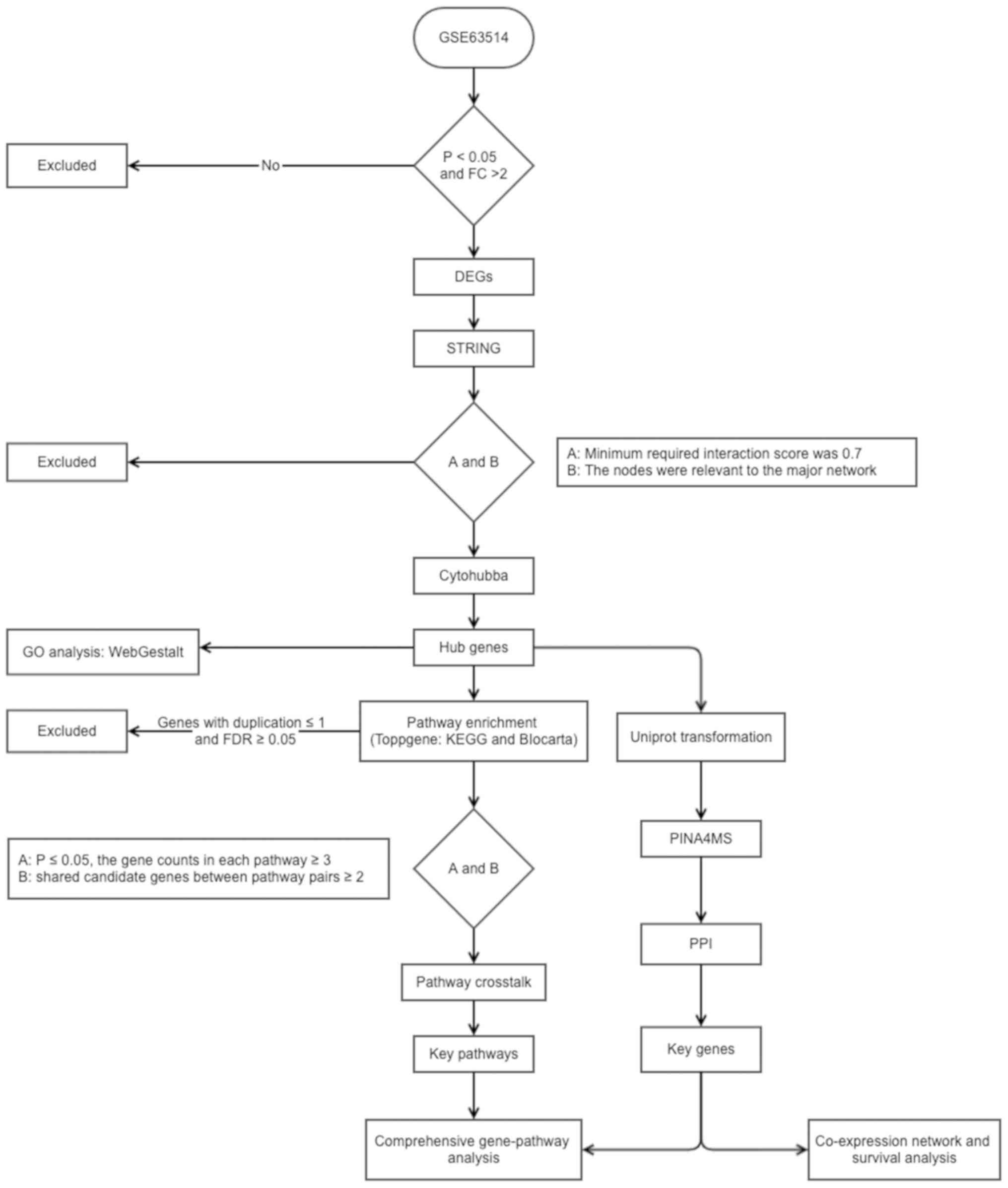
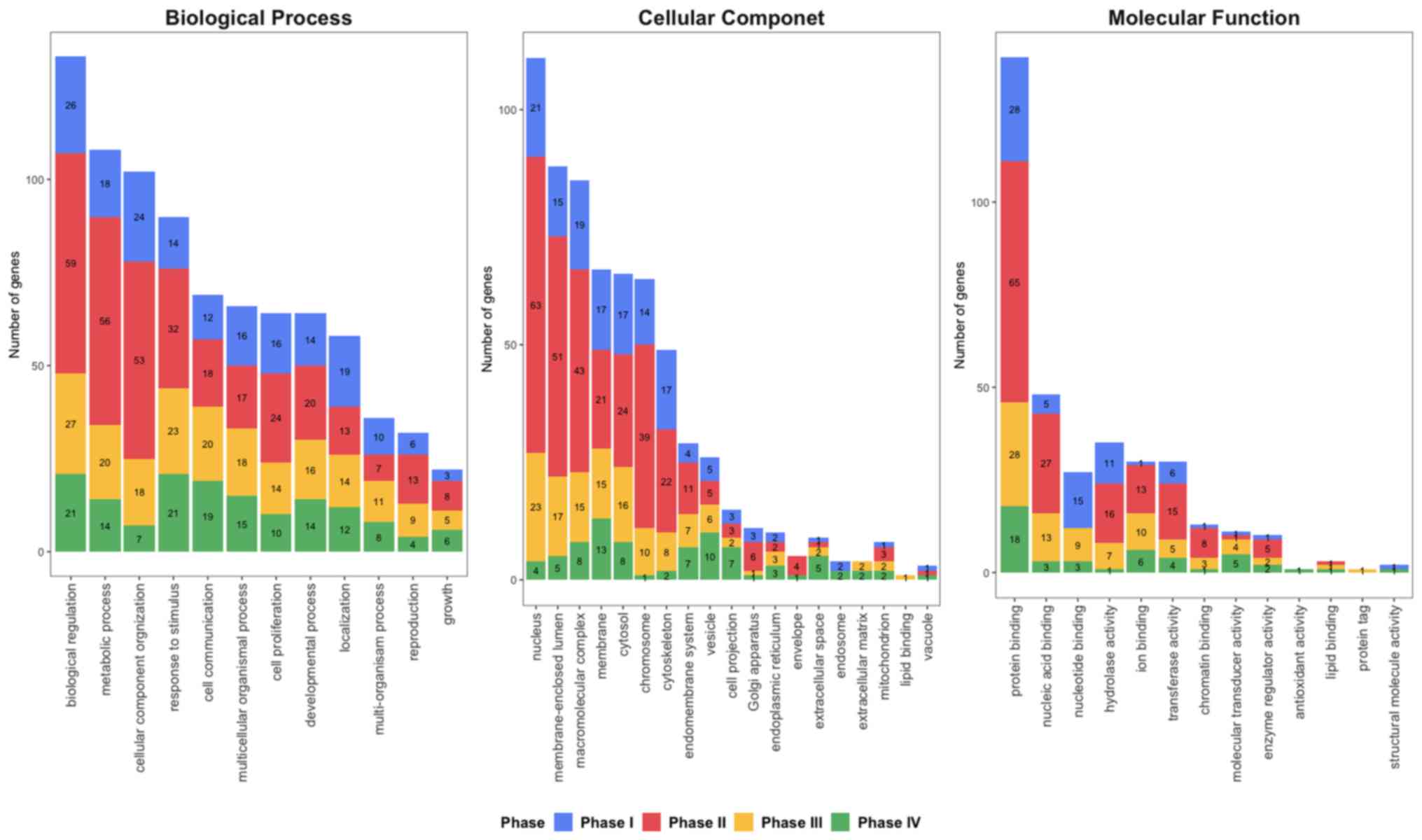
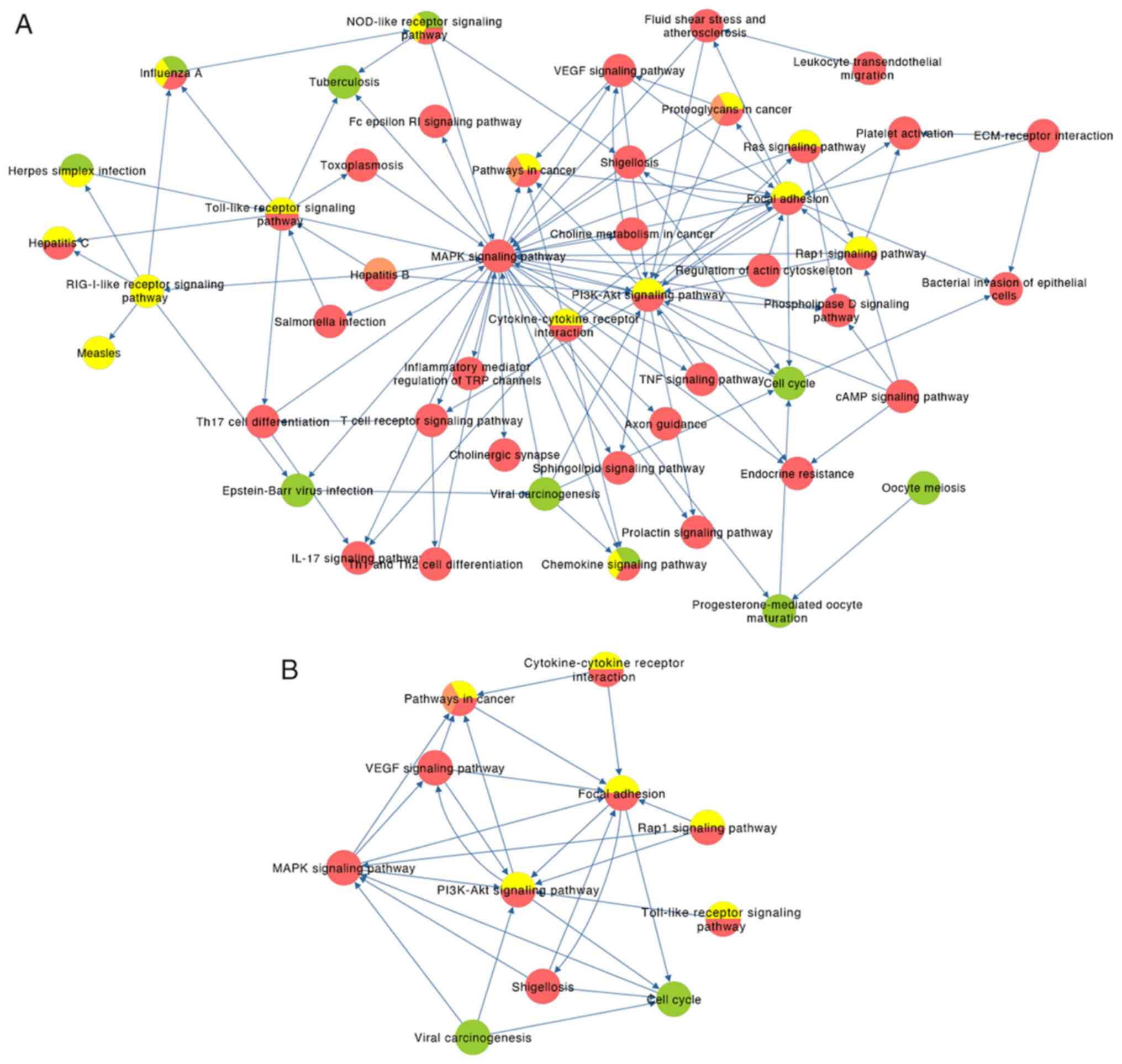
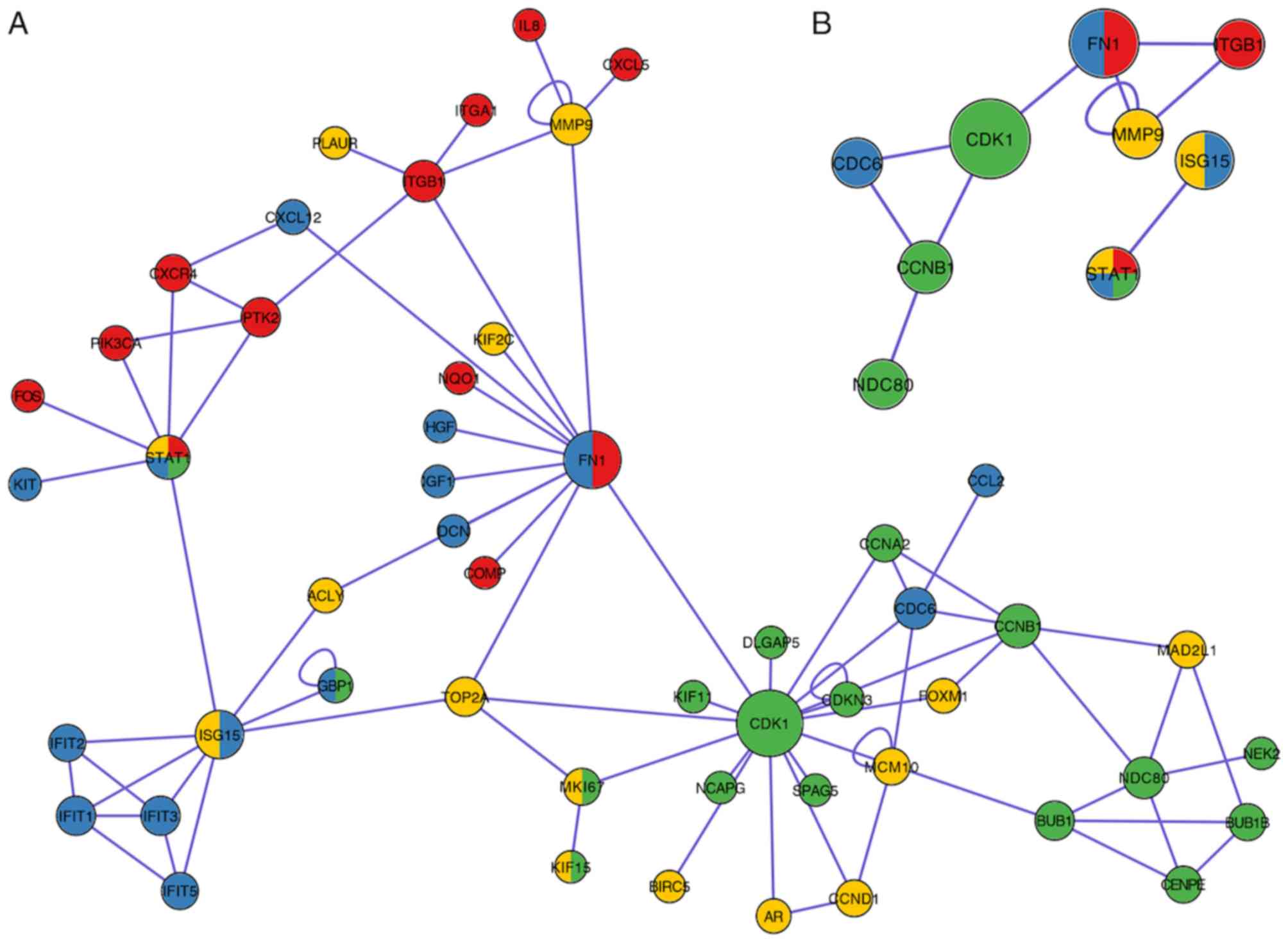
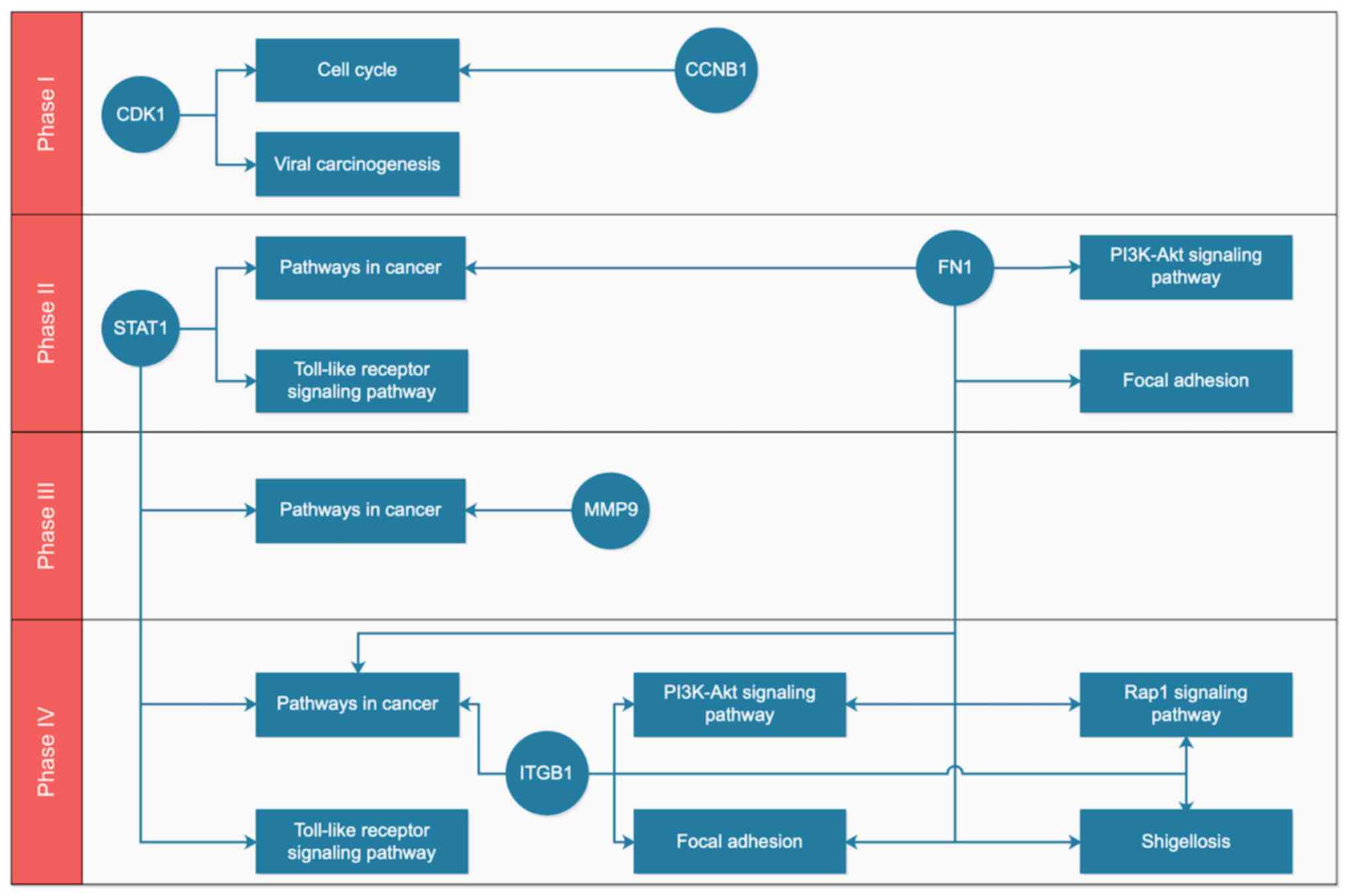
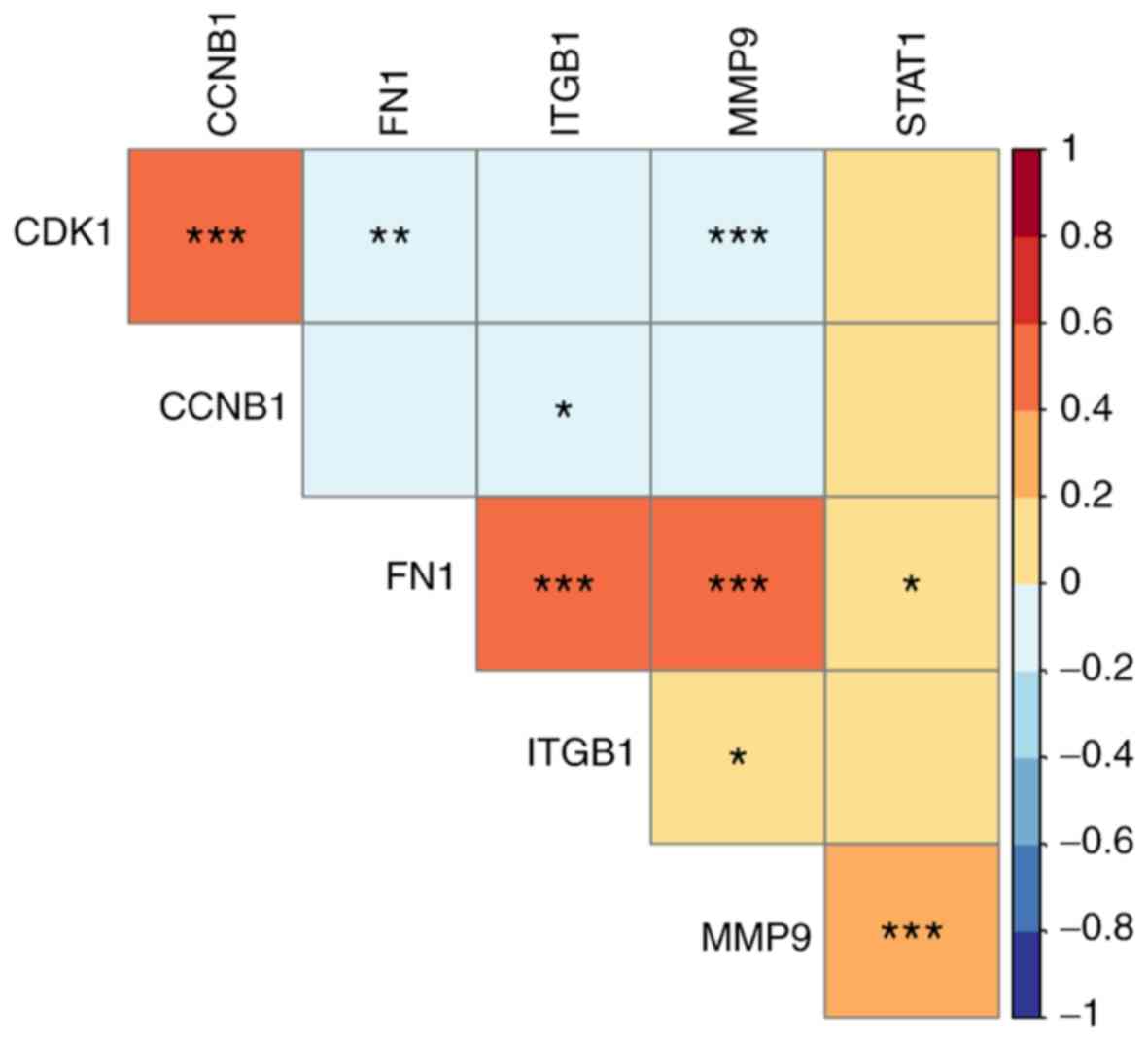
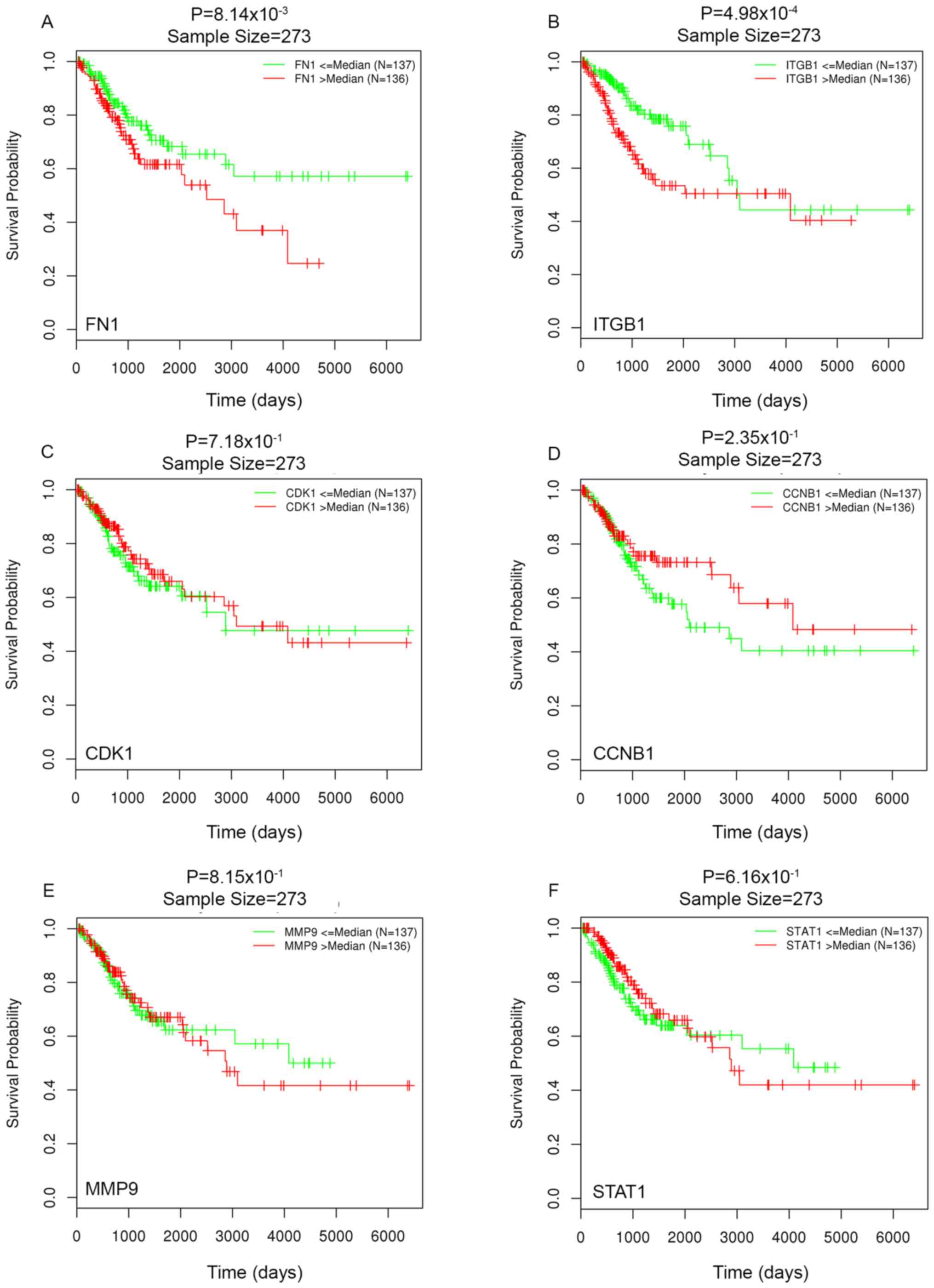
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Muliira RS, Salas AS and O'Brien B:
Quality of life among female cancer survivors in Africa: An
integrative literature review. Asia Pac J Oncol Nurs. 4:6–17. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gravitt PE and Winer RL: Natural history
of HPV infection across the lifespan: Role of viral latency.
Viruses. 9:E2672017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Walboomers JM, Jacobs MV, Manos MM, Bosch
FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ and Muñoz N:
Human papillomavirus is a necessary cause of invasive cervical
cancer worldwide. J Pathol. 189:12–19. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Louie KS, de Sanjose S, Diaz M,
Castellsagué X, Herrero R, Meijer CJ, Shah K, Franceschi S, Muñoz N
and Bosch FX; International Agency for Research on Cancer
Multicenter Cervical Cancer Study Group, : Early age at first
sexual intercourse and early pregnancy are risk factors for
cervical cancer in developing countries. Br J Cancer.
100:1191–1197. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu ZC, Liu WD, Liu YH, Ye XH and Chen SD:
Multiple sexual partners as a potential independent risk factor for
cervical cancer: A meta-analysis of epidemiological studies. Asian
Pac J Cancer Prev. 16:3893–3900. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Smith JS, Green J, Berrington de Gonzalez
A, Appleby P, Peto J, Plummer M, Franceschi S and Beral V: Cervical
cancer and use of hormonal contraceptives: A systematic review.
Lancet. 361:1159–1167. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ma YY, Wei SJ, Lin YC, Lung JC, Chang TC,
Whang-Peng J, Liu JM, Yang DM, Yang WK and Shen CY: PIK3CA as an
oncogene in cervical cancer. Oncogene. 19:2739–2744. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zheng L, Li T, Zhang Y, Guo Y, Yao J, Dou
L and Guo K: Oncogene ATAD2 promotes cell proliferation, invasion
and migration in cervical cancer. Oncol Rep. 33:2337–2344. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bai X, Wang W, Zhao P, Wen J, Guo X, Shen
T, Shen J and Yang X: LncRNA CRNDE acts as an oncogene in cervical
cancer through sponging miR-183 to regulate CCNB1 expression.
Carcinogenesis. Oct 12–2019.(Epub ahead of print).
|
|
11
|
Bremer GL, Tieboschb AT, van der Putten
HW, de Haan J and Arends JW: p53 tumor suppressor gene protein
expression in cervical cancer: Relationship to prognosis. Eur J
Obstet Gynecol Reprod Biol. 63:55–59. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cohen Y, Singer G, Lavie O, Dong SM,
Beller U and Sidransky D: The RASSF1A tumor suppressor gene is
commonly inactivated in adenocarcinoma of the uterine cervix. Clin
Cancer Res. 9:2981–2984. 2003.PubMed/NCBI
|
|
13
|
Hasina R, Pontier AL, Fekete MJ, Martin
LE, Qi XM, Brigaudeau C, Pramanik R, Cline EI, Coignet LJ and
Lingen MW: NOL7 is a nucleolar candidate tumor suppressor gene in
cervical cancer that modulates the angiogenic phenotype. Oncogene.
25:588–598. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Roura E, Castellsagué X, Pawlita M,
Travier N, Waterboer T, Margall N, Bosch FX, de Sanjosé S, Dillner
J, Gram IT, et al: Smoking as a major risk factor for cervical
cancer and pre-cancer: Results from the EPIC cohort. Int J cancer.
135:453–466. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Husain RS and Ramakrishnan V: Global
variation of human papillomavirus genotypes and selected genes
involved in cervical malignancies. Ann Glob Health. 81:675–683.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Soto D, Song C and McLaughlin-Drubin ME:
Epigenetic alterations in human papillomavirus-associated cancers.
Viruses. 9:E2482017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mincheva A, Gissmann L and zur Hausen H:
Chromosomal integration sites of human papillomavirus DNA in three
cervical cancer cell lines mapped by in situ hybridization. Med
Microbiol Immunol. 176:245–256. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Senapati R, Senapati NN and Dwibedi B:
Molecular mechanisms of HPV mediated neoplastic progression. Infect
Agent Cancer. 11:592016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Münger K, Baldwin A, Edwards KM, Hayakawa
H, Nguyen CL, Owens M, Grace M and Huh K: Mechanisms of human
papillomavirus-induced oncogenesis. J Virol. 78:11451–11460. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Martinez-Zapien D, Ruiz FX, Poirson J,
Mitschler A, Ramirez J, Forster A, Cousido-Siah A, Masson M, Vande
Pol S, Podjarny A, et al: Structure of the E6/E6AP/p53 complex
required for HPV-mediated degradation of p53. Nature. 529:541–545.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Scheffner M, Huibregtse JM, Vierstra RD
and Howley PM: The HPV-16 E6 and E6-AP complex functions as a
ubiquitin-protein ligase in the ubiquitination of p53. Cell.
75:495–505. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sherr CJ: The Pezcoller lecture: Cancer
cell cycles revisited. Cancer Res. 60:3689–3695. 2000.PubMed/NCBI
|
|
23
|
Martin LG, Demers GW and Galloway DA:
Disruption of the G1/S transition in human papillomavirus type 16
E7-expressing human cells is associated with altered regulation of
cyclin E. J Virol. 72:975–985. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Löffler H, Fechter A, Matuszewska M,
Saffrich R, Mistrik M, Marhold J, Hornung C, Westermann F, Bartek J
and Krämer A: Cep63 recruits Cdk1 to the centrosome: Implications
for regulation of mitotic entry, centrosome amplification, and
genome maintenance. Cancer Res. 71:2129–2139. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Skyldberg B, Fujioka K, Hellström AC,
Sylvén L, Moberger B and Auer G: Human papillomavirus infection,
centrosome aberration, and genetic stability in cervical lesions.
Mod Pathol. 14:279–284. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Duensing S and Münger K: The human
papillomavirus type 16 E6 and E7 oncoproteins independently induce
numerical and structural chromosome instability. Cancer Res.
62:7075–7082. 2002.PubMed/NCBI
|
|
27
|
Chaiwongkot A, Niruthisard S, Kitkumthorn
N and Bhattarakosol P: Quantitative methylation analysis of human
papillomavirus 16 L1 gene reveals potential biomarker for cervical
cancer progression. Diagn Microbiol Infect Dis. 89:265–270. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sharma A, De R, Javed S, Srinivasan R, Pal
A and Bhattacharyya S: Sonic hedgehog pathway activation regulates
cervical cancer stem cell characteristics during epithelial to
mesenchymal transition. J Cell Physiol. Feb 4–2019.(Epub ahead of
print). View Article : Google Scholar
|
|
29
|
Li YP, Zhang L, Zou YL and Yu Y:
Association between FGFR4 gene polymorphism and high-risk HPV
infection cervical cancer. Asian Pac J Trop Med. 10:680–684. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang J and Gao Y: CCAT-1 promotes
proliferation and inhibits apoptosis of cervical cancer cells via
the Wnt signaling pathway. Oncotarget. 8:68059–68070.
2017.PubMed/NCBI
|
|
31
|
Mora-García ML, Ávila-Ibarra LR,
García-Rocha R, Weiss-Steider B, Hernández-Montes J, Don-López CA,
Gutiérrez-Serrano V, Titla-Vilchis IJ, Fuentes-Castañeda MC,
Monroy-Mora A, et al: Cervical cancer cells suppress effector
functions of cytotoxic T cells through the adenosinergic pathway.
Cell Immunol. 320:46–55. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Song ZC, Ding L, Ren ZY, Sun XS, Yang Q,
Wang L, Feng MJ, Liu CL and Wang JT: Effects of Src on cervical
cancer cells proliferation and apoptosis through ERK signal
transduction pathway. Zhonghua Liu Xing Bing Xue Za Zhi.
38:1246–1251. 2017.(In Chinese; Abstract available in Chinese from
the publisher). PubMed/NCBI
|
|
33
|
Xu Z, Zhou Y, Shi F, Cao Y, Dinh TLA, Wan
J and Zhao M: Investigation of differentially-expressed microRNAs
and genes in cervical cancer using an integrated bioinformatics
analysis. Oncol Lett. 13:2784–2790. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen SZ, Ma WL and Zheng WL: Screening for
cervical cancer-related genes and their bioinformatics analysis.
Nan Fang Yi Ke Da Xue Xue Bao. 28:585–588. 2008.(In Chinese).
PubMed/NCBI
|
|
35
|
Nayar R and Wilbur DC: The bethesda system
for reporting cervical cytology: A historical perspective. Acta
Cytol. 61:359–372. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shoji T, Takatori E, Takeuchi S, Yoshizaki
A, Uesugi N, Sugai T and Sugiyama T: Clinical significance of
atypical glandular cells in the bethesda system 2001: A comparison
with the histopathological diagnosis of surgically resected
specimens. Cancer Invest. 32:105–109. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
O'Connor M, Gallagher P, Waller J, Martin
CM, O'Leary JJ and Sharp L; Irish Cervical Screening Research
Consortium (CERVIVA), : Adverse psychological outcomes following
colposcopy and related procedures: A systematic review. BJOG.
123:24–38. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Haque N, Uddin AFMK, Dey BR, Islam F and
Goodman A: Challenges to cervical cancer treatment in Bangladesh:
The development of a women's cancer ward at Dhaka Medical College
Hospital. Gynecol Oncol Rep. 21:67–72. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Roque DR, Wysham WZ and Soper JT: The
surgical management of cervical cancer: An overview and literature
review. Obstet Gynecol Surv. 69:426–441. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Marin JJ, Romero MR, Blazquez AG, Herraez
E, Keck E and Briz O: Importance and limitations of chemotherapy
among the available treatments for gastrointestinal tumours.
Anticancer Agents Med Chem. 9:162–184. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen HHW and Kuo MT: Improving
radiotherapy in cancer treatment: Promises and challenges.
Oncotarget. 8:62742–62758. 2017.PubMed/NCBI
|
|
42
|
Lecavalier-Barsoum M, Chaudary N, Han K,
Pintilie M, Hill RP and Milosevic M: Targeting CXCL12/CXCR4 and
myeloid cells to improve the therapeutic ratio in patient-derived
cervical cancer models treated with radio-chemotherapy. Br J
Cancer. 121:249–256. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang F, Ren CC, Liu L, Chen YN, Yang L,
Zhang XA, Wang XM and Yu FJ: SHH gene silencing suppresses
epithelial-mesenchymal transition, proliferation, invasion, and
migration of cervical cancer cells by repressing the hedgehog
signaling pathway. J Cell Biochem. 119:3829–3842. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gu J, Zhang X, Yang Z and Wang N:
Expression of cyclin D1 protein isoforms and its prognostic
significance in cervical cancer. Cancer Manag Res. 11:9073–9083.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chay DB, Han GH, Nam S, Cho H, Chung JY
and Hewitt SM: Forkhead box protein O1 (FOXO1) and paired box gene
3 (PAX3) overexpression is associated with poor prognosis in
patients with cervical cancer. Int J Clin Oncol. 24:1429–1439.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tang H, Wang Y, Zhang B, Xiong S, Liu L,
Chen W, Tan G and Li H: High brain acid soluble protein 1(BASP1) is
a poor prognostic factor for cervical cancer and promotes tumor
growth. Cancer Cell Int. 17:972017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yang X, Cheng Y and Li C: The role of TLRs
in cervical cancer with HPV infection: A review. Signal Transduct
Target Ther. 2:170552017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Frumovitz M and Sood AK: Vascular
endothelial growth factor (VEGF) pathway as a therapeutic target in
gynecologic malignancies. Gynecol Oncol. 104:768–778. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
den Boon JA, Pyeon D, Wang SS, Horswill M,
Schiffman M, Sherman M, Zuna RE, Wang Z, Hewitt SM, Pearson R, et
al: Molecular transitions from papillomavirus infection to cervical
precancer and cancer: Role of stromal estrogen receptor signaling.
Proc Natl Acad Sci USA. 112:E3255–E3264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang J, Vasaikar S, Shi Z, Greer M and
Zhang B: WebGestalt 2017: A more comprehensive, powerful, flexible
and interactive gene set enrichment analysis toolkit. Nucleic Acids
Res. 45:W130–W137. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chen J, Bardes EE, Aronow BJ and Jegga AG:
ToppGene Suite for gene list enrichment analysis and candidate gene
prioritization. Nucleic Acids Res. 37:W305–W311. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hu Y, Pan Z, Hu Y, Zhang L and Wang J:
Network and pathway-based analyses of genes associated with
Parkinson's disease. Mol Neurobiol. 54:4452–4465. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Scardoni G, Petterlini M and Laudanna C:
Analyzing biological network parameters with CentiScaPe.
Bioinformatics. 25:2857–2859. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Han JD, Berlin N, Hao T, Goldberg DS,
Berriz GF, Zhang LV, Dupuy D, Walhout AJ, Cusick ME, Roth FP and
Vidal M: Evidence for dynamically organized modularity in the yeast
protein-protein interaction network. Nature. 430:88–93. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kanwal A and Fazal S: Construction and
analysis of protein-protein interaction network correlated with
ankylosing spondylitis. Gene. 638:41–51. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wu J, Vallenius T, Ovaska K, Westermarck
J, Mäkelä TP and Hautaniemi S: Integrated network analysis platform
for protein-protein interactions. Nat Methods. 6:75–77. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kerrien S, Alam-Faruque Y, Aranda B,
Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen
A, Huntley R, et al: IntAct-source resource for molecular
interaction data. Nucleic Acids Res. 35:D561–D565. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chatr-aryamontri A, Ceol A, Palazzi LM,
Nardelli G, Schneider MV, Castagnoli L and Cesareni G: MINT: The
Molecular INTeraction database. Nucleic Acids Res. 35:D572–D574.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Breitkreutz BJ, Stark C, Reguly T, Boucher
L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bähler J,
Wood V, et al: The BioGRID interaction database: 2008 update.
Nucleic Acids Res. 36:D637–D640. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Salwinski L, Miller CS, Smith AJ, Pettit
FK, Bowie JU and Eisenberg D: The database of interacting proteins:
2004 update. Nucleic Acids Res. 32:D449–D451. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Peri S, Navarro JD, Amanchy R, Kristiansen
TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B,
Gandhi TK, Gronborg M, et al: Development of human protein
reference database as an initial platform for approaching systems
biology in humans. Genome Res. 13:2363–2371. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Güldener U, Münsterkötter M, Oesterheld M,
Pagel P, Ruepp A, Mewes HW and Stümpflen V: MPact: The MIPS protein
interaction resource on yeast. Nucleic Acids Res. 34:D436–D441.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46:D956–D963. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Cancer Genome Atlas Research Network, ;
Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA,
Ellrott K, Shmulevich I, Sander C and Stuart JM: The cancer genome
atlas pan-cancer analysis project. Nat Genet. 45:1113–1120. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Gasca S, Pellestor F, Assou S, Loup V,
Anahory T, Dechaud H, De Vos J and Hamamah S: Identifying new human
oocyte marker genes: A microarray approach. Reprod Biomed Online.
14:175–183. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Schwab MS, Roberts BT, Gross SD, Tunquist
BJ, Taieb FE, Lewellyn AL and Maller JL: Bub1 is activated by the
protein kinase p90(Rsk) during Xenopus oocyte maturation. Curr
Biol. 11:141–150. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Ouandaogo ZG, Frydman N, Hesters L, Assou
S, Haouzi D, Dechaud H, Frydman R and Hamamah S: Differences in
transcriptomic profiles of human cumulus cells isolated from
oocytes at GV, MI and MII stages after in vivo and in vitro oocyte
maturation. Hum Reprod. 27:2438–2447. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Li J, Qian WP and Sun QY: Cyclins
regulating oocyte meiotic cell cycle progression†. Biol Reprod.
101:878–881. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Kaplan MH: STAT signaling in inflammation.
JAKSTAT. 2:e241982013.PubMed/NCBI
|
|
75
|
Qiu X, Guo H, Yang J, Ji Y, Wu CS and Chen
X: Down-regulation of guanylate binding protein 1 causes
mitochondrial dysfunction and cellular senescence in macrophages.
Sci Rep. 8:16792018. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Bros M, Haas K, Moll L and Grabbe S: RhoA
as a key regulator of innate and adaptive immunity. Cells.
8:E7332019. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Erdely A, Antonini JM, Salmen-Muniz R,
Liston A, Hulderman T, Simeonova PP, Kashon ML, Li S, Gu JK, Stone
S, et al: Type I interferon and pattern recognition receptor
signaling following particulate matter inhalation. Part Fibre
Toxicol. 9:252012. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Luu K, Greenhill CJ, Majoros A, Decker T,
Jenkins BJ and Mansell A: STAT1 plays a role in TLR signal
transduction and inflammatory responses. Immunol Cell Biol.
92:761–769. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Smolen KK, Ruck CE, Fortuno ES III, Ho K,
Dimitriu P, Mohn WW, Speert DP, Cooper PJ, Esser M, Goetghebuer T,
et al: Pattern recognition receptor-mediated cytokine response in
infants across 4 continents. J Allergy Clin Immunol.
133:818–826.e4. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Xiang C, Chen J and Fu P: HGF/Met
signaling in cancer invasion: The impact on cytoskeleton
remodeling. Cancers (Basel). 9:E442017. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Lei T and Ling X: IGF-1 promotes the
growth and metastasis of hepatocellular carcinoma via the
inhibition of proteasome-mediated cathepsin B degradation. World J
Gastroenterol. 21:10137–10149. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Cho S, Kitadai Y, Yoshida S, Tanaka S,
Yoshihara M, Yoshida K and Chayama K: Deletion of the KIT gene is
associated with liver metastasis and poor prognosis in patients
with gastrointestinal stromal tumor in the stomach. Int J Oncol.
28:1361–1367. 2006.PubMed/NCBI
|
|
83
|
Li B, Shen W, Peng H, Li Y, Chen F, Zheng
L, Xu J and Jia L: Fibronectin 1 promotes melanoma proliferation
and metastasis by inhibiting apoptosis and regulating EMT. Onco
Targets Ther. 12:3207–3221. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Liu Y, Ren CC, Yang L, Xu YM and Chen YN:
Role of CXCL12-CXCR4 axis in ovarian cancer metastasis and
CXCL12-CXCR4 blockade with AMD3100 suppresses tumor cell migration
and invasion in vitro. J Cell Physiol. 234:3897–3909. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Chen Y, Jiang J, Zhao M, Luo X, Liang Z,
Zhen Y, Fu Q, Deng X, Lin X, Li L, et al: microRNA-374a suppresses
colon cancer progression by directly reducing CCND1 to inactivate
the PI3K/AKT pathway. Oncotarget. 7:41306–41319. 2016.PubMed/NCBI
|
|
86
|
Thomas SJ, Snowden JA, Zeidler MP and
Danson SJ: The role of JAK/STAT signalling in the pathogenesis,
prognosis and treatment of solid tumours. Br J Cancer. 113:365–371.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Stacker SA and Achen MG: The VEGF
signaling pathway in cancer: The road ahead. Chin J Cancer.
32:297–302. 2013.PubMed/NCBI
|
|
88
|
Shi X, Wang J, Lei Y, Cong C, Tan D and
Zhou X: Research progress on the PI3K/AKT signaling pathway in
gynecological cancer (Review). Mol Med Rep. 19:4529–4535.
2019.PubMed/NCBI
|
|
89
|
Li X, Jiang S and Tapping RI: Toll-like
receptor signaling in cell proliferation and survival. Cytokine.
49:1–9. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Gene Ontology Consortium: Gene ontology
consortium: Going forward. Nucleic Acids Res. 43:D1049–D1056. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Kawai T and Akira S: Toll-like receptor
and RIG-1-like receptor signaling. Ann N Y Acad Sci. 1143:1–20.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Gkretsi V and Stylianopoulos T: Cell
adhesion and matrix stiffness: Coordinating cancer cell invasion
and metastasis. Front Oncol. 8:1452018. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Zhang YL, Wang RC, Cheng K, Ring BZ and Su
L: Roles of Rap1 signaling in tumor cell migration and invasion.
Cancer Biol Med. 14:90–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Campbell PM and Der CJ: Oncogenic Ras and
its role in tumor cell invasion and metastasis. Semin Cancer Biol.
14:105–114. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Sun F, Wang J, Sun Q, Li F, Gao H, Xu L,
Zhang J, Sun X, Tian Y, Zhao Q, et al: Interleukin-8 promotes
integrin β3 upregulation and cell invasion through PI3K/Akt pathway
in hepatocellular carcinoma. J Exp Clin Cancer Res. 38:4492019.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Sanderson RD: Heparan sulfate
proteoglycans in invasion and metastasis. Semin Cell Dev Biol.
12:89–98. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Loo YM and Gale M Jr: Immune signaling by
RIG-I-like receptors. Immunity. 34:680–692. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Nyati KK and Prasad KN: Role of cytokines
and Toll-like receptors in the immunopathogenesis of Guillain-Barré
syndrome. Mediators Inflamm. 2014:7586392014. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Saxena M and Yeretssian G: NOD-Like
receptors: Master regulators of inflammation and cancer. Front
Immunol. 5:3272014. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Crosbie EJ, Einstein MH, Franceschi S and
Kitchener HC: Human papillomavirus and cervical cancer. Lancet.
382:889–899. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Balasubramaniam SD, Balakrishnan V, Oon CE
and Kaur G: Key molecular events in cervical cancer development.
Medicina (Kaunas). 55:E3842019. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Lin M, Ye M, Zhou J, Wang ZP and Zhu X:
Recent advances on the molecular mechanism of cervical
carcinogenesis based on systems biology technologies. Comput Struct
Biotechnol J. 17:241–250. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Ye J, Yin L, Xie P, Wu J, Huang J, Zhou G,
Xu H, Lu E and He X: Antiproliferative effects and molecular
mechanisms of troglitazone in human cervical cancer in vitro. Onco
Targets Ther. 8:1211–1218. 2015.PubMed/NCBI
|
|
104
|
Yugawa T and Kiyono T: Molecular
mechanisms of cervical carcinogenesis by high-risk human
papillomaviruses: Novel functions of E6 and E7 oncoproteins. Rev
Med Virol. 19:97–113. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Jones MC, Askari JA, Humphries JD and
Humphries MJ: Cell adhesion is regulated by CDK1 during the cell
cycle. J Cell Biol. 217:3203–3218. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Hochegger H, Dejsuphong D, Sonoda E,
Saberi A, Rajendra E, Kirk J, Hunt T and Takeda S: An essential
role for Cdk1 in S phase control is revealed via chemical genetics
in vertebrate cells. J Cell Biol. 178:257–268. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Vassilev LT: Cell cycle synchronization at
the G2/M phase border by reversible inhibition of CDK1. Cell Cycle.
5:2555–2556. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Shaikh F, Sanehi P and Rawal R: Molecular
screening of compounds to the predicted protein-protein interaction
site of Rb1-E7 with p53-E6 in HPV. Bioinformation. 8:607–612. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Yun J, Chae HD, Choy HE, Chung J, Yoo HS,
Han MH and Shin DY: p53 negatively regulates cdc2 transcription via
the CCAAT-binding NF-Y transcription factor. J Biol Chem.
274:29677–29682. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Lindqvist A, van Zon W, Karlsson Rosenthal
C and Wolthuis RM: Cyclin B1-Cdk1 activation continues after
centrosome separation to control mitotic progression. PLoS Biol.
5:e1232007. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Crasta K, Huang P, Morgan G, Winey M and
Surana U: Cdk1 regulates centrosome separation by restraining
proteolysis of microtubule-associated proteins. EMBO J.
25:2551–2563. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Fang Y, Yu H, Liang X, Xu J and Cai X:
Chk1-induced CCNB1 overexpression promotes cell proliferation and
tumor growth in human colorectal cancer. Cancer Biol Ther.
15:1268–1279. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Krek W and Nigg EA: Differential
phosphorylation of vertebrate p34cdc2 kinase at the G1/S and G2/M
transitions of the cell cycle: Identification of major
phosphorylation sites. EMBO J. 10:305–316. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Morgan DO: Principles of CDK regulation.
Nature. 374:131–134. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Banerjee NS, Wang HK, Broker TR and Chow
LT: Human papillomavirus (HPV) E7 induces prolonged G2 following S
phase reentry in differentiated human keratinocytes. J Biol Chem.
286:15473–15482. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Morgan EL, Wasson CW, Hanson L, Kealy D,
Pentland I, McGuire V, Scarpini C, Coleman N, Arthur JSC, Parish
JL, et al: STAT3 activation by E6 is essential for the
differentiation-dependent HPV18 life cycle. PLoS Pathog.
14:e10069752018. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Stewart DA, Cooper CR and Sikes RA:
Changes in extracellular matrix (ECM) and ECM-associated proteins
in the metastatic progression of prostate cancer. Reprod Biol
Endocrinol. 2:22004. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Chiu CF, Ho MY, Peng JM, Hung SW, Lee WH,
Liang CM and Liang SM: Raf activation by Ras and promotion of
cellular metastasis require phosphorylation of prohibitin in the
raft domain of the plasma membrane. Oncogene. 32:777–787. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Matter ML and Ruoslahti E: A signaling
pathway from the alpha5beta1 and alpha(v)beta3 integrins that
elevates bcl-2 transcription. J Biol Chem. 276:27757–27763. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Egeblad M and Werb Z: New functions for
the matrix metalloproteinases in cancer progression. Nat Rev
Cancer. 2:161–174. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
121
|
Sand JM, Larsen L, Hogaboam C, Martinez F,
Han M, Røssel Larsen M, Nawrocki A, Zheng Q, Karsdal MA and Leeming
DJ: MMP mediated degradation of type IV collagen alpha 1 and alpha
3 chains reflects basement membrane remodeling in experimental and
clinical fibrosis-validation of two novel biomarker assays. PLoS
One. 8:e849342013. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Bokhari AA, Baker TM, Dorjbal B, Waheed S,
Zahn CM, Hamilton CA, Maxwell GL and Syed V: Nestin suppression
attenuates invasive potential of endometrial cancer cells by
downregulating TGF-β signaling pathway. Oncotarget. 7:69733–69748.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
da Silva Cardeal LB, Brohem CA, Corrêa TC,
Winnischofer SM, Nakano F, Boccardo E, Villa LL, Sogayar MC and
Maria-Engler SS: Higher expression and activity of
metalloproteinases in human cervical carcinoma cell lines is
associated with HPV presence. Biochem Cell Biol. 84:713–719. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Cardeal LB, Boccardo E, Termini L,
Rabachini T, Andreoli MA, di Loreto C, Longatto Filho A, Villa LL
and Maria-Engler SS: HPV16 oncoproteins induce MMPs/RECK-TIMP-2
imbalance in primary keratinocytes: Possible implications in
cervical carcinogenesis. PLoS One. 7:e335852012. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Shiau MY, Fan LC, Yang SC, Tsao CH, Lee H,
Cheng YW, Lai LC and Chang YH: Human papillomavirus up-regulates
MMP-2 and MMP-9 expression and activity by inducing interleukin-8
in lung adenocarcinomas. PLoS One. 8:e544232013. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Ding Y, Pan Y, Liu S, Jiang F and Jiao J:
Elevation of MiR-9-3p suppresses the epithelial-mesenchymal
transition of nasopharyngeal carcinoma cells via down-regulating
FN1, ITGB1 and ITGAV. Cancer Biol Ther. 18:414–424. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Wang J, Deng L, Huang J, Cai R, Zhu X, Liu
F, Wang Q, Zhang J and Zheng Y: High expression of Fibronectin 1
suppresses apoptosis through the NF-κB pathway and is associated
with migration in nasopharyngeal carcinoma. Am J Transl Res.
9:4502–4511. 2017.PubMed/NCBI
|
|
128
|
Rajkumar T, Sabitha K, Vijayalakshmi N,
Shirley S, Bose MV, Gopal G and Selvaluxmy G: Identification and
validation of genes involved in cervical tumourigenesis. BMC
Cancer. 11:802011. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Koromilas AE and Sexl V: The tumor
suppressor function of STAT1 in breast cancer. JAKSTAT.
2:e233532013.PubMed/NCBI
|
|
130
|
Zhang Y and Liu Z: STAT1 in cancer: Friend
or foe? Discov Med. 24:19–29. 2017.PubMed/NCBI
|
|
131
|
Akram M, Kim KA, Kim ES, Shin YJ, Noh D,
Kim E, Kim JH, Majid A, Chang SY, Kim JK and Bae ON: Selective
inhibition of JAK2/STAT1 signaling and iNOS expression mediates the
anti-inflammatory effects of coniferyl aldehyde. Chem Biol
Interact. 256:102–110. 2016. View Article : Google Scholar : PubMed/NCBI
|