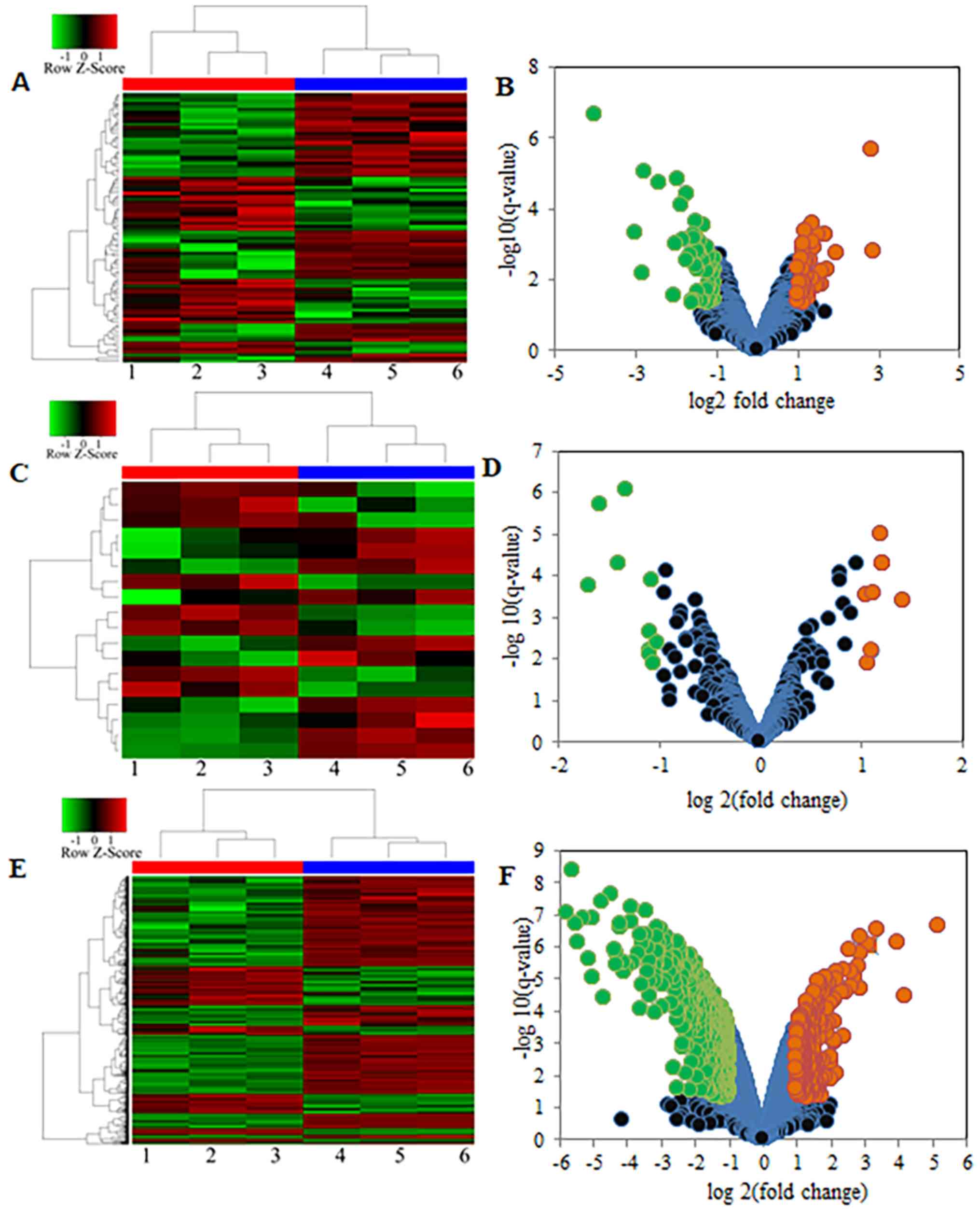
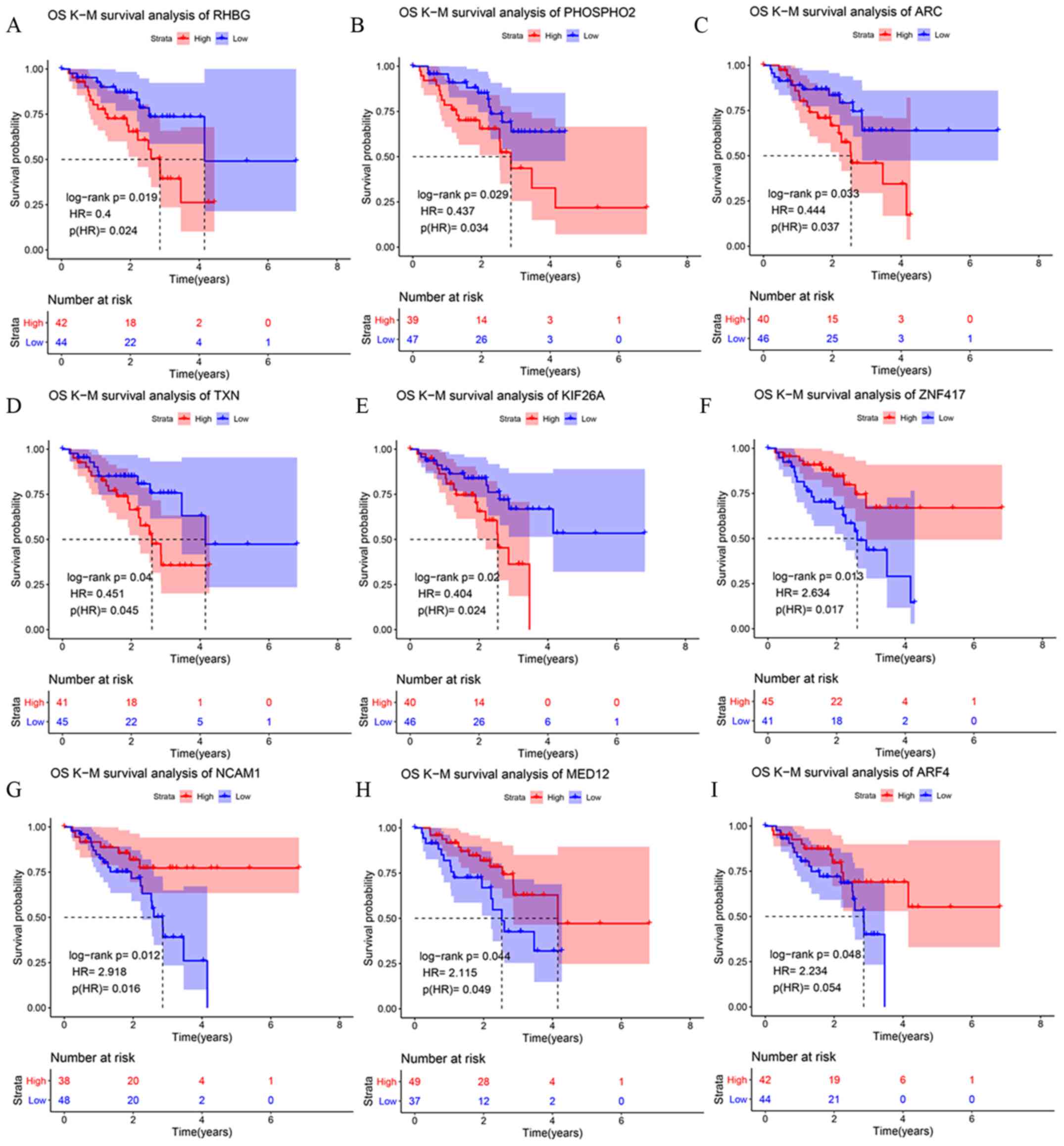
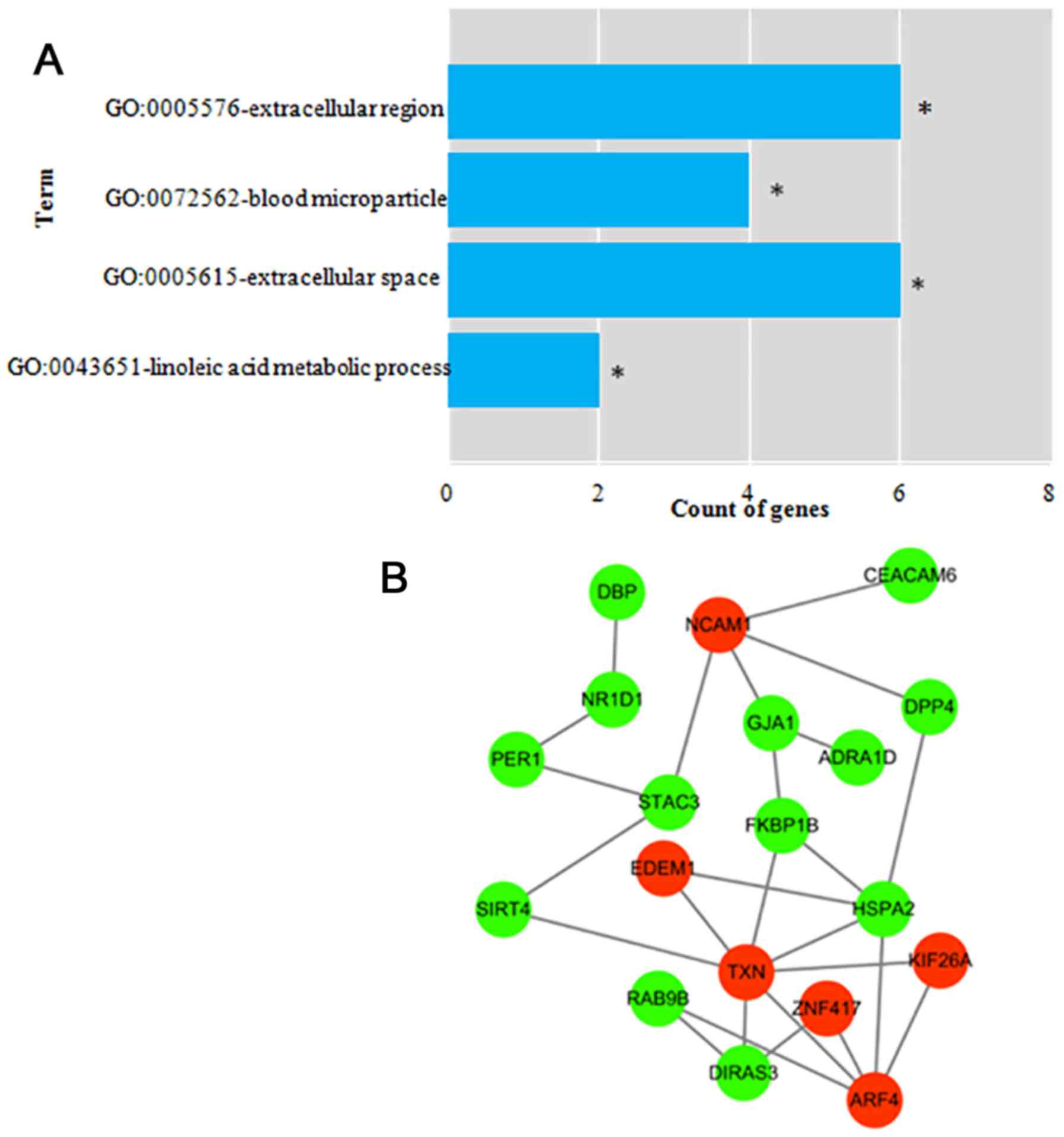
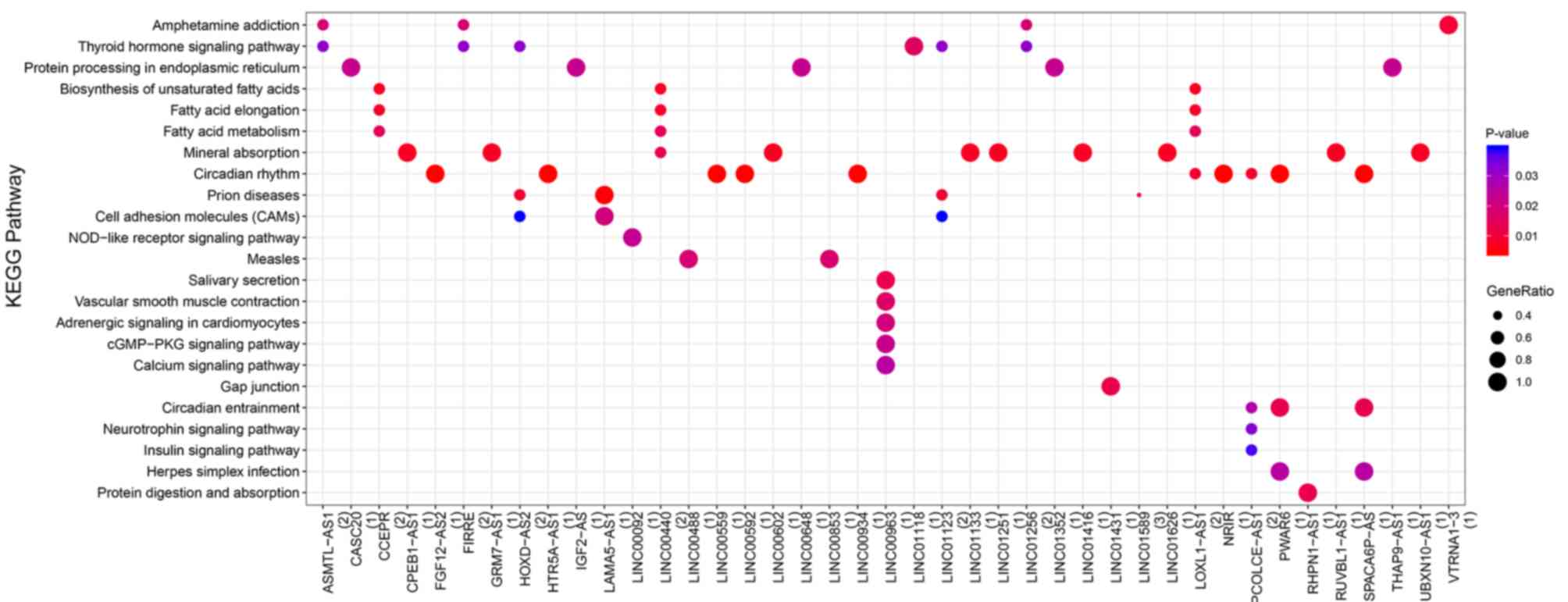
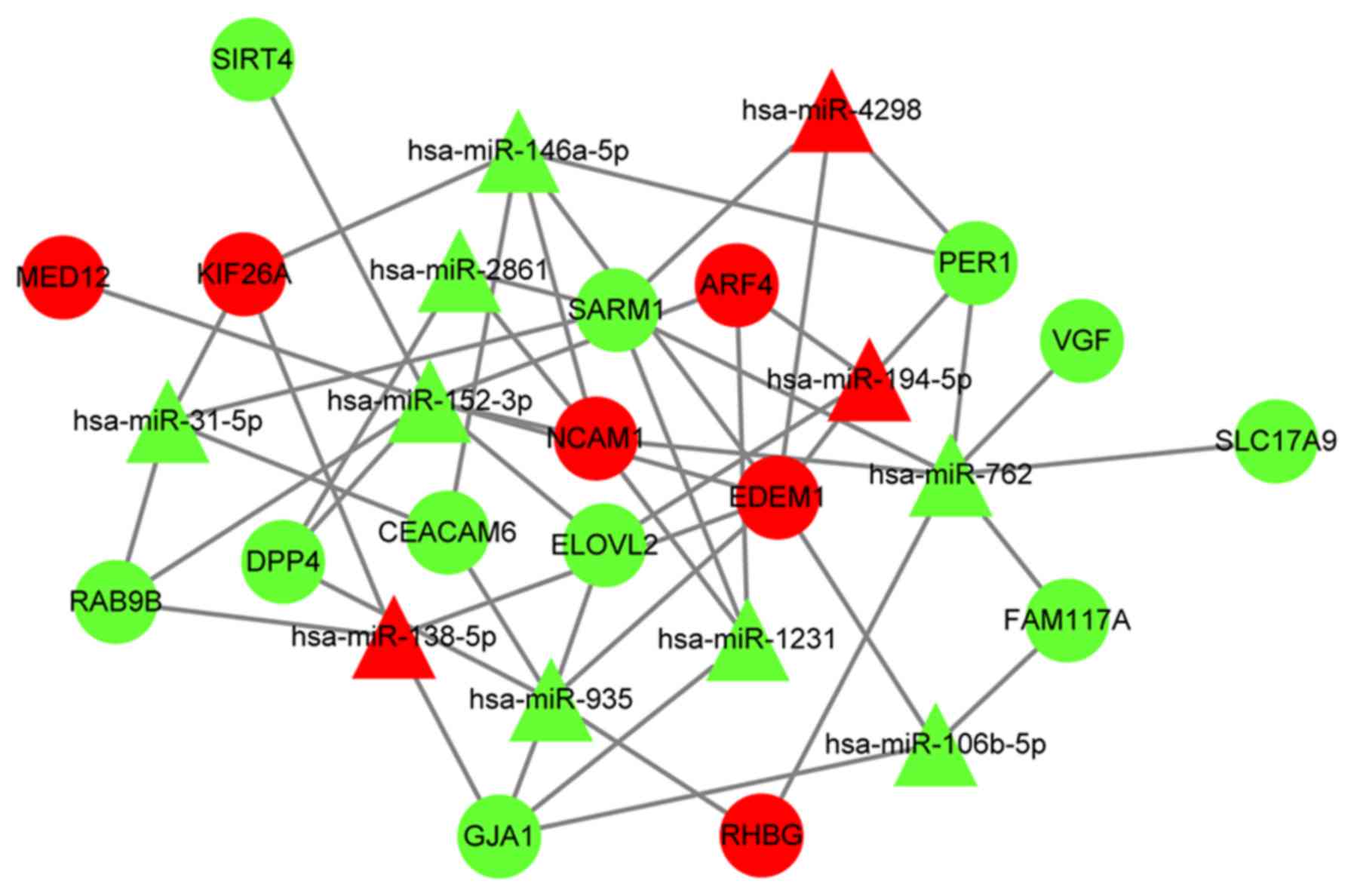
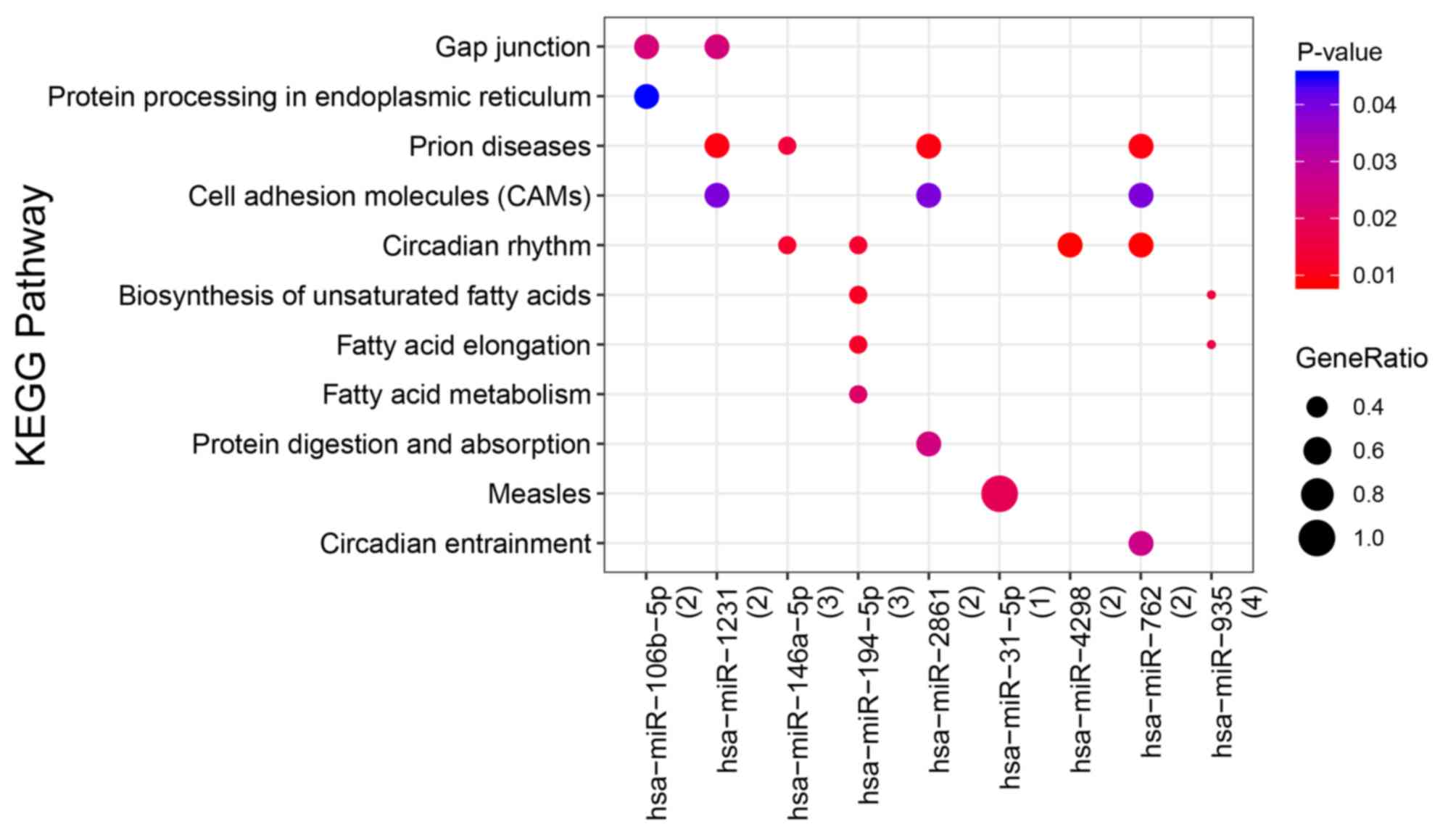
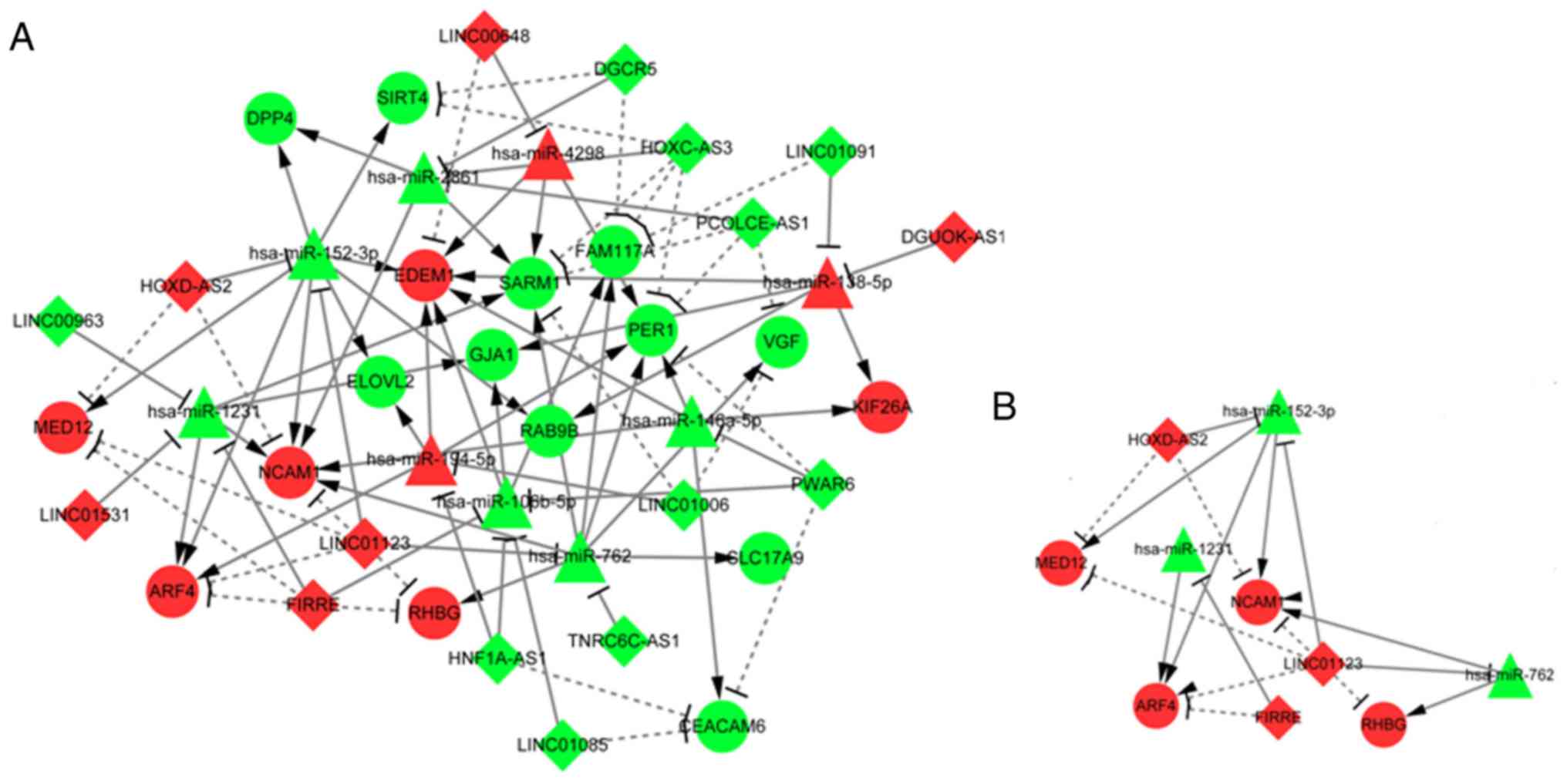
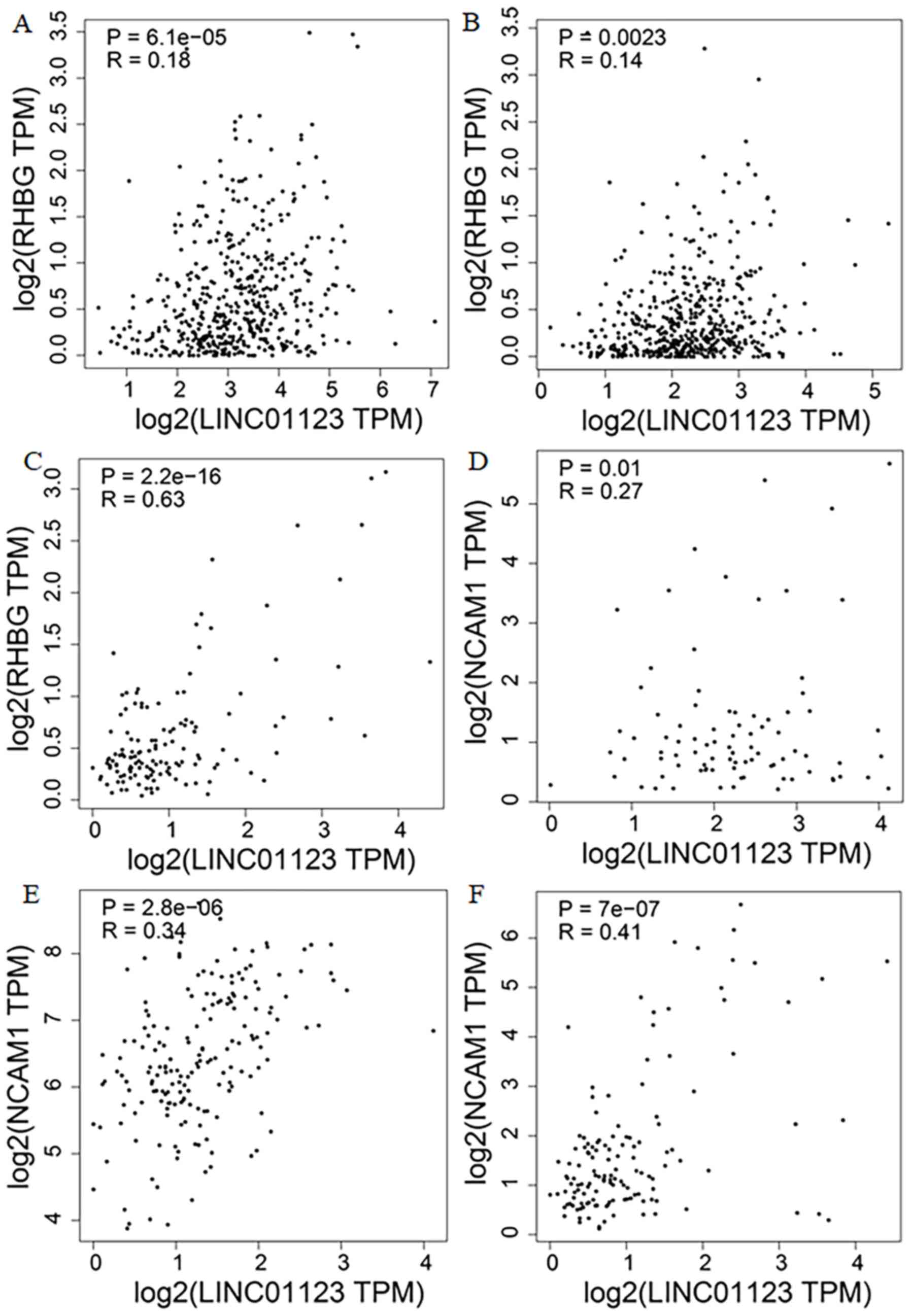
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-Small cell lung cancer: Epidemiology, risk
factors, treatment, and survivorship. Mayo Clin Proc. 83:584–594.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu L, Wu S, Yang Y, Cai J, Zhu X, Wu J,
Wu J, Li M and Guan H: SOSTDC1 is down-regulated in non-small cell
lung cancer and contributes to cancer cell proliferation. Cell
Biosci. 6:242016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li X, Shi Y, Yin Z, Xue X and Zhou B: An
eight-miRNA signature as a potential biomarker for predicting
survival in lung adenocarcinoma. J Transl Med. 12:1592014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Riaz SP, Lüchtenborg M, Coupland VH,
Spicer J, Peake MD and Møller H: Trends in incidence of small cell
lung cancer and all lung cancer. Lung Cancer. 75:280–284. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Murray N and Turrisi AT III: A review of
first-line treatment for small-cell lung cancer. J Thorac Oncol.
1:270–278. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sarvi S, Mackinnon AC, Avlonitis N,
Bradley M, Rintoul RC, Rassl DM, Wang W, Forbes SJ, Gregory CD and
Sethi T: CD133+ cancer stem-like cells in small cell lung cancer
are highly tumorigenic and chemoresistant but sensitive to a novel
neuropeptide antagonist. Cancer Res. 74:1554–1565. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Voigt W, Dietrich A and Schmoll HJ:
Overview of development status and clinical action. Cisplatin and
its analogues. Pharm Unserer Zeit. 35:134–143. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Silver DP, Richardson AL, Eklund AC, Wang
ZC, Szallasi Z, Li Q, Juul N, Leong CO, Calogrias D, Buraimoh A, et
al: Efficacy of neoadjuvant cisplatin in triple-negative breast
cancer. J Clin Oncol. 28:1145–1153. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Santabarbara G, Maione P, Rossi A and
Gridelli C: Pharmacotherapeutic options for treating adverse
effects of cisplatin chemotherapy. Expert Opin Pharmacother.
17:561–570. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yimit A, Adebali O, Sancar A and Jiang Y:
Differential damage and repair of DNA-adducts induced by
anti-cancer drug cisplatin across mouse organs. Nat Commun.
10:3092019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Eastman A: Improving anticancer drug
development begins with cell culture: Misinformation perpetrated by
the misuse of cytotoxicity assays. Oncotarget. 8:8854–8866. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao X, Li X, Zhou L, Ni J, Yan W, Ma R,
Wu J, Feng J and Chen P: LncRNA HOXA11-AS drives cisplatin
resistance of human LUAD cells via modulating miR-454-3p/Stat3.
Cancer Sci. 109:3068–3079. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Barrett T, Troup DB, Wilhite SE, Ledoux P,
Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M and
Edgar R: NCBI GEO: Mining tens of millions of expression
profiles-database and tools update. Nucleic Acids Res.
35:D760–D765. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang Y, Li H, Liu J and Wang J: Noncoding
RNA expression profiling of cisplatin-resistant cells derived from
the A549 lung cell line (miRNA). NCBI. 2013, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE43249January
2–2013
|
|
18
|
Yang Y, Li H, Liu J and Wang J: Noncoding
RNA expression profiling of cisplatin-resistant cells derived from
the A549 lung cell line (lncRNA and mRNA). NCBI. 2013, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE43493January
14–2013
|
|
19
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-Analysis of affymetrix genechip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatic. 19:185–193. 2003. View Article : Google Scholar
|
|
21
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guidelines of HUGO Gene Nomenclature
Committee (HGNC). European Bioinformatics Institute (EMBL-EBI).
https://www.genenames.org/about/guidelines/
|
|
23
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wickham H: ggplot2: Elegant graphics for
data analysis. New York. Springer–Verlag. 2016.
|
|
25
|
Zhao S, Yin L, Guo Y, Sheng Q and Shyr Y:
heatmap3: An Improved Heatmap Package. R package version 1.1.6.
https://CRAN.R-project.org/package=heatmap3December
1–2018
|
|
26
|
Huang DW, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS and Eppig
JT: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kanehisa M: KEGG: Kyoto encyclopedia of
genes and genomes. Nucleic Acids Res. 28:27–30. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dweep H and Gretz N: MiRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:6972015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Paraskevopoulou MD, Vlachos IS, Karagkouni
D, Georgakilas G, Kanellos I, Vergoulis T, Zagganas K, Tsanakas P,
Floros E, Dalamagas T, et al: DIANA-LncBase v2: Indexing microRNA
targets on non-coding transcripts. Nucleic Acids Res. 44:D231–D238.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liao Y, Xie B, Zhang H, He Q, Guo L,
Subramaniapillai M, Fan B, Lu C and Mclntyer RS: Efficacy of
omega-3 PUFAs in depression: A meta-analysis. Transl Psychiatr.
9:1902019. View Article : Google Scholar
|
|
36
|
Merhi A, De Mees C, Abdo R, Victoria
Alberola J and Marini AM: Wnt/β-Catenin signaling regulates the
expression of the ammonium permease gene RHBG in human cancer
cells. PLoS One. 10:e01286832015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Johansson FK, Brodd J, Eklof C, Ferletta
M, Hesselager G, Tiger CF, Uhrbom L and Westermark B:
Identification of candidate cancer-causing genes in mouse brain
tumors by retroviral tagging. Proc Natl Acad Sci USA.
101:11334–11337. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jensen M and Berthold F: Targeting the
neural cell adhesion molecule in cancer. Cancer Lett. 258:9–21.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kashiwagi K, Ishii J, Sakaeda M, Arimasu
Y, Shimoyamada H, Sato H, Sato H, Miyata C, Kamma H, Aoki I and
Yazawa T: Differences of molecular expression mechanisms among
neural cell adhesion molecule 1, synaptophysin, and chromogranin A
in lung cancer cells. Pathol Int. 62:232–245. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Vander Borght A, Duysinx M, Broers JLV,
Ummelen M, Falkenberg FW, Hahnel C and van der Zeijst BAM: The 180
splice variant of NCAM-containing exon 18-is specifically expressed
in small cell lung cancer cells. Transl Lung Cancer Res. 7:376–388.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sasca D, Szybinski J, Schüler A, Shah V,
Heidelberger J, Haehnel PS, Dolnik A, Kriege O, Fehr EM, Gebhardt
WH, et al: NCAM1 (CD56) promotes leukemogenesis and confers drug
resistance in AML. Blood. 133:2305–2319. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cho H, Kim JY and Oh YL: Diagnostic value
of HBME-1, CK19, galectin 3, and CD56 in the subtypes of follicular
variant of papillary thyroid carcinoma. Pathol Int. 68:605–613.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ghaderi F, Ahmadvand S, Ramezan A,
Montazer M and Ghaderi A: Production and characterization of
monoclonal antibody against a triple negative breast cancer cell
line. Biochem Biophys Res Commun. 505:181–186. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhao X, Tang DY, Zuo X, Zhang TD and Wang
C: Identification of lncRNA-miRNA-mRNA regulatory network
associated with epithelial ovarian cancer cisplatin-resistant. J
Cell Physiol. 234:19886–19894. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Fang L, Wang H and Li P: Systematic
analysis reveals a lncRNA-mRNA co-expression network associated
with platinum resistance in high-grade serous ovarian cancer.
Invest New Drugs. 36:187–194. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yoshida T, Ri M, Kinoshita S, Narita T,
Totani H, Ashour R, Ito A, Kusumoto S, Ishida T, Komatsu H and Iida
S: Low expression of neural cell adhesion molecule, CD56, is
associated with low efficacy of bortezomib plus dexamethasone
therapy in multiple myeloma. PLoS One. 13:e01967802018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kämpjärvi K, Mäkinen N, Kilpivaara O,
Arola J, Heinonen HR, Böhm J, Abdel-Wahab O, Lehtonen HJ, Pelttari
LM, Mehine M, et al: Somatic MED12 mutations in uterine
leiomyosarcoma and colorectal cancer. Br J Cancer. 107:1761–1765.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kämpjärvi K, Kim NH, Keskitalo S, Clark
AD, von Nandelstadh P, Turunen M, Heikkinen T, Park MJ, Mäkinen N,
Kivinummi K, et al: Somatic MED12 mutations in prostate cancer and
uterine leiomyomas promote tumorigenesis through distinct
mechanisms. Prostate. 76:22–31. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Huang S, Hölzel M, Knijnenburg T,
Schlicker A, Roepman P, McDermott U, Garnett M, Grernrum W, Sun C,
Prahallad A, et al: MED12 controls the response to multiple cancer
drugs through regulation of TGF-β receptor signaling. Cell.
151:937–950. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Feliciano P: MED12 in cancer drug
resistance. Nat Genet. 45:112012. View Article : Google Scholar
|
|
51
|
Howley BV, Link LA, Grelet S, El-Sabban M
and Howe PH: A CREB3-regulated ER-Golgi trafficking signature
promotes metastatic progression in breast cancer. Oncogene.
37:1308–1325. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ramalho-Carvalho J, Gonçalves CS, Graça I,
Bidarra D, Pereira-Silva E, Salta S, Godinho MI, Gomez A, Esteller
M, Costa BM, et al: A multiplatform approach identifies miR-152-3p
as a common epigenetically regulated onco-suppressor in prostate
cancer targeting TMEM97. Clin Epigenetics. 10:402018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ge S, Wang D, Kong Q, Gao W and Sun J:
Function of miR-152 as a tumor suppressor in human breast cancer by
targeting PIK3CA. Oncol Res. 25:1363–1371. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li LW, Xiao HQ, Ma R, Yang M, Li W and Lou
G: MiR-152 is involved in the proliferation and metastasis of
ovarian cancer through repression of ERBB3. Int J Mol Med.
41:1529–1535. 2018.PubMed/NCBI
|
|
55
|
Wang Y, Bao W, Liu Y, Wang S, Xu S, Li X,
Li Y and Wu S: MiR-98-5p contributes to cisplatin resistance in
epithelial ovarian cancer by suppressing miR-152 biogenesis via
targeting dicer1. Cell Death Dis. 9:4472018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Huang S, Li X and Zhu H: MicroRNA-152
targets phosphatase and tensin homolog to inhibit apoptosis and
promote cell migration of nasopharyngeal carcinoma cells. Med Sci
Monit. 22:4330–4337. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hou R, Yang Z, Wang S, Chu D, Liu Q, Liu J
and Jiang L: MiR-762 can negatively regulate menin in ovarian
cancer. Onco Targets Ther. 10:2127–2137. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li Y, Huang R, Wang L, Hao J, Zhang Q,
Ling R and Yun J: MicroRNA-762 promotes breast cancer cell
proliferation and invasion by targeting IRF7 expression. Cell
Prolif. 48:643–649. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Shi Y, Jia Y, Zhao W, Zhou L, Xie X and
Tong Z: Histone deacetylase inhibitors alter the expression of
molecular markers in breast cancer cells via microRNAs. Int J Mol
Med. 42:435–442. 2018.PubMed/NCBI
|
|
60
|
Wang H, Wu J, Luo WJ and Hu JL: Low
expression of miR-1231 in patients with glioma and its prognostic
significance. Eur Rev Med Pharmacol Sci. 22:8399–8405.
2018.PubMed/NCBI
|
|
61
|
Zhang J, Zhang J, Qiu W, Zhang J, Li Y,
Kong E, Lu A, Xu J and Lu X: MicroRNA-1231 exerts a tumor
suppressor role through regulating the EGFR/PI3K/AKT axis in
glioma. J Neurooncol. 139:547–562. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chen SL, Ma M, Yan L, Xiong SH, Liu Z, Li
S, Liu T, Shang S, Zhang YY, Zeng H, et al: Clinical significance
of exosomal miR-1231 in pancreatic cancer. Zhonghua Zhong Liu Za
Zhi. 41:46–49. 2019.(In Chinese; Abstract available in Chinese from
the publisher). PubMed/NCBI
|
|
63
|
Pan Y, Xu T, Liu Y, Li W and Zhang W:
Upregulated circular RNA circ_0025033 promotes papillary thyroid
cancer cell proliferation and invasion via sponging miR-1231 and
miR-1304. Biochem Biophys Res Commun. 510:334–338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yang W, Li Y, Song X, Xu J and Xie J:
Genome-Wide analysis of long noncoding RNA and mRNA co-expression
profile in intrahepatic cholangiocarcinoma tissue by RNA
sequencing. Oncotarget. 8:26591–26599. 2017.PubMed/NCBI
|
|
65
|
Ma W, Chen X, Ding L, Ma J, Jing W, Lan T,
Sattar H, Wei Y, Zhou F and Yuan Y: The prognostic value of long
noncoding RNAs in prostate cancer: A systematic review and
meta-analysis. Oncotarget. 8:57755–57765. 2017.PubMed/NCBI
|
|
66
|
Qi Y, Wang Z, Wu F, Yin B, Jiang T, Qiang
B, Yuan J, Han W and Peng X: Long noncoding RNA HOXD-AS2 regulates
cell cycle to promote glioma progression. J Cell Biochem.
120:281172018.
|
|
67
|
Shi X, Cui Z, Liu X, Wu S, Wu Y, Fang F
and Zhao H: LncRNA FIRRE is activated by MYC and promotes the
development of diffuse large B-cell lymphoma via Wnt/β-catenin
signaling pathway. Biochem Biophys Res Commun. 510:594–600. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Li M, Zhao LM, Li SL, Li J, Gao B, Wang
FF, Wang SP, Hu XH, Cao J and Wang GY: Differentially expressed
lncRNAs and mRNAs identified by NGS analysis in colorectal cancer
patients. Cancer Med. 7:4650–4664. 2018. View Article : Google Scholar : PubMed/NCBI
|