
SOCS family of proteins, also known as cytokine-induced
SH-2-containing proteins, inhibit STAT-3 binding and decrease JAK
activity via competitive binding to phosphorylated tyrosine
residues on activated cytokine receptors or JAK (19). As the receptor lacks its own
enzymatic activity, STAT-3 is primarily activated via the
JAK-dependent pathway (37).
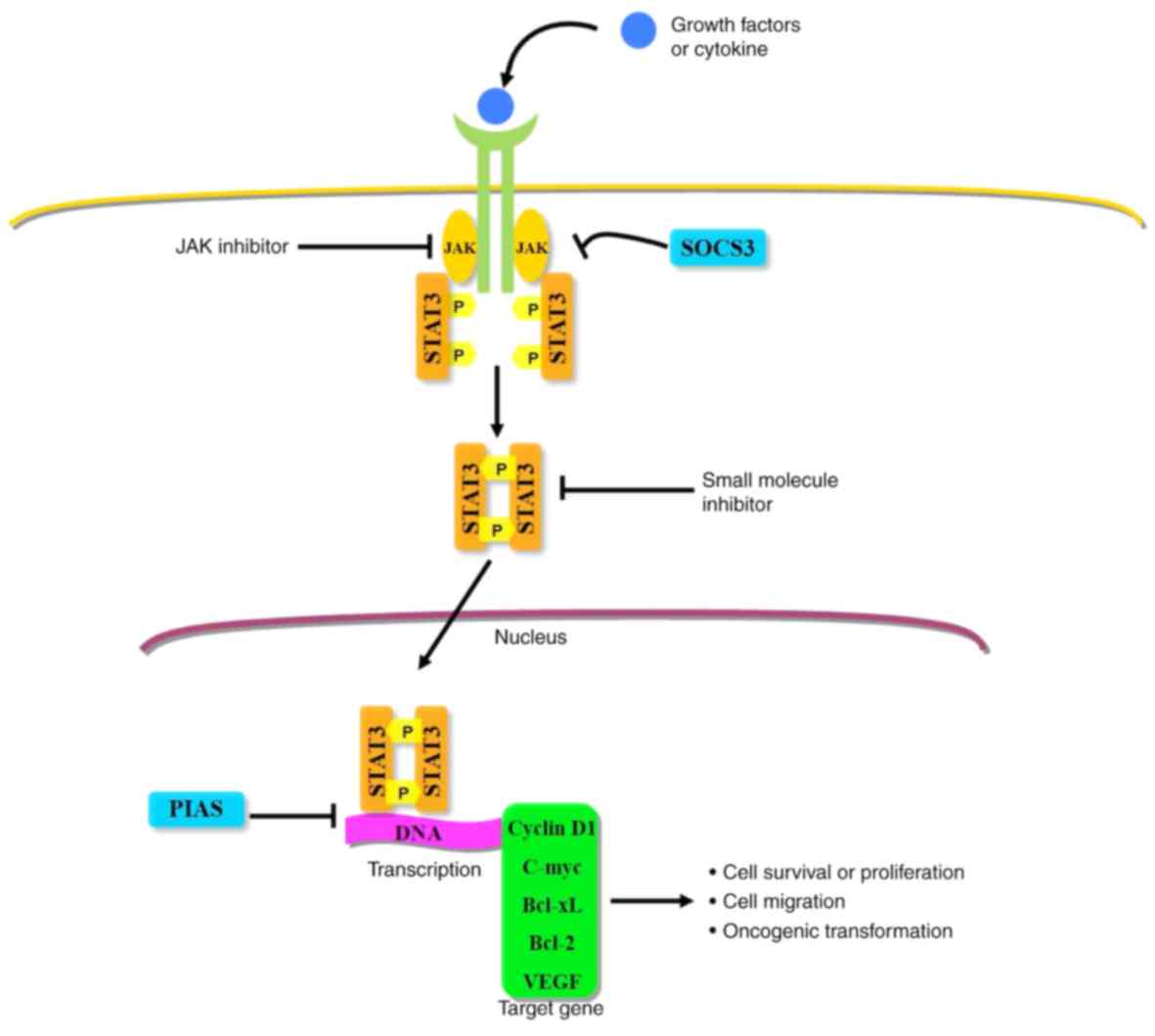
The JAK/STAT-3 pathway is typically considered the
most crucial signaling pathway in the activation of STAT-3
(18). Notably, interleukin (IL)-6
is the most well-characterized upstream cytokine, which binds
specifically to membrane-bound receptors in the JAK/STAT-3 pathway
(38–40). IL-6 binds IL-6 receptor-α on the cell
surface, inducing conformational changes and resulting in the
formation of a homodimer and heterodimer. This initiates the
activation of JAK, followed by recruitment and activation of
cytosolic STAT-3 (18).
Consequently, activated STAT-3 translocates to the nucleus and
binds to its target gene (Fig.
2).
Certain other IL family members can induce STAT-3
translocation to the nucleus, including IL-11 and −31, leukemia
inhibitory factor, ciliary neurotrophic factor and oncostatin M.
Moreover, it has been reported that IL-6-class cytokines serve
crucial roles in the progression of numerous tumor types, including
breast, lung and prostate cancer, and also multiple hematopoietic
malignancies (41–50). Although other IL-6-class cytokines
contribute to tumor development, IL-6 is considered the most
significant regulator of the JAK/STAT-3 pathway in tumors.
Furthermore, certain reports have demonstrated that IL-6 is
regulated by specific oncogenes, such as breakpoint cluster
region-ABL protooncogene and RAS (51,52).
Recent studies have reported that IL-6 promotes cell migration via
the induction of epithelial-mesenchymal transition (EMT), and
IL-6-mediated EMT in breast cancer may be a consequence of STAT-3
activation (53–55). These findings indicate that STAT-3
may represent a target that can inhibit tumor development mediated
by the aforementioned oncogenes (53).
G-protein-coupled receptors (GPCRs) are not
traditional receptors in the JAK/STAT-3 pathway; however, several
findings suggested that GPCRs may be a STAT-3 activator
contributing to tumor progression (56). Sphingosine-1-phosphate receptor-1
(S1PR-1) is a type of GPCR that is upregulated in malignant, immune
and endothelial cells. Previously, S1PR-1 was revealed to be
associated with tumor cell survival and resistance to chemotherapy
in a variety of cancer cells (57–62).
Moreover, sphingosine-1-phosphate receptor 1 (S1PR-1) activates
STAT-3 via JAK-2, resulting in the cyclical upregulation of S1PR-1
expression (63). In general,
toll-like receptors (TLRs) participate in innate immune system
responses, connecting specific immunity to non-specific immunity.
Nevertheless, recent studies have demonstrated the crucial role of
TLRs in the activation of STAT-3 and tumor progression. It has been
reported that TLR-4 induces STAT-3 activation via upregulation of
IL-6 and microRNA (miR)-21, resulting in the neoplastic progression
of colon cancer in vivo (64). Furthermore, TLR-2, −7 and −9 were all
identified to correlate with STAT-3 activation and tumor
progression (65–67). These findings indicate that GPCRs and
TLRs activate the JAK/STAT-3 signaling pathway and support the
potential of targeting GPCRs and TLRs to inhibit STAT-3-induced
tumor growth. Although numerous STAT-3-associated regulatory
mechanisms mediating cancer progression have been revealed, the
targeting of STAT3 in oncotherapy remains a challenge. This is due
to the shallow surface pockets of STAT3 molecules, which make it
difficult to form effective binding.
Additionally, inactivation can also occur via two
pathways: i) The RAS/MAPK pathway; and ii) the non-receptor
tyrosine kinase pathway. Mitogen-activated protein kinase (MAPK) is
a serine/threonine-protein kinase and a downstream signaling
molecule of the RAS pathway, which influences cell proliferation
and differentiation, the inflammatory response and cell pathology.
Various reports have demonstrated that RAS mediates STAT-3-induced
autophagy and tumorigenesis via regulation of MAPK signaling
(68–70), and that the influence of STAT-3 on
gene transcription is significantly decreased following inhibition
of MAPK (71,72). This is due to phosphorylation of
tyrosine residues during signal transduction of STAT-3 and
phosphorylation of serine residues.
Independent of the JAK/STAT-3 and RAS/MAPK pathways,
STAT-3 influences numerous other cytokine signal transduction
pathways by interacting with molecules such as cardiotrophin-1,
angiotensin II and epidermal growth factor receptor. Moreover,
certain non-receptor tyrosine kinases also activate STAT-3, such as
Src (37). Oncogenic Src can
activate STAT-3, while BCR-ABL fusion protein can co-activate STATs
−1, −3 and −5 (73). A recent study
revealed aberrant activation of STAT-3 in normal and neoplastic
colorectal epithelial cells and tumor tissues with upregulated Src
(74). Src homology region 2
domain-containing phosphatase 1 (SHP-1) is a non-receptor protein
tyrosine phosphatase and serves as a tumor suppressor gene in
numerous cancer types. Liu et al (75) demonstrated that SHP-1 expression
levels are downregulated in the majority of tumor types and
correlate with high expression levels of p-STAT3 expression. Thus,
the SHP-1/p-STAT3 signaling axis may represent a potential
therapeutic target and a clinical prognostic indicator in patients
with cancer.
Activation of STAT-3 is transiently and rapidly
sustained for a few minutes in the normal physiological state.
However, persistent activation of STAT-3 can induce abnormal
expression of various genes associated with cell proliferation,
differentiation and apoptosis (76).
Due to its significant carcinogenic properties, STAT-3 has been
recognized as an oncogene. Numerous genes downstream of STAT-3 have
been identified, including Mcl-1, cyclin D1, MYC proto-oncogene
bHLH transcription factor (c-Myc) and vascular endothelial growth
factor (77). Bcl-xL and Mcl-1 are
both members of the Bcl-2 anti-apoptotic family. Bcl-xL and Bcl-2
bind Bax via BH-1 and BH-2, forming homologous and heterologous
dimmers that influence cellular apoptosis (78). In addition, Mcl-1 inhibits the
release of cytochrome c, which may induce the intrinsic
apoptosis pathway. A recent study have demonstrated that the Bcl-xL
promoter initiates transcription during the activation of STAT-3,
resulting in a malignant transformation (79). Typically, cancer cells originate from
healthy cells due to the combinatorial effects of various factors
at different phases of cell division and growth, resulting in
abnormal cell proliferation and differentiation. Notably, the cell
cycle is a key aspect of this malignant transformation. STAT-3
binds Src proto-oncogene non-receptor tyrosine kinase via its SH-2
domain, activating c-myc and inducing the upregulation of cyclin
D1. However, cyclin D1 and c-myc participate in the regulation of
cell cycle progression, and their upregulation results in
dysfunction of the cell cycle and uncontrolled cell proliferation
(80). Angiogenesis is essential for
cancer cell proliferation and metastasis, as it provides tumor
cells with the nutrients and oxygen required for survival.
Increasing evidence has indicated that persistently activated
STAT-3 stimulates tumor angiogenesis (81). It has been reported that STAT-3 not
only regulates the expression of VEGF in a variety of human cancer
types, but that it also influences other critical angiogenic
factors, including angiopoietin, matrix metallopeptidase-9,
chemokine (C-X-C motif) ligand 16 and insulin-like growth factor
binding protein (81–84).
STAT-3 is essential in various cellular processes,
including the cell cycle, cell proliferation, cellular apoptosis,
tumorigenesis and the regulation of the tumor niche. In healthy
cells, STAT-3 activation is regulated to prevent uncontrolled gene
regulation; however, abnormal activation of STAT-3 can result in
the occurrence of numerous disease types (18). Increasing evidence has indicated that
high-frequency abnormal activation of STAT-3 is associated with a
variety of cancer types, including brain, lung, pancreatic, renal,
colorectal, endometrial, cervical, ovarian, breast and prostate
cancer, melanoma, glioma, head and neck squamous cell carcinoma,
lymphoma and leukemia (85–87). Grivennikov et al (43) constructed a colitis-associated cancer
model using mice with intestinal epithelial cell STAT-3-specific
deletion and demonstrated that STAT-3-specific deletion
significantly inhibits the occurrence of tumors and their
progression (44). In addition,
STAT-3 inhibits p53 synthesis and reduces its protective effect on
genomic stability. Following the stimulation of inflammatory
mediators, the probability of DNA damage and gene mutation in
parenchymal cells increases significantly, and STAT-3 is also able
to reduce the tolerance of ovarian cancer cells to stress and
damage (67). Another study revealed
that STAT-3 activates miR-608, which inhibits the proliferation,
migration and invasiveness of lung cancer cells (88). Moreover, STAT3 also serves a critical
role in the regulation of tumor niche. Sun et al (89) reported that Annexin10 promotes
extrahepatic cholangiocarcinoma metastasis by stimulating EMT via
the STAT-3 pathway. Taken together, the aforementioned evidence
indicates that persistent activation of STAT-3 contributes to cell
proliferation, differentiation, migration and survival, and
consequently, researchers have attempted to inhibit the STAT-3
signaling pathway as a method of cancer treatment (11–16).
In previous research, attempts were made to inhibit
the effect of receptor tyrosine kinase (RTK), but mechanistic
studies indicated that the inhibition of specific RTKs initiated
the activation of STAT-3. Although certain small molecules
targeting RTKs were used clinically, the therapeutic efficacy was
limited by the development of drug resistance (90). Drug resistance represents a
significant challenge for effective antitumor therapy, as it often
ultimately results in treatment failure. Thus, activation of STAT-3
may contribute to the development of drug resistance; therefore,
inhibition of the STAT-3 pathway can restore the efficacy of
chemotherapeutics agents (91).
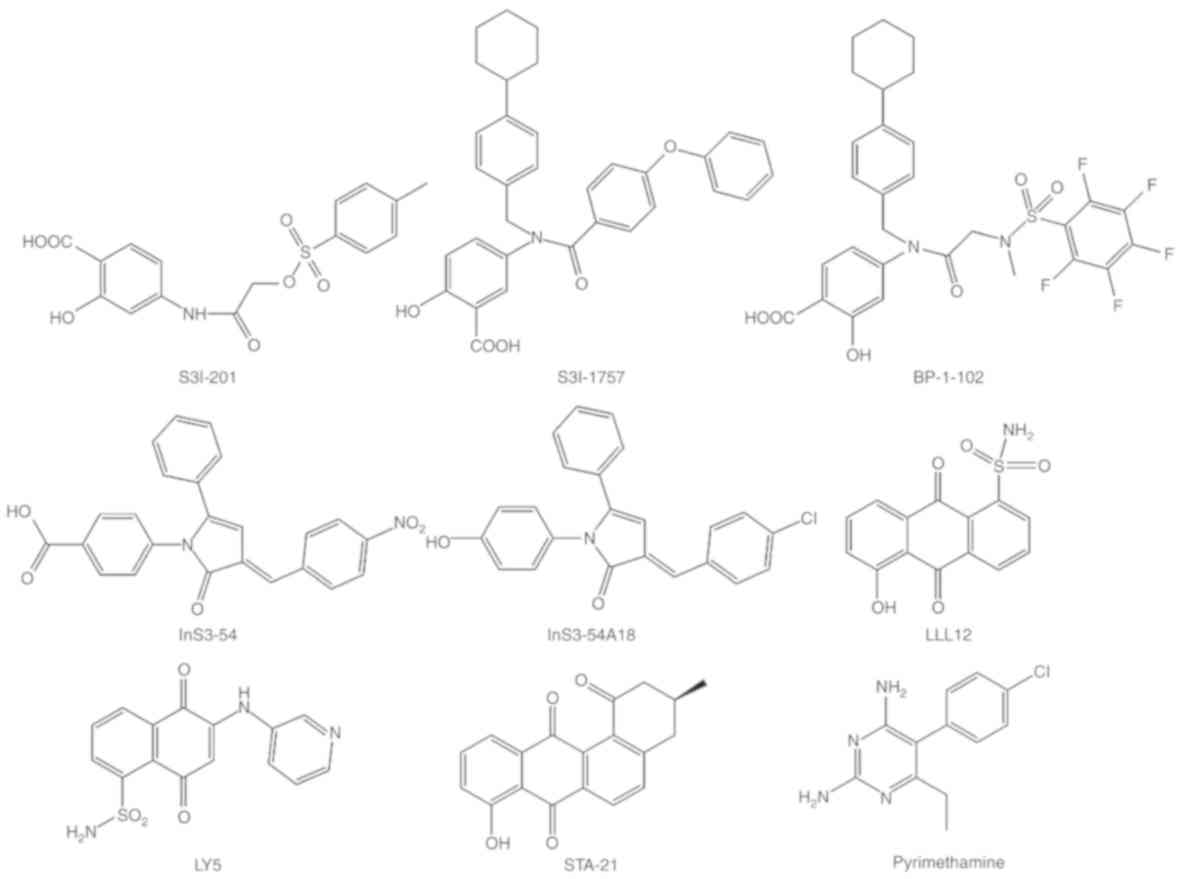
Notably, only one compound (BBI-608) targeting STAT-3 has been
approved by the Federal Drug Administration for clinical use.
However, a few small molecules have been demonstrated to antagonize
the STAT-3 signaling pathway (Fig.
3).
In order to target STAT-3 tyrosine phosphorylation,
researchers have attempted to identify a small inhibitor molecule
that directly binds the SH-2 domain of STAT-3, and prevents
tyrosine phosphorylation, protein dimerization and transcriptional
activity (92,93). Recently, structure-based drug design
and computational docking techniques have been widely used for the
identification of small molecules. For example, STA-21
(deoxytetrangomycin) is an analog of tetrangomycin (a non-peptide
small molecule STAT-3 inhibitor) that was discovered using
structure-based drug design and has successfully completed phase
I/II clinical trials (94,95). Furthermore, a variety of STA-21
analogs with improved potency, including LLL-12, S-3I-201, BP-1-102
and S-3I-1757, have been demonstrated to inhibit malignant
transformation, tumor cell proliferation, migration and invasion.
LLL-12 is a structurally optimized analog of STA-21 and inhibits
the activation of STAT-3 in a similar manner to STA-21 (96–99).
Additionally, LLL-12 exerts no inhibitory effects on STAT-1 and
other RTKs, indicating its specificity to STAT-3, and is more
sensitive to a variety of cancer cell lines (100,101).
S-31-201 is another specific inhibitor of STAT-3 that inhibits
STAT-3 phosphorylation and dimerization. However, molecular
modeling indicated that S31-201 selectively binds to the SH-2
domain (102). Consequently, a
library of S31-201 analogs has been developed and, of these,
S31-201 and −1–066 exhibit potent inhibition of STAT-3
dimerization, both in vitro and in vivo (103,104).
Furthermore, via structure modification, BP-1-102 (an analog of
SF-1-066) demonstrated improved specificity and oral
bioavailability. BP-1-102 binds three locations of the STAT-3-SH-2
domain and inhibits STAT-3 activation at concentrations of 4.1–6.8
µM (105,106). In addition, certain analogs of
BP-1-102 have been synthesized and evaluated to improve potency,
such as SH-5-07 and −4-54 (107).
These features indicate that BP-1-102 and its analogs may represent
promising anticancer agents.
LY-5 is another small molecule that inhibits STAT-3
by selectively binding the major pTyr-705 region, as well as a
sub-pocket of the STAT-3-SH-2 domain. LY-5 was designed by computer
models using docking simulation and evaluated for inhibitory
effects on STAT-3 activation and functions in human medulloblastoma
cells (108,109). Further studies demonstrated that
LY-5 not only suppressed various cancer cells with an
IC50 range of 0.5–1.4 µM, but that it also inhibited
tumor growth in an in vivo mouse model. Furthermore,
previous reports have demonstrated that a combination of a MEK
inhibitor and LY-5 may represent a potential therapeutic strategy
for overcoming resistance to MEK inhibitors in multiple human
cancer cell lines (108,109). Notably, further evidence has
demonstrated that the SH-2 domain is an effective target for small
molecule STAT-3 inhibitors. For example, it has been demonstrated
that OPB-31121 and −51602 represent two potent STAT-3 inhibitors
(110,111). OPB-31121 and −51602 also bind to
the SH-2 domain; however, molecular docking and dynamic simulations
indicate that their binding site do not overlap with any other
STAT-3 inhibitors (110,111).
Typically, the STAT-3 SH-2 domain is considered a
prime target for various STAT-3 inhibitors, due to the lack of
selectivity for other domains. However, previous studies have
reported that InS3-54 (designed by an improved in silico
approach) effectively inhibits the STAT-3 DBD. ‘In silico’
refers to the use of computers to solve biological problems. In the
present study, optimization and design of the lead compound were
performed by simulating and calculating the interaction between
receptor and ligand. This approach is known to notably improve the
discovery of novel drugs (112).
Notably, an optimized compound (InS3-54A18) has been identified
with improved specificity and pharmacological properties, which not
only inhibits STAT-3 activation via targeting the DBD of STAT-3,
but also significantly inhibits the downstream target gene of
STAT-3 (113–115). The aforementioned findings indicate
that InS3-54A18 may be a starting point for the further development
of anticancer therapeutics targeting the DBD of STAT-3.
Currently, only a few small-molecule inhibitors
targeting STAT-3 are undergoing the early phases of clinical trials
(Table I), and there is no inhibitor
of STAT-3 approved by the FDA except BBI-608, which is a STAT-3 and
cancer cell stemness inhibitor. STA-21, OPB-31121 and OPB-51602
have already completed phase I/II clinical trials in leukemia.
OPB-31121 and −51602 are currently in phase I/II clinical trials
for advanced solid tumors. STA-21 is not undergoing a clinical
trial at present. Phase I/II studies of OPB-31121 and −51602
revealed that these compounds exert potent antitumor activities
with an acceptable safety profile (116–119).
Pyrimethamine is another STAT-3 inhibitor used in the treatment of
chronic lymphocytic and small lymphocytic leukemia, and is
currently in phase I/II clinical trials (120).
Notably, JAKs serve a crucial role in JAK/STAT-3
signaling pathway; thus, inhibiting the activity of JAKs may be a
novel approach to inhibit STAT-3 activation. AG490, a JAK
inhibitor, reduces the proliferation of cancer cells by inhibiting
JAK-2 activity and blocking the activation of STAT-3. In addition,
AG-490 has been tested in models and in clinical trials (121–123).
A range of plant-derived compounds exhibit antitumor
activity against a variety of cancer types. Notably, during the
previous decade, a number of STAT-3 inhibitors derived from natural
sources have been employed and shown to exhibit significant
efficacy in regulating STAT-3 activation. Recent studies have
demonstrated that certain natural therapeutic agents serve an
inhibitory role in the genesis, progression and metastasis of
various cancer types (37).
Betulinic acid, a pentacyclic triterpene, extracted from
Zizyphus mauritiana, displayed potency in inhibiting STAT-3
activation, Src kinase, and JAK-1 and −2 (124,125).
Furthermore, it has been reported that betulinic acid induces
apoptosis in thyroid, breast, lung and colon carcinomas, indicating
its potential as a chemotherapeutic agent (124). Furthermore, caffeic acid is a
phenolic compound discovered in plants, and certain studies have
reported that caffeic acid exerts potent antioxidant and
anti-inflammatory properties (126–128).
However, a previous study demonstrated that caffeic acid exhibits
antitumor properties via the inhibition of STAT-3, preventing
STAT-3 recruitment and inhibiting the formation of a
transcriptional unit between STAT-3, HIF-1α and p-300 on the VEGF
promoter (129). Moreover,
celastol, obtained from Tripterygium wilfordii, is a Chinese
medicinal plant. Certain reports have indicated that celastrol can
inhibit proliferation, induce apoptosis and suppress
invasion/migration and angiogenesis via modulation of the
DNA-binding activity of STAT-3 in a wide variety of in vitro
and in vivo tumor models (130–133).
In addition, a variety of agents derived from natural plants have
also been demonstrated to exert antitumor effects, such as
curcumin, diosgenin and honokiol derived from carcuma longa, and
fenugreek and mangnolia officinalis, respectively. Furthermore, a
number of reports confirmed their antitumor effects were mediated
via modulation of constitutive STAT-3 activation in glioma cells,
HCC cells and HepG2 cells (134–136).
At present, specific inhibitors of STAT-3
predominantly target the disruption of the protein-protein
interactions or DNA-binding activity, such as inhibitors that
prevent the recruitment of STAT-3 to the IL-6/IL-6Rα/gp-130
complex, upstream kinase inhibitors and, primarily, JAK inhibitors.
Over the past decade, small-molecule drugs that directly target
STAT-3 have been identified; nevertheless, there are no
STAT-3-specific drugs available clinically. It has been
demonstrated that STAT-3 activation promotes oncogenesis via
phosphorylation or acetylation. STAT-3 inhibition has been revealed
to reverse acquired resistance, synergistically inhibit tumor
growth, induce apoptosis and stimulate an immune response.
Therefore, this signifies a requirement to reassess ongoing
strategies in order to develop clinically useful drugs. Future
research should focus on the development of therapeutic molecules
with STAT-3-inhibitory modalities, as this has the potential to
improve the treatment of a plethora of cancer types.
Not applicable.
The present study was funded by the First Affiliated
Hospital of Bengbu Medical College Science Fund for outstanding
Young Scholars (grant no. 2019byyfyyq06).
Data sharing is not applicable to this article, as
no datasets were generated or analyzed during the present
study.
The paper was conceived and designed by ZL. YCG
wrote the paper, and ISM revised the paper. All authors read and
approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Levy DE and Darnell JE Jr: Stats:
Transcriptional control and biological impact. Nat Rev Mol Cell
Biol. 3:651–662. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schindler C, Levy DE and Decker T:
JAK-STAT signaling: From interferons to cytokines. J Biol Chem.
282:20059–20063. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Reich NC and Liu L: Tracking STAT nuclear
traffic. Nat Rev Immunol. 6:602–612. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stark GR and Darnell JE Jr: The JAK-STAT
pathway at twenty. Immunity. 36:503–514. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu H, Pardoll D and Jove R: STATs in
cancer inflammation and immunity: A leading role for STAT-3. Nat
Rev Cancer. 9:798–809. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bromberg J and Darnell JE Jr: The role of
STATs in transcriptional control and their impact on cellular
function. Oncogene. 19:2468–2473. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Darnell JE Jr: STATs and gene regulation.
Science. 277:1630–1635. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Buettner R, Mora LB and Jove R: Activated
STAT signaling in human tumors provides novel molecular targets for
therapeutic intervention. Clin Cancer Res. 8:945–954.
2002.PubMed/NCBI
|
|
9
|
Yu H and Jove R: The STATs of cancer-new
molecular targets come of age. Nat Rev Cancer. 4:97–105. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Haura EB, Turkson J and Jove R: Mechanisms
of disease: Insights into the emerging role of signal transducers
and activators of transcription in cancer. Nat Clin Pract Oncol.
2:315–324. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Herrmann A, Kortylewski M, Kujawski M,
Zhang C, Reckamp K, Armstrong B, Wang L, Kowolik C, Deng J, Figlin
R and Yu H: Targeting STAT3 in the myeloid compartment drastically
improves the in vivo antitumor functions of adoptively transferred
T cells. Cancer Res. 70:7455–7464. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kortylewski M and Yu H: Role of STAT-3 in
suppressing anti-tumor immunity. Curr Opin Immunol. 20:228–233.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kujawski M, Kortylewski M, Lee H, Herrmann
A, Kay H and Yu H: STAT-3 mediates myeloid cell-dependent tumor
angiogenesis in mice. J Clin Invest. 118:3367–3377. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang L, Yi T, Kortylewski M, Pardoll DM,
Zeng D and Yu H: IL-17 can promote tumor growth through an
IL-6-STAT-3 signaling pathway. J Exp Med. 206:1457–1464. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kortylewski M, Kujawski M, Wang T, Wei S,
Zhang S, Pilon-Thomas S, Niu G, Kay H, Mulé J, Kerr WG, et al:
Inhibiting STAT3 signaling in the hematopoietic system elicits
multicomponent antitumor immunity. Nat Med. 11:1314–1321. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu H, Kortylewski M and Pardoll D:
Crosstalk between cancer and immune cells: Role of STAT-3 in the
tumour microenvironment. Nat Rev Immunol. 7:41–51. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nowak EM, Poczęta M, Bieg D and Bednarek
I: DNA methyltransferase inhibitors influence on the DIRAS3 and
STAT3 expression and in vitro migration of ovarian and breast
cancer cells. Ginekol Pol. 88:543–551. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu H, Lee H, Herrmann A, Buettner R and
Jove R: Revisiting STAT3 signalling in cancer: New and unexpected
biological functions. Nat Rev Cancer. 14:736–746. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mali SB: Review of STAT3 (Signal
Transducers and Activators of Transcription) in head and neck
cancer. Oral Oncol. 51:565–569. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Benekli M, Baer MR, Baumann H and Wetzler
M: Signal transducer and activator of transcription proteins in
leukemias. Blood. 101:2940–2954. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Calò V, Migliavacca M, Bazan V, Macaluso
M, Buscemi M, Gebbia N and Russo A: STAT proteins: From normal
control of cellular events to tumorigenesis. J Cell Physiol.
197:157–168. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sternberg DW and Gilliland DG: The role of
signal transducer and activator of transcription factors in
leukemogenesis. J Clin Oncol. 22:361–371. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Abroun S, Saki N, Ahmadvand M, Asghari F,
Salari F and Rahim F: STATs: An old story, yet mesmerizing. Cell J.
17:395–411. 2015.PubMed/NCBI
|
|
24
|
Yang E, Henriksen MA, Schaefer O,
Zakharova N and Darnell JE Jr: Dissociation time from DNA
determines transcriptional function in a STAT1 linker mutant. J
Biol Chem. 277:13455–13462. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wake MS and Watson CJ: STAT3 the
oncogene-still eluding therapy? FEBS J. 282:2600–2611. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang Y, Qiu J, Dong S, Redell MS, Poli V,
Mancini MA and Tweardy DJ: Stat3 isoforms, alpha and beta,
demonstrate distinct intracellular dynamics with prolonged nuclear
retention of Stat3beta mapping to its unique C-terminal end. J Biol
Chem. 282:34958–34967. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chakraborty A, Dyer KF, Cascio M, Mietzner
TA and Tweardy DJ: Identification of a novel STAT-3 recruitment and
activation motif within the granulocyte colony-stimulating factor
receptor. Blood. 93:15–24. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chakraborty A and Tweardy DJ: STAT-3 and
G-CSF-induced myeloid differentiation. Leuk Lymphoma. 30:433–442.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ilaria RL Jr: STAT isoforms: Mediators of
STAT specificity or leukemogenesis? Leuk Res. 25:483–484. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huynh J, Chand A, Gough D and Ernst M:
Therapeutically exploiting STAT3 activity in cancer-using tissue
repair as a road map. Nat Rev Cancer. 19:82–96. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chai EZ, Shanmugam MK, Arfuso F,
Dharmarajan A, Wang C, Kumar AP, Samy RP, Lim LH, Wang L, Goh BC,
et al: Targeting transcription factor STAT3 for cancer prevention
and therapy. Pharmacol Ther. 162:86–97. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
ten Hoeve J, de Jesus Ibarra-Sanchez M, Fu
Y, Zhu W, Tremblay M, David M and Shuai K: Identification of a
nuclear Stat1 protein tyrosine phosphatase. Mol Cell Biol.
22:5662–5668. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shuai K and Liu B: Regulation of
gene-activation pathways by PIAS proteins in the immune system. Nat
Rev Immunol. 5:593–605. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chung CD, Liao J, Liu B, Rao X, Jay P,
Berta P and Shuai K: Specific inhibition of STAT-3 signal
transduction by PIAS3. Science. 278:1803–1805. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu B, Gross M, ten Hoeve J and Shuai K: A
transcriptional corepressor of Stat1 with an essential LXXLL
signature motif. Proc Natl Acad Sci USA. 98:3203–3207. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu B, Liao J, Rao X, Kushner SA, Chung
CD, Chang DD and Shuai K: Inhibition of Stat1-mediated gene
activation by PIAS1. Proc Natl Acad Sci USA. 95:10626–10631. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Verhoeven Y, Tilborghs S, Jacobs J, De
Waele J, Quatannens D, Deben C, Prenen H, Pauwels P, Trinh XB,
Wouters A, et al: The potential and controversy of targeting STAT
family members in cancer. Semin Cancer Biol. Oct 9–2019.doi:
10.1016/j.semcancer.2019.10.002 (Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Catlett-Falcone R, Landowski TH, Oshiro
MM, Turkson J, Levitzki A, Savino R, Ciliberto G, Moscinski L,
Fernández-Luna JL, Nuñez G, et al: Constitutive activation of STAT3
signaling confers resistance to apoptosis in human U266 myeloma
cells. Immunity. 10:105–115. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Huynh J, Etemadi N, Hollande F, Ernst M
and Buchert M: The JAK/STAT3 axis: A comprehensive drug target for
solid malignancies. Semin Cancer Biol. 45:13–22. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Johnson DE, O'Keefe RA and Grandis JR:
Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev
Clin Oncol. 15:234–248. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Park EJ, Lee JH, Yu GY, He G, Ali SR,
Holzer RG, Osterreicher CH, Takahashi H and Karin M: Dietary and
genetic obesity promote liver inflammation and tumorigenesis by
enhancing IL-6 and TNF expression. Cell. 140:197–208. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jones SA, Scheller J and Rose-John S:
Therapeutic strategies for the clinical blockade of IL-6/gp130
signaling. J Clin Invest. 121:3375–3383. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Grivennikov S, Karin E, Terzic J, Mucida
D, Yu GY, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H,
Eckmann L and Karin M: IL-6 and STAT3 are required for survival of
intestinal epithelial cells and development of colitis-associated
cancer. Cancer Cell. 15:103–113. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bollrath J, Phesse TJ, von Burstin VA,
Putoczki T, Bennecke M, Bateman T, Nebelsiek T, Lundgren-May T,
Canli O, Schwitalla S, et al: Gp130-mediated STAT3 activation in
enterocytes regulates cell survival and cell-cycle progression
during colitis-associated tumorigenesis. Cancer Cell. 15:91–102.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Schiechl G, Bauer B, Fuss I, Lang SA,
Moser C, Ruemmele P, Rose-John S, Neurath MF, Geissler EK, Schlitt
HJ, et al: Tumor development in murine ulcerative colitis depends
on MyD88 signaling of colonic F4/80+CD11b(high)Gr1(low)
macrophages. J Clin Invest. 121:1692–1708. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang L, Yang J, Qian J, Li H, Romaguera
JE, Kwak LW, Wang M and Yi Q: Role of the microenvironment in
mantle cell lymphoma: IL-6 is an important survival factor for the
tumor cells. Blood. 120:3783–3792. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Schafer ZT and Brugge JS: IL-6 involvement
in epithelial cancers. J Clin Invest. 117:3660–3663. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sansone P, Storci G, Tavolari S, Guarnieri
T, Giovannini C, Taffurelli M, Ceccarelli C, Santini D, Paterini P,
Marcu KB, et al: IL-6 triggers malignant features in mammospheres
from human ductal breast carcinoma and normal mammary gland. J Clin
Invest. 117:3988–4002. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Pine SR, Mechanic LE, Enewold L,
Chaturvedi AK, Katki HA, Zheng YL, Bowman ED, Engels EA, Caporaso
NE and Harris CC: Increased levels of circulating interleukin 6,
interleukin 8, C-reactive protein, and risk of lung cancer. J Natl
Cancer Inst. 103:1112–1122. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Nakashima J, Tachibana M, Horiguchi Y, Oya
M, Ohigashi T, Asakura H and Murai M: Serum interleukin 6 as a
prognostic factor in patients with prostate cancer. Clin Cancer
Res. 6:2702–2706. 2000.PubMed/NCBI
|
|
51
|
Reynaud D, Pietras E, Barry-Holson K, Mir
A, Binnewies M, Jeanne M, Sala-Torra O, Radich JP and Passegué E:
IL-6 controls leukemic multipotent progenitor cell fate and
contributes to chronic myelogenous leukemia development. Cancer
Cell. 20:661–673. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ancrile B, Lim KH and Counter CM:
Oncogenic Ras-induced secretion of IL6 is required for
tumorigenesis. Genes Dev. 21:1714–1719. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Gyamfi J, Lee YH, Eom M and Choi J:
Interleukin-6/STAT3 signalling regulates adipocyte induced
epithelial-mesenchymal transition in breast cancer cells. Sci Rep.
8:88592018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Gao X, Liu X, Lu Y, Wang Y, Cao W, Liu X,
Hu H and Wang H: PIM1 is responsible for IL-6-induced breast cancer
cell EMT and stemness via c-myc activation. Breast Cancer.
26:663–671. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kim MS, Lee WS, Jeong J, Kim SJ and Jin W:
Induction of metastatic potential by TrkB via activation of
IL6/JAK2/STAT3 and PI3K/AKT signaling in breast cancer. Oncotarget.
6:40158–40171. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Xin H, Lu R, Lee H, Zhang W, Zhang C, Deng
J, Liu Y, Shen S, Wagner KU, Forman S, et al: G-protein-coupled
receptor agonist BV8/prokineticin-2 and STAT3 protein form a
feed-forward loop in both normal and malignant myeloid cells. J
Biol Chem. 288:13842–13849. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lee H, Deng J, Kujawski M, Yang C, Liu Y,
Herrmann A, Kortylewski M, Horne D, Somlo G, Forman S, et al:
STAT-3-induced S1PR1 expression is crucial for persistent STAT-3
activation in tumors. Nat Med. 16:1421–1428. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Visentin B, Vekich JA, Sibbald BJ, Cavalli
AL, Moreno KM, Matteo RG, Garland WA, Lu Y, Yu S, Hall HS, et al:
Validation of an anti-sphingosine-1-phosphate antibody as a
potential therapeutic in reducing growth, invasion, and
angiogenesis in multiple tumor lineages. Cancer Cell. 9:225–238.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kawamori T, Kaneshiro T, Okumura M,
Maalouf S, Uflacker A, Bielawski J, Hannun YA and Obeid LM: Role
for sphingosine kinase 1 in colon carcinogenesis. FASEB J.
23:405–414. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Sarkar S, Maceyka M, Hait NC, Paugh SW,
Sankala H, Milstien S and Spiegel S: Sphingosine kinase 1 is
required for migration, proliferation and survival of MCF-7 human
breast cancer cells. FEBS Lett. 579:5313–5317. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Liu Y, Deng J, Wang L, Lee H, Armstrong B,
Scuto A, Kowolik C, Weiss LM, Forman S and Yu H: S1PR1 is an
effective target to block STAT-3 signaling in activated B cell-like
diffuse large B-cell lymphoma. Blood. 120:1458–1465. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Ponnusamy S, Meyers-Needham M, Senkal CE,
Saddoughi SA, Sentelle D, Selvam SP, Salas A and Ogretmen B:
Sphingolipids and cancer: Ceramide and sphingosine-1-phosphate in
the regulation of cell death and drug resistance. Future Oncol.
6:1603–1624. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Priceman SJ, Shen S, Wang L, Deng J, Yue
C, Kujawski M and Yu H: S1PR1 is crucial for accumulation of
regulatory T cells in tumors via STAT3. Cell Rep. 6:992–999. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Eyking A, Ey B, Rünzi M, Roig AI, Reis H,
Schmid KW, Gerken G, Podolsky DK and Cario E: Toll-like receptor 4
variant D299G induces features of neoplastic progression in Caco-2
intestinal cells and is associated with advanced human colon
cancer. Gastroenterology. 141:2154–2165. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Tye H, Kennedy CL, Najdovska M, McLeod L,
McCormack W, Hughes N, Dev A, Sievert W, Ooi CH, Ishikawa TO, et
al: STAT-3-driven upregulation of TLR2 promotes gastric
tumorigenesis independent of tumor inflammation. Cancer Cell.
22:466–478. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Ochi A, Graffeo CS, Zambirinis CP, Rehman
A, Hackman M, Fallon N, Barilla RM, Henning JR, Jamal M, Rao R, et
al: Toll-like receptor 7 regulates pancreatic carcinogenesis in
mice and humans. J Clin Invest. 122:4118–4129. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wild CA, Brandau S, Lindemann M, Lotfi R,
Hoffmann TK, Lang S and Bergmann C: Toll-like receptors in
regulatory T cells of patients with head and neck cancer. Arch
Otolaryngol Head Neck Surg. 136:1253–1259. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Liang F, Ren C, Wang J, Wang S, Yang L,
Han X, Chen Y, Tong G and Yang G: The crosstalk between STAT3 and
p53/RAS signaling controls cancer cell metastasis and cisplatin
resistance via the Slug/MAPK/PI3K/AKTmediated regulation of EMT and
autophagy. Oncogenesis. 8:592019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wang JR, Shen GN, Luo YH, Piao XJ, Zhang
Y, Wang H, Li JQ, Xu WT, Zhang Y, Wang SN, et al:
2-(4-methoxyphenylthio)-5,8-dimethoxy-1,4-naphthoquinone induces
apoptosis via ROS-mediated MAPK and STAT3 signaling pathway in
human gastric cancer cells. J Chemother. 31:214–226. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Lakshmanachetty S, Balaiya V, High WA and
Koster MI: Loss of TP63 promotes the metastasis of head and neck
squamous cell carcinoma by activating MAPK and STAT3 signaling. Mol
Cancer Res. 17:1279–1293. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ahmed ST and Ivashkiv LB: Inhibition of
IL-6 and IL-10 signaling and Stat activation by inflammatory and
stress pathways. J Immunol. 165:5227–5237. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Yan K, Xu X, Wu T, Li J, Cao G, Li Y and
Ji Z: Knockdown of PYCR1 inhibits proliferation, drug resistance
and EMT in colorectal cancer cells by regulating STAT3-Mediated p38
MAPK and NF-kB signalling pathway. Biochem Biophys Res Commun.
520:486–491. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Benekli M, Baumann H and Wetzler M:
Targeting signal transducer and activator of transcription
signaling pathway in leukemias. J Clin Oncol. 27:4422–4432. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhang S, Yang Z, Bao W, Liu L, You Y, Wang
X, Shao L, Fu W, Kou X, Shen W, et al: SNX10 (sorting nexin 10)
inhibits colorectal cancer initiation and progression by
controlling autophagic degradation of SRC. Autophagy. 4:1–15. 2019.
View Article : Google Scholar
|
|
75
|
Liu CY, Huang TT, Chu PY, Huang CT, Lee
CH, Wang WL, Lau KY, Tsai WC, Chao TI, Su JC, et al: The tyrosine
kinase inhibitor nintedanib activates SHP-1 and induces apoptosis
in triple-negative breast cancer cells. Exp Mol Med. 49:e3662017.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Yu H, Pardoll D and Jove R: STATs in
cancer inflammation and immunity: A leading role for STAT3. Nat Rev
Cancer. 9:798–809. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Jarnicki A, Putoczki T and Ernst M: Stat3:
Linking inflammation to epithelial cancer-more than a ‘gut’
feeling? Cell Div. 5:142010. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Siveen KS, Sikka S, Surana R, Dai X, Zhang
J, Kumar AP, Tan BK, Sethi G and Bishayee A: Targeting the STAT3
signaling pathway in cancer: Role of synthetic and natural
inhibitors. Biochim Biophys Acta. 1845:136–154. 2014.PubMed/NCBI
|
|
79
|
Huang DC, Adams JM and Cory S: The
conserved N-terminal BH4 domain of Bcl-2 homologues is essential
for inhibition of apoptosis and interaction with CED-4. EMBO J.
17:1029–1039. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Reed JC: Mechanisms of apoptosis avoidance
in cancer. Curr Opin Oncol. 11:68–75. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Welcker M, Lukas J, Strauss M and Bartek
J: Enhanced protein stability: A novel mechanism of D-type cyclin
over-abundance identified in human sarcoma cells. Oncogene.
13:419–425. 1996.PubMed/NCBI
|
|
82
|
Wei D, Le X, Zheng L, Wang L, Frey JA, Gao
AC, Peng Z, Huang S, Xiong HQ, Abbruzzese JL and Xie K: STAT-3
activation regulates the expression of vascular endothelial growth
factor and human pancreatic cancer angiogenesis and metastasis.
Oncogene. 22:319–329. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Dechow TN, Pedranzini L, Leitch A, Leslie
K, Gerald WL, Linkov I and Bromberg JF: Requirement of matrix
metalloproteinase-9 for the transformation of human mammary
epithelial cells by STAT-3-C. Proc Natl Acad Sci USA.
101:10602–10607. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Niu G, Wright KL, Huang M, Song L, Haura
E, Turkson J, Zhang S, Wang T, Sinibaldi D, Coppola D, et al:
Constitutive STAT-3 activity up-regulates VEGF expression and tumor
angiogenesis. Oncogene. 21:2000–2008. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Lieblein JC, Ball S, Hutzen B, Sasser AK,
Lin HJ, Huang TH, Hall BM and Lin J: STAT3 can be activated through
paracrine signaling in breast epithelial cells. BMC Cancer.
8:3022008. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Lin J, Jin X, Rothman K, Lin HJ, Tang H
and Burke W: Modulation of signal transducer and activator of
transcription 3 activities by p53 tumor suppressor in breast cancer
cells. Cancer Res. 62:376–380. 2002.PubMed/NCBI
|
|
87
|
Li L, Tang W, Wu X, Karnak D, Meng X,
Thompson R, Hao X, Li Y, Qiao XT, Lin J, et al: HAb18G/CD147
promotes pSTAT-3-mediated pancreatic cancer development via CD44s.
Clin Cancer Res. 19:6703–6715. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Xu W, Sun D, Wang Y, Zheng X, Li Y, Xia Y
and Teng Y: Inhibitory effect of microRNA-608 on lung cancer cell
proliferation, migration, and invasion by targeting BRD4 through
the JAK2/STAT3 pathway. Bosn J Basic Med Sci. Oct 17–2019.doi:
10.17305/bjbms.2019.4216 (Epub ahead of print). View Article : Google Scholar
|
|
89
|
Sun R, Liu Z, Qiu B, Chen T, Li Z, Zhang
X, Xu Y and Zhang Z: Annexin10 promotes extrahepatic
cholangiocarcinoma metastasis by facilitating EMT via
PLA2G4A/PGE2/STAT3 pathway. EBioMedicine. 47:142–155. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Sun C and Bernards R: Feedback and
redundancy in receptor tyrosine kinase signaling: Relevance to
cancer therapies. Trends Biochem Sci. 39:465–474. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Li G, Zhao L, Li W, Fan K, Qian W, Hou S,
Wang H, Dai J, Wei H and Guo Y: Feedback activation of STAT3
mediates trastuzumab resistance via upregulation of MUC1 and MUC4
expression. Oncotarget. 5:8317–8329. 2014.PubMed/NCBI
|
|
92
|
Song H, Wang R, Wang S and Lin J: A
low-molecular-weight compound discovered through virtual database
screening inhibits STAT-3 function in breast cancer cells. Proc
Natl Acad Sci USA. 102:4700–4705. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Chen CL, Cen L, Kohout J, Hutzen B, Chan
C, Hsieh FC, Loy A, Huang V, Cheng G and Lin J: Signal transducer
and activator of transcription 3 activation is associated with
bladder cancer cell growth and survival. Mol Cancer. 7:782008.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Miyoshi K, Takaishi M, Nakajima K, Ikeda
M, Kanda T, Tarutani M, Iiyama T, Asao N, DiGiovanni J and Sano S:
STAT3 as a therapeutic target for the treatment of psoriasis: A
clinical feasibility study with STA-21, a STAT3 inhibitor. J Invest
Dermatol. 131:108–117. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Park JS, Kwok SK, Lim MA, Kim EK, Ryu JG,
Kim SM, Oh HJ, Ju JH, Park SH, Kim HY and Cho ML: STA-21, a
promising STAT-3 inhibitor that reciprocally regulates Th17 and
Treg cells, inhibits osteoclastogenesis in mice and humans and
alleviates autoimmune inflammation in an experimental model of
rheumatoid arthritis. Arthritis Rheumatol. 66:918–929. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Lin L, Liu A, Peng Z, Lin HJ, Li PK, Li C
and Lin J: STAT-3 is necessary for proliferation and survival in
colon cancer-initiating cells. Cancer Res. 71:7226–7237. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Lin L, Benson DM Jr, DeAngelis S, Bakan
CE, Li PK, Li C and Lin J: A small molecule, LLL12 inhibits
constitutive STAT-3 and IL-6-induced STAT-3 signaling and exhibits
potent growth suppressive activity in human multiple myeloma cells.
Int J Cancer. 130:1459–1469. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Liu Y, Li PK, Li C and Lin J: Inhibition
of STAT-3 signaling blocks the anti-apoptotic activity of IL-6 in
human liver cancer cells. J Biol Chem. 285:27429–27439. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Jain R, Kulkarni P, Dhali S, Rapole S and
Srivastava S: Quantitative proteomic analysis of global effect of
LLL12 on U87 cell's proteome: An insight into the molecular
mechanism of LLL12. J Proteomics. 113:127–142. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Zuo M, Li C, Lin J and Javle M: LLL12, a
novel small inhibitor targeting STAT-3 for hepatocellular carcinoma
therapy. Oncotarget. 6:10940–10949. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Bid HK, Kibler A, Phelps DA, Manap S, Xiao
L, Lin J, Capper D, Oswald D, Geier B, DeWire M, et al:
Development, characterization, and reversal of acquired resistance
to the MEK1 inhibitor selumetinib (AZD6244) in an in vivo model of
childhood astrocytoma. Clin Cancer Res. 19:6716–6729. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Siddiquee K, Zhang S, Guida WC, Blaskovich
MA, Greedy B, Lawrence HR, Yip ML, Jove R, McLaughlin MM, Lawrence
NJ, et al: Selective chemical probe inhibitor of STAT3, identified
through structure-based virtual screening, induces antitumor
activity. Proc Natl Acad Sci USA. 104:7391–7396. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Zhang X, Yue P, Fletcher S, Zhao W,
Gunning PT and Turkson J: A novel small-molecule disrupts STAT-3
SH2 domain-phosphotyrosine interactions and STAT-3-dependent tumor
processes. Biochem Pharmacol. 79:1398–1409. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Fletcher S, Singh J, Zhang X, Yue P, Page
BD, Sharmeen S, Shahani VM, Zhao W, Schimmer AD, Turkson J and
Gunning PT: Disruption of transcriptionally active Stat3 dimers
with non-phosphorylated, salicylic acid-based small molecules:
Potent in vitro and tumor cell activities. Chembiochem.
10:1959–1964. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Zhang X, Yue P, Page BD, Li T, Zhao W,
Namanja AT, Paladino D, Zhao J, Chen Y, Gunning PT and Turkson J:
Orally bioavailable small-molecule inhibitor of transcription
factor Stat-3 regresses human breast and lung cancer xenografts.
Proc Natl Acad Sci USA. 109:9623–9628. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Resetca D, Haftchenary S, Gunning PT and
Wilson DJ: Changes in signal transducer and activator of
transcription 3 (STAT-3) dynamics induced by complexation with
pharmacological inhibitors of Src homology 2 (SH2) domain
dimerization. J Biol Chem. 289:32538–32547. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Haftchenary S, Luchman HA, Jouk AO, Veloso
AJ, Page BD, Cheng XR, Dawson SS, Grinshtein N, Shahani VM, Kerman
K, et al: Potent targeting of the STAT 3 protein in brain cancer
stem cells: A promising route for treating glioblastoma. ACS Med
Chem Lett. 4:1102–1107. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Zhao C, Xiao H, Wu X, Li C, Liang G, Yang
S and Lin J: Rational combination of MEK inhibitor and the STAT-3
pathway modulator for the therapy in K-Ras mutated pancreatic and
colon cancer cells. Oncotarget. 6:14472–14487. 2015.PubMed/NCBI
|
|
109
|
Xiao H, Bid HK, Jou D, Wu X, Yu W, Li C,
Houghton PJ and Lin J: A novel small molecular STAT-3 inhibitor,
LY5, inhibits cell viability, cell migration, and angiogenesis in
medulloblastoma cells. J Biol Chem. 290:3418–3429. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Hayakawa F, Sugimoto K, Harada Y,
Hashimoto N, Ohi N, Kurahashi S and Naoe T: A novel STAT inhibitor,
OPB-31121, has a significant antitumor effect on leukemia with
STAT-addictive oncokinases. Blood Cancer J. 3:e1662013. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Kim MJ, Nam HJ, Kim HP, Han SW, Im SA, Kim
TY, Oh DY and Bang YJ: OPB-31121, a novel small molecular
inhibitor, disrupts the JAK2/STAT-3 pathway and exhibits an
antitumor activity in gastric cancer cells. Cancer Lett.
335:145–152. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Yu W, Xiao H, Lin J and Li C: Discovery of
novel STAT3 small molecule inhibitors via in silico site-directed
fragment-based drug design. J Med Chem. 56:4402–4412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Huang W, Dong Z, Chen Y, Wang F, Wang CJ,
Peng H, He Y, Hangoc G, Pollok K, Sandusky G, et al: Small-molecule
inhibitors targeting the DNA-binding domain of STAT-3 suppress
tumor growth, metastasis and STAT-3 target gene expression in vivo.
Oncogene. 35:8022016. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Huang W, Dong Z, Wang F, Peng H, Liu JY
and Zhang JT: A small molecule compound targeting STAT-3
DNA-binding domain inhibits cancer cell proliferation, migration,
and invasion. ACS Chem Biol. 9:1188–1196. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Ogura M, Uchida T, Terui Y, Hayakawa F,
Kobayashi Y, Taniwaki M, Takamatsu Y, Naoe T, Tobinai K, Munakata
W, et al: Phase I study of OPB-51602, an oral inhibitor of signal
transducer and activator of transcription 3, in patients with
relapsed/refractory hematological malignancies. Cancer Sci.
106:896–901. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Wong AL, Soo RA, Tan DS, Lee SC, Lim JS,
Marban PC, Kong LR, Lee YJ, Wang LZ, Thuya WL, et al: Phase I and
biomarker study of OPB-51602, a novel signal transducer and
activator of transcription (STAT) 3 inhibitor, in patients with
refractory solid malignancies. Ann Oncol. 26:998–1005. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Brambilla L, Genini D, Laurini E, Merulla
J, Perez L, Fermeglia M, Carbone GM, Pricl S and Catapano CV:
Hitting the right spot: Mechanism of action of OPB-31121, a novel
and potent inhibitor of the Signal Transducer and Activator of
Transcription 3 (STAT-3). Mol Oncol. 9:1194–1206. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Oh DY, Lee SH, Han SW, Kim MJ, Kim TM, Kim
TY, Heo DS, Yuasa M, Yanagihara Y and Bang YJ: Phase I study of
OPB-31121, an Oral STAT-3 inhibitor, in patients with advanced
solid tumors. Cancer Res Treat. 47:607–615. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Okusaka T, Ueno H, Ikeda M, Mitsunaga S,
Ozaka M, Ishii H, Yokosuka O, Ooka Y, Yoshimoto R, Yanagihara Y and
Okita K: Phase 1 and pharmacological trial of OPB-31121, a signal
transducer and activator of transcription-3 inhibitor in patients
with advanced hepatocellular carcinoma. Hepatol Res. 45:1283–1291.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Takakura A, Nelson EA, Haque N, Humphreys
BD, Zandi-Nejad K, Frank DA and Zhou J: Pyrimethamine inhibits
adult polycystic kidney disease by modulating STAT signaling
pathways. Hum Mol Genet. 20:4143–4154. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Meydan N, Grunberger T, Dadi H, Shahar M,
Arpaia E, Lapidot Z, Leeder JS, Freedman M, Cohen A, Gazit A, et
al: Inhibition of acute lymphoblastic leukaemia by a Jak-2
inhibitor. Nature. 379:645–648. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Jin J, Guo Q, Xie J, Jin D and Zhu Y:
Combination of MEK inhibitor and the JAK2-STAT3 pathway inhibition
for the therapy of colon cancer. Pathol Oncol Res. 25:769–775.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Joung YH, Na YM, Yoo YB, Darvin P, Sp N,
Kang DY, Kim SY, Kim HS, Choi YH, Lee HK, et al: Combination of
AG490, a Jak2 inhibitor, and methylsulfonylmethane synergistically
suppresses bladder tumor growth via the Jak2/STAT3 pathway. Int J
Oncol. 44:883–895. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Pandey MK, Sung B and Aggarwal BB:
Betulinic acid suppresses STAT-3 activation pathway through
induction of protein tyrosine phosphatase SHP-1 in human multiple
myeloma cells. Int J Cancer. 127:282–292. 2010.PubMed/NCBI
|
|
125
|
Su D, Gao YQ, Dai WB, Hu Y, Wu YF and Mei
QX: Helicteric acid, oleanic acid, and betulinic acid, three
triterpenes from helicteres angustifolia L., Inhibit proliferation
and induce apoptosis in HT-29 colorectal cancer cells via
suppressing NF-κB and STAT3 signaling. Evid Based Complement
Alternat Med. 2017:51807072017. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Nardini M, Pisu P, Gentili V, Natella F,
Di Felice M, Piccolella E and Scaccini C: Effect of caffeic acid on
tert-butyl hydroperoxide-induced oxidative stress in U937. Free
Radic Biol Med. 25:1098–1105. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Tapiero H, Tew KD, Ba GN and Mathe G:
Polyphenols: Do they play a role in the prevention of human
pathologies? Biomed Pharmacother. 56:200–207. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Nardini M, Leonardi F, Scaccini C and
Virgili F: Modulation of ceramide-induced NF-kappaB binding
activity and apoptotic response by caffeic acid in U937 cells:
Comparison with other antioxidants. Free Radic Biol Med.
30:722–733. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Jung JE, Kim HS, Lee CS, Park DH, Kim YN,
Lee MJ, Lee JW, Park JW, Kim MS, Ye SK and Chung MH: Caffeic acid
and its synthetic derivative CADPE suppress tumor angiogenesis by
blocking STAT-3-mediated VEGF expression in human renal carcinoma
cells. Carcinogenesis. 28:1780–1787. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Rajendran P, Li F, Shanmugam MK, Kannaiyan
R, Goh JN, Wong KF, Wang W, Khin E, Tergaonkar V, Kumar AP, et al:
Celastrol suppresses growth and induces apoptosis of human
hepatocellular carcinoma through the modulation of STAT-3/JAK2
signaling cascade in vitro and in vivo. Cancer Prev Res (Phila).
5:631–643. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Kannaiyan R, Hay HS, Rajendran P, Li F,
Shanmugam MK, Vali S, Abbasi T, Kapoor S, Sharma A, Kumar AP, et
al: Celastrol inhibits proliferation and induces chemosensitization
through down-regulation of NF-KB and STAT-3 regulated gene products
in multiple myeloma cells. Br J Pharmacol. 164:1506–1521. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Pang X, Yi Z, Zhang J, Lu B, Sung B, Qu W,
Aggarwal BB and Liu M: Celastrol suppresses angiogenesis-mediated
tumor growth through inhibition of AKT/mammalian target of
rapamycin pathway. Cancer Res. 70:1951–1959. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Lu Z, Jin Y, Qiu L, Lai Y and Pan J:
Celastrol, a novel HSP90 inhibitor, depletes Bcr-Abl and induces
apoptosis in imatinib-resistant chronic myelogenous leukemia cells
harboring T315I mutation. Cancer Lett. 290:182–191. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Weissenberger J, Priester M, Bernreuther
C, Rakel S, Glatzel M, Seifert V and Kögel D: Dietary curcumin
attenuates glioma growth in a syngeneic mouse model by inhibition
of the JAK1,2/STAT-3 signaling pathway. Clin Cancer Res.
16:5781–5795. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Li F, Fernandez PP, Rajendran P, Hui KM
and Sethi G: Diosgenin, a steroidal saponin, inhibits STAT-3
signaling pathway leading to suppression of proliferation and
chemosensitization of human hepatocellular carcinoma cells. Cancer
Lett. 292:197–207. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Rajendran P, Li F, Shanmugam MK, Vali S,
Abbasi T, Kapoor S, Ahn KS, Kumar AP and Sethi G: Honokiol inhibits
signal transducer and activator of transcription-3 signaling,
proliferation, and survival of hepatocellular carcinoma cells via
the protein tyrosine phosphatase SHP-1. J Cell Physiol.
227:2184–2195. 2012. View Article : Google Scholar : PubMed/NCBI
|