Introduction
Malignant gliomas account for ~70% of human primary
malignant brain tumors annually in the United States and can lead
to a median survival of only 12–15 months, even with optimal
treatment such as strategies to increase the radiation dose and
novel chemotherapeutic agents (1).
Gliomas are histologically heterogeneous and invasive tumors, which
are derived from abnormal proliferative glial cells and gliomas are
characterized by the sequential accumulation of genetic aberrations
such as Olig2 and the deregulation of the growth-factor signaling
pathway (2). Treatment options at
diagnosis often consist of a combination of surgery, radiotherapy
and chemotherapy (3). However, the
benefit of surgical resection in prolonging overall survival rate
remains limited due to the infiltrative nature of glioma, which
usually leads to recurrence (1).
Despite the application of standard treatment, including
radio-chemotherapy with the alkylating agent temozolomide (4), the median survival time is <2 years
due to treatment resistance (5,6), and
little progress has been made in the field of glioma treatment
(7). With invasion and metastasis
being the most devastating difficulties in the clinical management
of glioma, understanding the underlying pathogenic mechanisms of
gliomas is important, including an understanding of the cytokines
potentially involved in the development and progression of
gliomas.
The regulatory effects of cytokines on tumors have
been intensively investigated (8–10).
Neuregulins (NRGs) are cytokines associated with a variety of
different malignant tumors, which function as signaling molecules
that mediate cellular interactions. For example, NRG1 was found
increase the risk of bladder cancer development (11), uveal melanoma (12) and node-negative invasive breast
cancer (13). NRG1, as an epidermal
growth factor (EGF)-like family member, is one of the most active
malignant tumor-related cytokines, with ≥6 identified isoforms
resulting from alternative splicing of the NRG1 gene,
including Type I to III NRG1α and NRG1β (14). After binding to the receptor
tyrosine-protein kinase erbB-2 (also called NEU), NRG1 initiates
intracellular responses, including proliferation stimulation and
inhibition, apoptosis, migration, differentiation and adhesion
(13). Ritch et al (15) demonstrated that glioma cell survival
is enhanced via either an autocrine or a paracrine NRG1/ErbB
receptor signaling pathway.
Cell adhesion molecule L1 like (CHL1) encodes a
single-pass transmembrane cell adhesion molecule (CAM) that is
capable of both homophilic and heterophilic interactions (16). CHL1 is a member of the neural CAM L1
family (16), which comprises 4
structurally related transmembrane proteins in vertebrates: L1,
CHL1, neuronal cell adhesion molecule (NrCAM) and neurofascin
(17). Similar to L1, CHL1 is also
involved in cell adhesion, axon guidance and synaptic plasticity
(18). Accumulating evidence has
also indicated that CHL1 serves a significant role in tumor
metastasis and progression (19,20).
Our previous research indicated that NRG1 promotes
glioma cell migration by enhancing the protein expression levels of
L1 (21,22). In the present study, the correlation
between CHL1 and glioma grade and the role of NRG1 in modulating L1
was investigated. The results demonstrated that both NRG1α and
NRG1β promote glioma cell migration by enhancing CHL1 expression
levels, possibly by controlling the ERK pathway. This may provide
insight into the development of inhibitors to antagonize
NRG1-induced CHL1 expression in the treatment of glioma.
Materials and methods
Reagents and microarray
A total of 2 recombinant E. coli-derived
NRG1α1 (rNRG1α, RP-317-P0A; Thermo Fisher Scientific, Inc.) and
NRG1β1 (rNRG1β, RP-318-P0A; Thermo Fisher Scientific, Inc.)
isoforms, corresponding to the extracellular domains of the 2
molecules were used. A rat anti-human CHL1 antibody specifically
targeting the extracellular domain of CHL1 was purchased from
R&D Systems, Inc. (1:200 for both immunohistochemistry and
immunuofluorescence, and 1:500 for western blot analysis; cat. no.
MAB2126) and an anti-rabbit phosphorylated(p)-Neu antibody (1:200;
cat. no. sc-101695) was purchased from Santa Cruz Biotechnology,
Inc. A donkey anti-rat secondary antibody conjugated to Alexa Fluor
488 (1:500; cat. no. 712-545-153), a donkey anti-rat secondary
antibody conjugated to Dylight™ 594 (1:1,000; cat. no.
712-515-150), a donkey anti-mouse secondary antibody conjugated to
Dylight™ 488 (1:500; cat. no. 715-485-150) and a donkey anti-rabbit
secondary antibody conjugated to Dylight™ 594 (1:1,000; cat. no.
711-515-152) were all purchased from Jackson ImmunoResearch
Laboratories, Inc. A mouse monoclonal anti-NRG1 antibody (1:500;
cat. no. MA5-12896; Thermo Fisher Scientific, Inc.), a mouse
monoclonal anti-PCNA antibody (1:200; cat. no. sc-25280; Santa Cruz
Biotechnology, Inc.), a mouse monoclonal anti-ERK1/2 antibody
(1:1,000; cat. no. sc-135900; Santa Cruz Biotechnology, Inc.), a
mouse monoclonal anti-p-ERK1/2 antibody (1:1,000; cat. no. sc-7383;
Santa Cruz Biotechnology, Inc.), a mouse monoclonal anti-GAPDH
antibody (1:1,000; cat. no. sc-3650620; Santa Cruz Biotechnology,
Inc.) and horseradish peroxidase-conjugated goat anti-mouse
(1:1,000; cat. no. BA1050; Wuhan Boster Biological Technology,
Ltd.) and rabbit anti-rat secondary antibodies (1:1,000; cat. no.
BA1058; Wuhan Boster Biological Technology, Ltd.) were used.
Dulbecco's modified Eagle's medium (HyClone; Thermo Fisher
Scientific, Inc.), penicillin/streptomycin mixture (Beijing
Solarbio Science and Technology Co., Ltd.), 10% FBS (Hangzhou
Sijiqing Biological Engineering Materials Co., Ltd.), 10% normal
goat serum and 10% normal donkey serum (Beijing Solarbio Science
and Technology Co., Ltd.) were used. A human glioma microarray,
containing 184 glioma tissues, which can be graded from I to IV and
18 cancer adjacent normal (CAN) tissues, was purchased from US
Biomax, Inc. (cat. no. GL 2083). The tumors were graded by a
licensed pathologist on a scale of I to IV according to their
degree of malignancy as defined by the World Health Organization
classification of tumors of the central nervous system (23): I, Pilocytic astrocytoma; II, fairly
differentiated astrocytoma; III, anaplastic astrocytoma; and IV,
glioblastoma.
Hematoxylin and eosin (H&E) and
immunohistochemical staining
Immunohistochemical staining of paraffinized
sections was performed as previously described (22). Human glioma tissue microarray
sections (4-µm thick and deparaffinized) were rehydrated using a
graded ethanol series (from 100, 90, 80, 70, 60 to 50%) that
culminated with PBS. Antigen retrieval was performed using 10 mM
citrate buffer (pH 6.0) at 99°C for 1 h and endogenous peroxidase
was cleared by incubation in 3% H2O2 at room
temperature for 20 min. The sections were then blocked with 10%
normal goat serum in PBS at room temperature for 30 min, and the
samples were incubated at 4°C overnight with a rat-anti-human
primary antibody targeting human CHL1 (1:100). The antigen-antibody
complexes were visualized using the avidin-biotin-peroxidase
complex method using an AEC kit (cat. no. ZLI-9036, Beijing
Zhongshan Jinqiao Biotechnology Co., Ltd.). For H&E staining,
tissues were stained with eosin solution (cat. no. BSBA-4022,
Beijing Zhongshan Jinqiao Biotechnology Co., Ltd.) for 5 min at
room temperature. Counterstaining was performed with Mayer's
hematoxylin (cat. no. BSBA-4021A, Beijing Zhongshan Jinqiao
Biotechnology Co., Ltd.) for 1 min at room temperature. Images of
the H&E and immunohistochemical staining results were captured
using a light microscope (magnification, ×200; http://www.jnoec.com). The percentage of the area that
was positive for CHL1 staining on each whole tissue point was
evaluated using ImageTool (IT) software [version 3.0; Department of
Dental Diagnostic Science at the University of Texas Health Science
Center (UTHSCSA); magnification, ×200], which indicated the
expression levels of CHL1 in the CAN tissue and glioma tissues
graded from I to IV.
Cell culture
The human glioblastoma U-87 MG cell line (cat. no.
CL-0238; unknown origin) and the human glioma U251 cell line (cat.
no. CL-0237) were purchased from Procell Life Science and
Technology Co., Ltd. The human glioma cell line SHG-44 (cat. no.
SHG44) was purchased from Guangzhou Jennio Bioech Co., Ltd;
http://www.jennio-bio.com.). All cell
lines were authenticated using short tandem repeat analysis and
tested for mycoplasma contamination. Cell lines were maintained in
Dulbecco's modified Eagle's medium (HyClone; Thermo Fisher
Scientific, Inc.) supplemented with 50 U/ml penicillin/streptomycin
mixture (Beijing Solarbio Science and Technology Co., Ltd.) and 10%
fetal bovine serum (Hangzhou Sijiqing Biological Engineering
Materials Co., Ltd.). The cells were routinely cultured in 75
cm2 cell culture plates (Corning Inc.) at 37°C in a
humidified incubator with 5% CO2.
Small interferring (si)RNA
treatment
One control siRNA and siRNA against CHL1 (Table I), and one control siRNA and three
different siRNAs against NRG1 (Table
II), as well as the transfection reagent siRNA-Mate were
purchased from Shanghai GenePharma Co., Ltd.. To test the effect of
NRG1 on CHL1 expression, cells (5×104 cells/well) in
normal culture medium were allowed to adhere overnight in 48-well
plates. When 80% confluence was achieved, the media were aspirated
and replaced with fresh medium containing RNA/siRNA-Mate complexes
(10 nM of either the random control siRNA or the targeting
siRNAs/well) and the cells were further cultured at 37°C in a
humidified incubator with 5% CO2 for 48 h before western
blot analysis.
 | Table I.Sequences of control siRNA and siRNA
targeting CHL1. |
Table I.
Sequences of control siRNA and siRNA
targeting CHL1.
| siRNA | Sequence,
5′-3′ |
|---|
| Random control |
|
|
Sense |
UUCUCCGAACGUGUCACGUTT |
|
Antisense |
ACGUGACACGUUCGGAGAATT |
| CHL1 |
|
|
Sense |
GGAGCUAAUUUGACCAUAUTT |
|
Antisense |
AUAUGGUCAAAUUAGCUCCTT |
 | Table II.Sequences of control siRNA and siRNA
targeting NRG1. |
Table II.
Sequences of control siRNA and siRNA
targeting NRG1.
| siRNA | Sequence,
5′-3′ |
|---|
| Random control |
|
|
Sense |
UUCUCCGAACGUGUCACGUTT |
|
Antisense |
ACGUGACACGUUCGGAGAATT |
| No. 1 (NRG1) |
|
|
Sense |
GGCUGAUUCUGGAGAGUAUTT |
|
Antisense |
AUACUCUCCAGAAUCAGCCTT |
| No. 2 (NRG1) |
|
|
Sense |
GAGUCUCCCAUUAGAAUAUTT |
|
Antisense |
AUAUUCUAAUGGGAGACUCTT |
| No. 3 (NRG1) |
|
|
Sense |
GCCACUCUGUAAUCGUGAUTT |
|
Antisense |
AUCACGAUUACAGAGUGGCTT |
To determine the effects of CHL1 on the level of
proliferation cell nuclear antigen (PCNA), cells
(2×104/well) in normal culture medium were allowed to
adhere overnight in an 8-well Lab-Tek Chamber Slide™ (cat. no.
154941PK; Nalge Nunc International). The media were then aspirated
and replaced with fresh medium containing RNA/siRNA-Mate complexes
(10 nM of either the random control siRNA or the siRNA against
CHL1/well) and the cells were further cultured for 48 h. Cells were
fixed with 4% paraformaldehyde in PBS (pH 7.3) at 4ºC
for 15 min, and were then subjected to double immunofluorescence
staining of both CHL1 and PCNA.
Cell senescence assay
The effects of CHL1 on cell senescence was assessed
with the application of CHL1 siRNA in human glioblastoma U-87 MG
cells. Cells (1×105 cells/well) in culture medium were
allowed to adhere to 24-well plates overnight. When 80% confluence
was achieved, the medium was aspirated and replaced with fresh
medium containing RNA-siRNA-Mate complexes (10 nM of either the
control or the CHL1 siRNA/well). The cells were further cultured at
37°C in a humidified incubator with 5% CO2 for 48 h.
After 48 h of treatment, the cells were fixed for 20 min at 4°C
with 4% paraformaldehyde in PBS. A β-galactosidase/X-Gal complex
was added to each well and incubated overnight at 37°C according to
the manufacturer's protocol (Beyotime Institute of Biotechnology).
Cell senescence was determined by the development of a deep blue
color in the cytoplasm, which reflected the activation of
β-galactosidase.
Treatment with NRG1s
To investigate the effects of NRG1α and β on CHL1
expression levels and NRG1-induced activation of ERK1/2 in
vitro, U-87 MG, U251 and SHG-44 cells (1×105
cells/well) were allowed to adhere to 24-well plates (Costar;
Corning Inc.). After an overnight incubation, the medium was
aspirated and replaced with serum-free culture medium containing
NRG1α or NRG1β at concentrations of 0, 0.5, 1.0, 1.5, 2.5 and 5.0
nM for 48 h. Cells were lysed using RIPA lysis buffer mixture
supplemented with PMSF (1:200; both Beijing Solarbio Science and
Technology Co., Ltd.). The cell lysates were centrifuged at 14,000
× g for 15 min at 4°C, and the supernatants were collected for
western blot analysis.
For immunofluorescence staining, 2×104
cells/well were seeded overnight on an 8-well Lab-Tek Chamber
Slide™ (Nunc). After the overnight incubation, the medium was
aspirated and replaced with serum-free culture medium containing
NRG1α or NRG1β at concentrations of 2.5 nM for 48 h. The cells were
then subjected to fixation with 4% paraformaldehyde in PBS (pH 7.3)
at 4°C for 15 min, which was followed by immunofluorescence
staining for both CHL1 and p-Neu, as aforementioned.
Western blotting
Cells were then lysed with RIPA buffer (Solarbio
Life Sciences). The cell lysate was then subjected to concentration
determination with a BCA kit (Solarbio Life Sciences). A total of
20 µg of protein from each cell lysate were heated at 95°C in 20%
sample loading buffer (0.125 M Tris-HCl, pH 6.8, 20% glycerol, 10%
SDS, 0.1% bromophenol blue and 5% β-mercaptoethanol), then resolved
using SDS-PAGE (with an 8% gel) and transferred onto polyvinylidene
difluoride membranes (EMD Millipore). Non-specific protein binding
sites were blocked with 5% skimmed milk diluted in Tris-buffered
saline (pH 7.4) buffer containing 0.05% Tween-20 (TBST). The
membranes were incubated overnight at 4°C with a mouse monoclonal
anti-NRG1 antibody, a rat anti-human CHL1 antibody, a mouse
monoclonal anti-ERK1/2 antibody and a mouse monoclonal
anti-p-ERK1/2 antibody. To control protein loading, a mouse
monoclonal anti-GAPDH antibody was used to blot the membrane
overnight at 4°C. After 3 washes of 5 min each with TBST at room
temperature, horseradish peroxidase-conjugated goat anti-mouse and
rabbit anti-rat secondary antibodies diluted in TBST were incubated
with the membranes for 1 h at room temperature, which was followed
by 3 washes with TBST for 5 min each at room temperature. The
antigens were visualized using enhanced chemiluminescence (Beyotime
Institute of Biotechnology). The signal intensity was quantified
using IT software as the average densitometry multiplied by the
area (measured as the number of pixels) (22).
Immunofluorescence analysis
Deparaffinized human glioma tissue microarray
sections (4-µm thick and deparaffinized) were rehydrated through
incubation with a graded ethanol series (from 100, 90, 80, 70, 60
to 50%) that culminated with PBS. Antigen retrieval was performed
using 10 mM citrate buffer (pH 6.0) at 100°C for 1 h. The samples
were blocked with 10% normal donkey serum (Beijing Solarbio Science
and Technology Co., Ltd.) in PBS at room temperature for 60 min.
The sections were incubated with the following primary antibodies:
Rat anti-human CHL1 (1:200) and rabbit anti-PCNA antibodies
(1:200). The cultured cells were incubated with rat anti-human CHL1
(1:200) antibody together with either mouse anti-PCNA antibody
(1:200) or rabbit anti-p-Neu antibody (1:100). After being washed
in PBS 3 times for 5 min, the cells were incubated at room
temperature for 90 min with a donkey anti-rat secondary antibody
conjugated to Dylight™ 488 (1:500) and a donkey anti-rabbit
secondary antibody conjugated to Dylight™ 594 (1:1,000). After
being washed in PBS 3 times for 5 min, the tissue samples were
incubated at room temperature for 90 min with a donkey anti-mouse
secondary antibody conjugated to Dylight™ 488 (1:500) and a donkey
anti-rat secondary antibody conjugated to Dylight™ 594 (1:1,000).
The tissues and cells were co-stained with DAPI at room temperature
for 20 min and mounted using an anti-fade mounting solution
(Beyotime Institute of Biotechnology). Confocal images were
captured using an Olympus confocal system (FV-1000; Olympus
Corporation). DAPI, Dylight™ 488 and Dylight™ 594 were excited at
405, 488 and 594 nm, respectively.
Correlation analysis between CHL1 and
PCNA
CHL1 and PCNA immunofluorescence images for each
sample were rearranged onto one panel based on the tumor grade. The
protein level (measured as the integrated immunofluorescence
intensity) was evaluated on the basis of gray scale ranging from 0
to 255, which was obtained using Image Tool II software. The
correlation between CHL1 and PCNA was evaluated via Pearson's
correlation analysis, using SPSS software (version 17.0; SPSS
Inc.).
Statistical analysis
In vitro experiments were repeated at least 3
times using independent culture preparations. All numerical data
are presented as group mean ± standard error of the mean.
Statistical analyses were performed using SPSS version 10.0 (SPSS,
Inc.). The analysis of CHL1 levels in glioma samples was undertaken
using one-way ANOVA followed by Tukey's post hoc test. The effects
of NRG1s on CHL1 were analyzed using one-way ANOVA followed by
Dunnett's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Immunohistochemical staining of CHL1
in the glioma tissue microarray
Human tissues from a commercial glioma tissue
microarray were subjected to H&E staining and
immunohistochemical staining of CHL1. Representative staining from
the CAN tissue and tissues of different grades is shown in Fig. 1A. H&E staining showed regularly
arranged cells in the CAN tissue, whereas abnormally proliferating
cells were observed in the structurally disordered glioma tissues
graded from I to IV (Fig. 1A), CHL1
was weakly stained in the CAN tissue, whereas it was strongly
expressed in glioma tissue graded from I to IV, with the highest
staining intensity detected in one grade II glioma tissue (Fig. 1A). The percentage of CHL1-positive
area in each tissue point was measured to evaluate the expression
levels of CHL1. As shown in Fig. 1B,
the percentages of CHL1 positivity in CAN, and grade I, I–II, II,
III and IV tumor tissues, were 32.33±5.00, 52.61±2.91, 66.67±4.43,
64.58±2.85, 46.67±5.92 and 42.90±5.33%, respectively. As was
indexed by the percentage of CHL1-positive area in each tissue
point, CHL1 expression levels in I–II and II grade gliomas were
significantly higher compared with that in CAN tissue samples
(P<0.05 for grade I–II and P<0.001 for grade II). By
contrast, CHL1 expression levels in grade IV tissues were
significantly lower than those in grade I–II samples (P<0.05).
In addition, the CHL1 expression levels in grade III and IV tissues
were significantly lower than those in grade II samples (P<0.05
for grade III and P<0.001 for grade IV). It was thus concluded
that CHL1 expression levels is associated with the malignancy of
glioma.
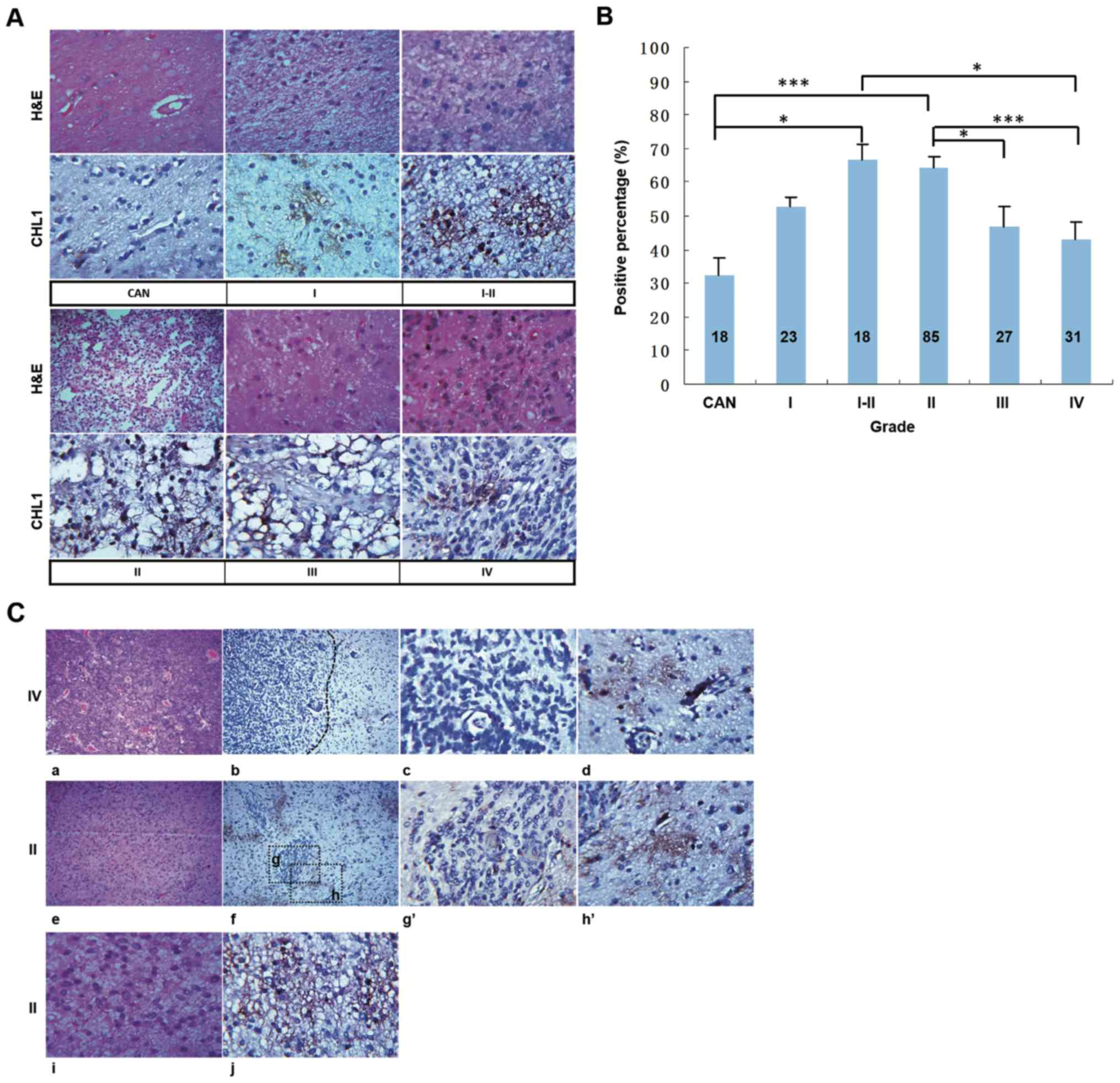 | Figure 1.CHL1 expression levels in human
glioma tissues. (A) Human glioma tissues were subjected to H&E
and immunohistochemical staining for CHL1. (B) Percentage of
CHL1-positive areas in each tissue point was measured to evaluate
the expression levels of CHL1. *P<0.05 and ***P<0.001. (C)
H&E staining of one grade IV glioma tissue (a) containing both
low-cell-density and high-cell-density areas, (b) in which CHL1
expression was shown. In contrast to the weak CHL1 staining
intensity in the (c) high-cell-density area of the grade IV glioma
tissue, the staining intensity of CHL1 was relatively high in the
(d) low-cell-density area of the tissue. H&E staining of one
grade II glioma tissue (e) containing both low-cell-density and
high-cell-density areas, (f) in which CHL1 expression was shown. In
contrast to the weak CHL1 staining intensity in the (g and g′)
high-cell-density area of the grade II glioma tissue, the staining
intensity of CHL1 was relatively high in the (h and h′)
low-cell-density area of the tissue. H&E staining of one grade
II glioma tissue that only exhibited a (i) low-cell-density area.
(j) CHL1 was highly expressed in the grade II glioma tissue only
with a low-cell-density area. g′, the enlarged boxed region g, and
h′, the enlarged boxed region h. CHL1, cell adhesion molecule L1
like; CAN, cancer adjacent normal tissue. H&E, hematoxylin and
eosin. |
To identify the differential expression of CHL1
between low-grade and high-grade gliomas, the distribution of CHL1
expression between gliomas graded II and IV were further compared.
Fig. 1C-a and -e showed one grade IV
and grade II tissue containing both low-cell-density and
high-cell-density areas, and Fig.
1C-i showed one grade II tissue only with a low-cell-density
area, as revealed by H&E staining. The limited distribution of
CHL1 in grade IV and II glioma tissues and the scattered
distribution of CHL1 in grade II are presented in Fig. 1C-b, -f and -j, respectively. Fig. 1C-b and -d showed a restricted CHL1
expression in one grade IV glioblastoma, and that CHL1 expression
level was associated with cell density within the glioma tissue.
Generally, CHL1 was distributed in low-cell-density areas (Fig. 1C-d), while it was not notably
expressed in high-cell-density areas (Fig. 1C-c). Similarly, CHL1 was largely
expressed in the cell sparse areas in grade II glioma tissues
(Fig. 1C-f, -h and -h′), with
limited distribution found in the cell dense area (Fig. 1C-f, -g and -g′). In grade II gliomas
with a scattered cell distribution, CHL1 was also dispersedly
localized (Fig. 1C-j). Taken
together, these results indicated that CHL1 was predominately
expressed in low-cell-density area relative to that in the
high-cell-density area within the glioma tissue.
Correlation between CHL1 and PCNA
expression levels in human glioma tissues, and the effects of CHL1
on glioma cell senescence
First, the colocalization of CHL1 and PCNA in human
glioma tissues was analyzed. Representative staining of CHL1 and
PCNA are shown, in CAN and grade I–II and II human glioma tissues,
in which the expression levels of CHL1 is highly expressed relative
to that in the other grades of gliomas (Fig. 2A). In CAN tissue, both PCNA and CHL1
were weakly colocalized in the nucleus. In one grade I–II sample,
CHL1 was solely localized on the cell membrane, while PCNA was
weakly detected in the nucleus. However, in one grade II sample,
CHL1 was not only detected on the cell membrane but it was also
detected in the nucleus, where intensive colocalization with PCNA
was observed. These data led to the hypothesis that CHL1 expression
levels may be associated with the proliferation level of glioma
cells. The correlation between CHL1 and PCNA on human gliomas of
each grade was then evaluated.
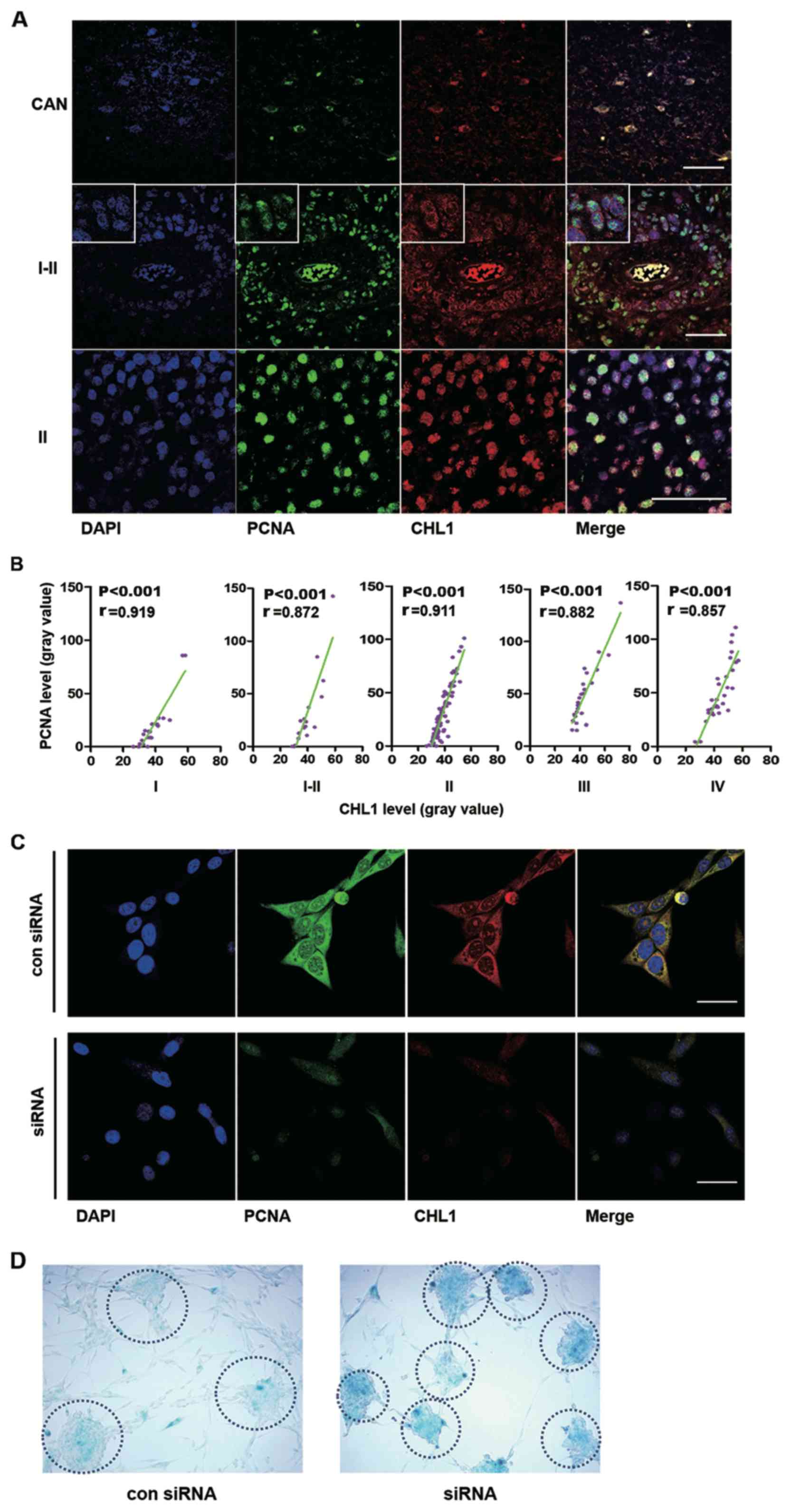 | Figure 2.CHL1 expression level is correlated
with PCNA expression levels in different grades of human gliomas,
and downregulation of CHL1 induces glioma cell senescence. (A)
Representative immunofluorescence staining of PCNA and CHL1 in CAN
and human glioma tissues (graded I–II and II). Scale bars, 50 µm.
(B) Correlation between CHL1 and PCNA expression levels (n=23, 18,
85, 27 and 11 for I, I–II, II, III and IV grades, respectively).
(C) Immunofluorescence staining of CHL1 and PCNA in U-87 MG cells
treated with either a control siRNA or an siRNA targeting CHL1 for
48 h. Downregulation of CHL1 resulted in the reduction of PCNA
expression levels. Scale bars, 25 µm. (D) Enhanced staining of
senescence-associated β-galactosidase was observed in cells with
CHL1 knockdown, suggesting that CHL1 may function by delaying the
senescence of glioblastoma cells. CHL1, cell adhesion molecule L1
like; PCNA, proliferation cell nuclear antigen; si, small
interfering; con, control. |
Correlations between CHL1 and PCNA expression levels
in the microarray data of human glioma tissues (n=23, 18, 85, 27
and 11 for grades I, I–II, II, III and IV, respectively) were then
analyzed using Pearson's correlation (Fig. 2B). R values derived from Pearson
correlation assay were 0.919, 0.872, 0.911, 0.882, and 0.857 in I,
I–II, II, III and IV-grade glioma tissues, respectively, suggesting
that CHL1 expression levels were highly positively correlated with
the expression of PCNA and that CHL1 may modulate the proliferation
of glioma cells, as indicated by PCNA. Immunofluorescence staining
of CHL1 and PCNA was undertaken in U-87 MG cells treated with
either control siRNA or siRNA targeting CHL1. The results
demonstrated that downregulation of CHL1 resulted in the reduction
of PCNA expression levels, suggesting that CHL1 regulates glioma
cell proliferation (Fig. 2C).
To investigate whether downregulation of CHL1 can
lead to cell senescence, senescence staining was conducted using a
β-galactosidase activity assay. Enhanced senescence was observed in
U-87 MG cells treated with an siRNA targeting CHL1. Compared with
the negative control siRNA, cells treated with an siRNA targeting
CHL1 demonstrated greater staining intensity by 48 h as shown in
the dotted circle, which is indicative of a higher β-galactosidase
activity and suggests that glioblastoma cells with less CHL1
activity are more prone to senescence (Fig. 2D).
Western blot analysis of CHL1 protein
expression levels in response to NRG1 in 3 human glioma cell
lines
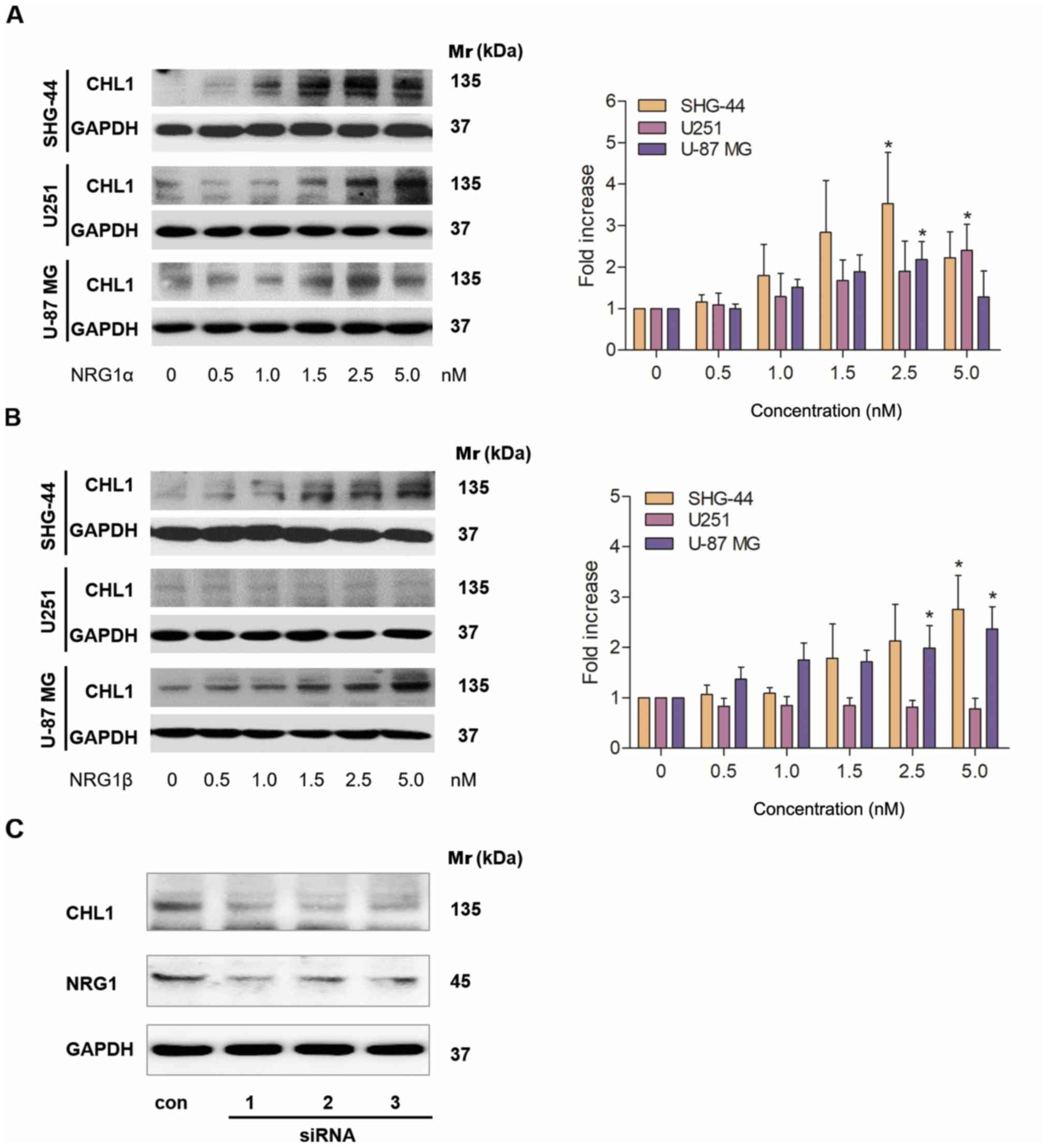
Different concentrations of NRG1α and β were added
to SHG-44 and U251 human glioma and U-87 MG human glioblastoma
cells, and western blotting was used to investigate the effects of
the different NRG1 isoforms on CHL1 protein expression levels. A
total of 48 h after the application, NRG1α treatment resulted in a
dose-dependent increase in the protein expression levels of CHL1 in
both SHG-44 and U251 cells, with the highest levels observed at 2.5
and 5 nM, respectively. In U-87 MG cells, CHL1 levels were
significantly increased in cells treated with 1.0–2.5 nM NRG1α,
with the highest level observed following 2.5 nM treatment
(Fig. 3A). Statistical significances
were found in SHG44 an U-87 MG cells treated with 2.5 nM of
NRG1αand in U251 cells treated with 5.0 nM of NRG1α (P<0.05;
Fig. 3A).
A total of 48 h after application, NRG1β treatment
resulted in a dose-dependent increase in the expression levels of
CHL1 in both SHG-44 and U-87 MG cells, with the highest level
observed at 5.0 nM (Fig. 3B). In
contrast, NRG1β showed no significant effects on CHL1 expression
levels at any of the concentrations (Fig. 3B).
Statistical significances were found in SHG44 cells
treated with both 2.5 and 5.0 nM of NRG1βand in U-87 MG cells
treated with 5.0 nM of NRG1β (P<0.05; Fig. 3B).
Subsequently U-87 MG glioblastoma cells, whose
malignancy is relatively high, were treated with a control siRNA
and 3 individual NRG1 siRNAs for 48 h. The siRNA results
demonstrated that all siRNAs targeting NRG1 markedly reduced the
protein level of NRG1, which was accompanied by a reduction in CHL1
(Fig. 3C). Since all the three
siRNAs worked in reducing the targeted protein NRG1, no statistical
analysis was applied.
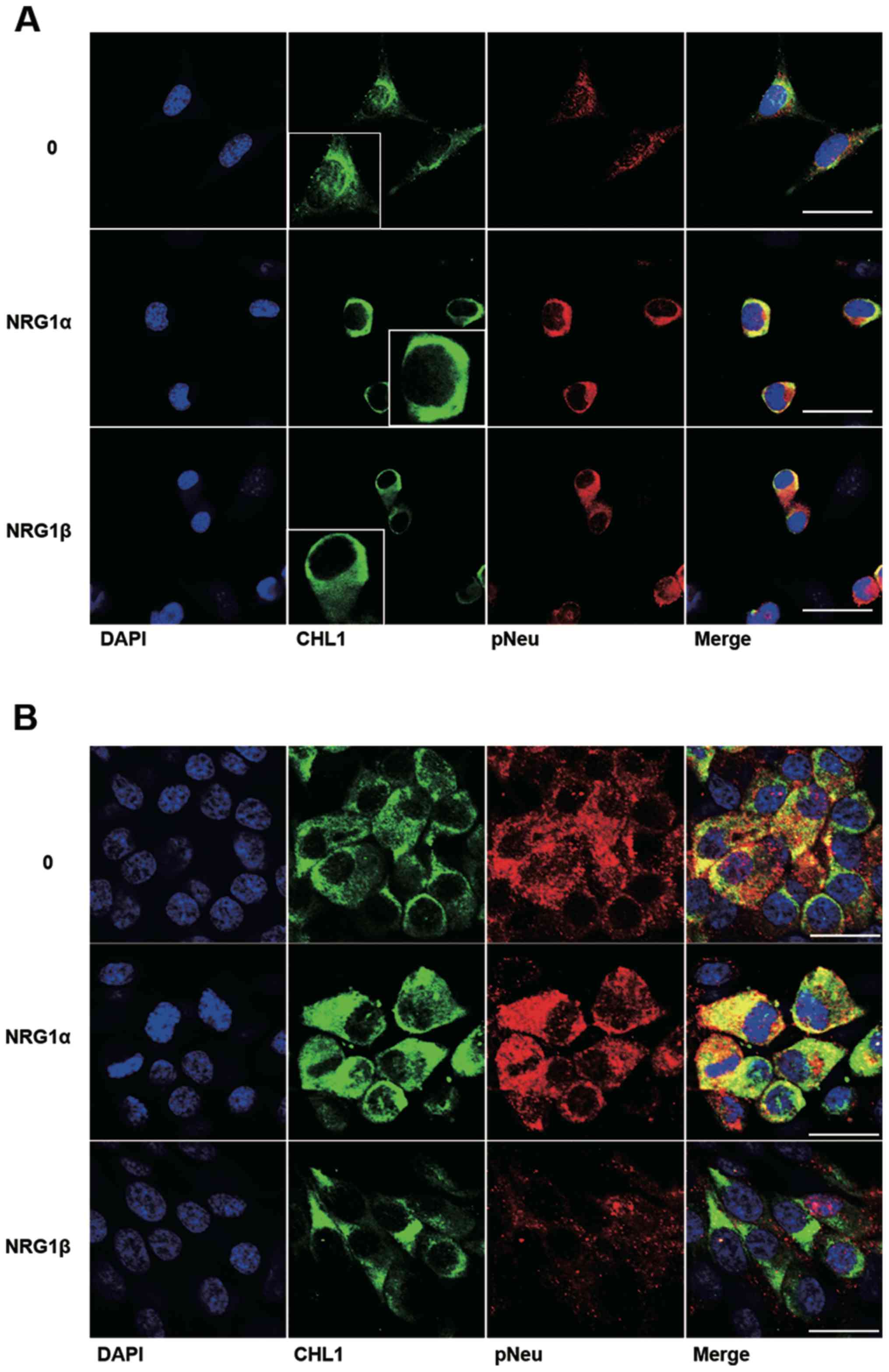
Colocalization of CHL1 and p-Neu in
U251 human glioma and U-87 MG human glioblastoma cells treated with
NRG1α and NRG1β
Among the ErbB receptors for NRG1, NEU is a commonly
shared receptor that can form heterodimers with either ErbB3 or
ErbB4 (14). The colocalization of
CHL1 and p-Neu was further investigated as a result of NRG1
activation in both U251 human glioma cells and U-87 MG human
glioblastoma cells. The cell lines were treated with both NRG1s at
2.5 nM, a concentration at which NRG1s can sufficiently work in
both cell lines. The immunofluorescence staining results
demonstrate that NRG1α increased the protein level of CHL1 in the
cytoplasm and at the plasma membrane of U251 cells. Consistent with
the western blot results, NRG1β failed to increase the expression
levels of CHL1 at 2.5 nM, although increased activation of p-Neu
was detected in U251 cells (Fig.
4A). The immunofluorescence staining results demonstrate that
both NRG1α and NRG1β increased CHL1 expression levels in the
cytoplasm and at the plasma membrane of U-87 MG cells. However, the
fluorescence signaling of p-Neu was not increased following
treatment with NRG1β at 2.5 nM (Fig.
4B).
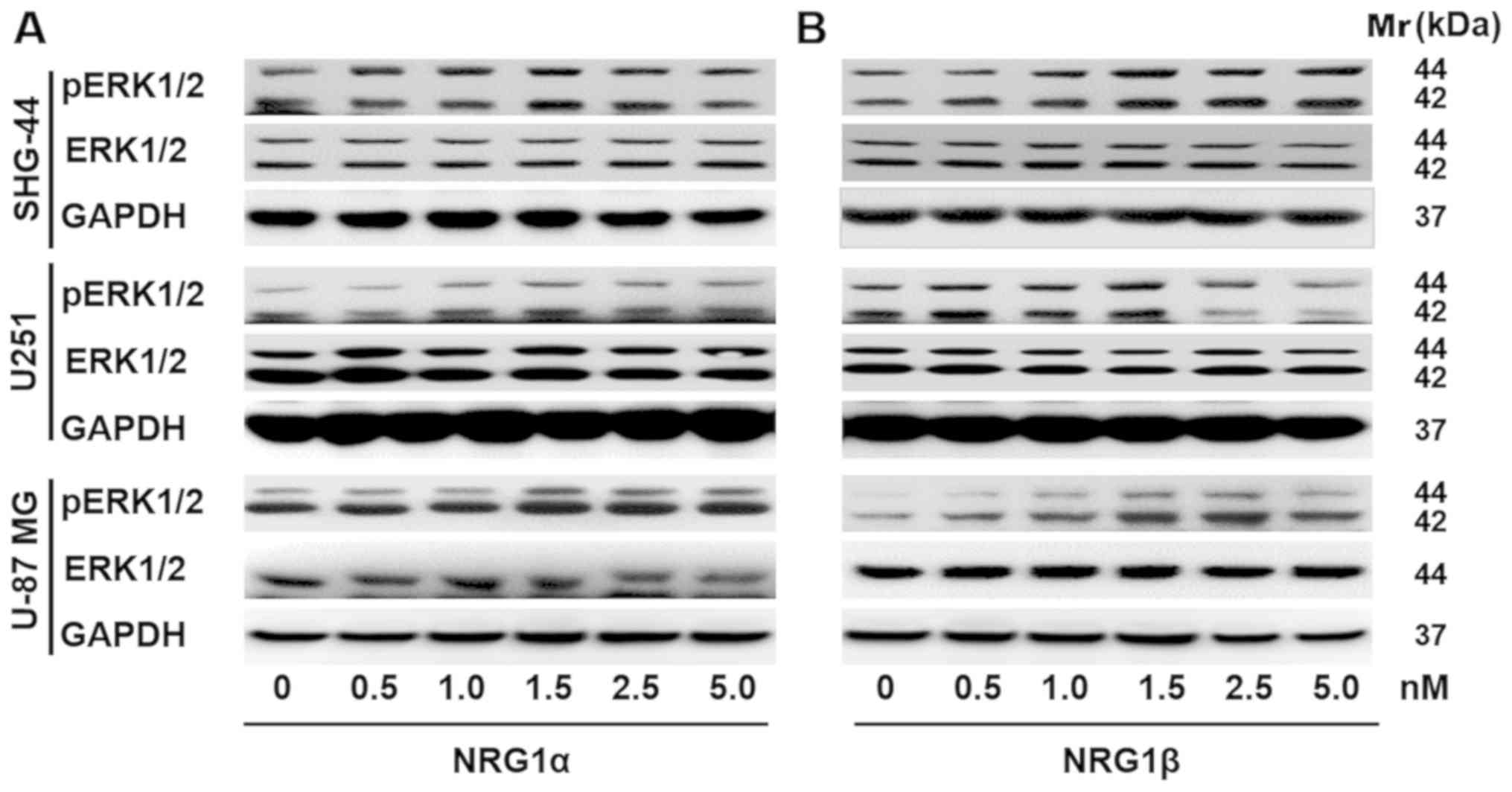
Effects of NRG1 on ERK1/2 signaling in
human glioma cells
Accumulating evidence has indicated that the
Ras/MAPK/ERK signaling pathways contribute to cell growth,
proliferation and survival (1).
Thus, western blot analysis was performed to assess p-ERK1/2
expression levels in U251, SHG-44 and U-87 MG cells in response to
treatment with NRG1s. It was demonstrated that p-ERK1/2 protein
expression levels were upregulated in U251 and U-87 MG human
glioma/glioblastoma cells in response to 48 h of rNRG1α treatment,
and that the response was dose-dependent; however, rNRG1α showed no
effect on the protein expression levels of p-ERK1/2 in the SHG-44
human glioma cell line (Fig. 5A).
Similarly, rNRG1β increased p-ERK1/2 expression levels in a
dose-dependent manner in SHG-44, U251 and U-87 MG cells. The
p-ERK1/2 expression levels peaked when the rNRG1β concentration was
1.5 nM and then p-ERK1/2 expression levels gradually returned to
the baseline level (Fig. 5B).
Discussion
Previous studies have demonstrated that both NRG1
and CHL1 serve roles in the development, proliferation and
metastasis of glioma cells (15,24).
However, the association between NRG1 signaling and CHL1 in these
processes has not been fully elucidated. The present study
demonstrated that, for the first time, the tumor-associated
cytokine NRG1 can regulate CHL1 expression levels in gliomas and
that this is associated with glioblastoma grade. In addition, these
data suggest that NRG1 may function in this underlying mechanism,
contributing to malignancy by upregulating CHL1 expression levels
in glioblastoma cells.
CHL1 is important for the normal development of the
visual and somatosensory cortex (25). A study in CHL1-deficient mice
revealed that CHL1 can facilitate the radial migration of neuronal
precursors to suitable cortical lamellas in deep-layered pyramid
neurons (25). In the adult brain,
CHL1 functions as a key regulator of synapse formation and synaptic
activity in the maintenance and rehabilitation of neural circuits
(26). CHL1 is also associated with
the metastasis and invasion of malignant tumors, such as glioma,
melanoma, ovarian cancer, colon carcinoma and breast cancer
(21). However, the roles of CHL1 in
tumor development and progression remain unclear. In previous
research, CHL1 was found to function as a tumor suppressor during
primary tumor growth (27) and it
was reported that CHL1 expression levels were decreased in human
breast cancer with a low malignancy grade (27), whereas downregulation of CHL1
expression levels by the targeting of microRNA (miR)-10a enhanced
the migration and invasion properties of human cervical cancer
cells (20). In contrast to these
diverse findings regarding CHL1 in most types of cancer, our
previous research revealed that CHL1 functions in promoting cell
proliferation, metastasis and migration of human glioma cells both
in vitro and in vivo, suggesting a positive
association between CHL1 level and glioma grade (28).
The present results from the glioma tissue
microarray staining demonstrated that CHL1 is highly expressed in
glioma tissues of each grade, with the highest expression level
detected in gliomas graded II. As indicated by correlation analysis
between CHL1 and PCNA, CHL1 was highly correlated with glioma cell
proliferation in grade I–II gliomas, which may facilitate the
determination of the grade of glioma malignancy. Delayed cellular
aging-related uncontrolled proliferation has been a major
drug-resistance factor in the treatment of glioma (29,30). The
extent of cellular senescence can be determined by the levels of
lysosomal-β-galactosidase activity (31). In the present study, glioma cells
treated with CHL1 siRNA exhibited significant signs of senescence,
suggesting that CHL1 may promote glioma growth by inhibiting cell
senescence; thus, CHL1 is associated with glioma malignancy
grade.
Both isoforms of NRG1 protein (α and β) contain
structurally distinct EGF-like domains that contain a common amino
terminal segment followed by α and β variant sequences,
respectively (14). The NRG1/ErbB
receptor interaction serves an important role in modulating the
development of human astrocytic glioma cells (15). Thus, the interaction between NRG1 and
its ErbB receptors may consequently enhance intercellular
communication through cell-cell or cell-extracellular matrix
interaction (22). In particular,
NRG1α induces marked proliferation in human transitional cell
carcinomas by binding to the ErbB4 receptor, indicating the
existence of an autocrine pathway (13,14).
NRG1α also stimulates cell proliferation mediated by multiple
matrix metalloproteases during tumor cell migration (10). Another study demonstrated that the
expression levels of NRG1β were significantly increased in melanoma
cell lines (32). These previous
studies indicated that both CHL1 and NRG1 are involved in the
invasion and reduced apoptosis of glioma cells.
Hyperactivation of the PI3K/AKT signaling pathway is
frequently reported in a variety of cancers, including glioblastoma
multiforme, where the PI3K/AKT signaling pathway regulates tumor
cell survival, growth, motility, angiogenesis and metabolism
(33,34). Glioblastoma has been found to
overexpress mutated epidermal growth factor receptor, which in turn
leads to the activation of several downstream signaling pathways,
such as the phosphatidylinositol PI3K/AKT/mammalian target of
rapamycin (mTOR) pathway (35).
Research on PI3K inhibitors, including pan-PI3K inhibitors,
isoform-selective inhibitors and dual PI3K/mTOR inhibitors has
provided valuable insight into the treatment of a variety of
malignancies, such as breast, prostate and lung cancers, and
mesothelioma, sarcoma and lymphoma (36). Furthermore, the combination of
inhibitors targeting PI3K and the PI3K/AKT/mTOR signaling pathway
can contribute to the suppression of tumor growth and improve
prognosis (36). In a previous
study, it was reported that activation of Akt1 is reduced by CHL1
downregulation, while no significant effects were observed
regarding ERK1/2 activation, indicating that the effects of CHL1 on
the behavior of glioma are possibly mediated by the Akt1 signaling
pathway (28). Other research
revealed that NRG1β promotes survival of glioma cells, while
inhibiting apoptosis of glioma cells via the PI3K/AKT signaling
pathway (15,37).
Similar to the PI3K/AKT signaling pathway, the
Ras/MAPK/ERK signaling pathway is also involved in the
proliferation and survival of malignant glioma cells (33). miR-126 regulates the ERK pathway by
targeting KRAS to inhibit the proliferation and invasion of glioma
cells (38). Blocking the MAPK/ERK
signaling pathway can inhibit the progression of glioma (39). Our previous study demonstrated a
positive association between the expression levels of CHL1 and Akt1
activation with no significant effect of CHL1 on ERK1/2 activation
(28). In the present study, both
NRG1α and β isoforms were reported to activate the ERK signaling
pathway. Specifically, NRG1α participates in the activation of the
ERK signaling pathway in U251 and U-87 MG glioma cells, but not in
SHG-44 human glioma cells. NRG1β activates the ERK signaling
pathway in U-87 MG and SHG-44 human glioma/glioblastoma cells, but
showed no observable effect in U251 human glioma cells. NRG1α and
NRG1β may act synergistically or compensate for each other to
increase the level of survival, proliferation, invasion and
metastasis via CHL1 enhancement.
In summary, the results of the present study
suggested that NRG1 modulates the growth and proliferation of
glioma cells, and may contribute to the malignancy of glioma, as an
upstream signaling molecule of CHL1. NRG1 may function by enhancing
CHL1 via the ERK1/2 signaling pathway. The variable effects of the
isoforms of NRG1 on the 3 different glioma cell lines may be
ascribed to variation in the expression levels of molecules that
interact with NRG1, a characteristic that currently complicates
glioma/glioblastoma treatment. Further investigation may provide
insights into the development of inhibitors to antagonize
NRG1-induced CHL1 expression in the clinical management of
glioma.
Acknowledgements
The authors would like to thank Mr. Huifan Shen from
the Center for Neuroscience at Shantou University Medical College
(Shantou, China) for his assistance in the maintenance of the cell
culture room.
Funding
The present study was supported by The National
Natural Science Foundation of China (grant nos. 81471279 and
81171138) and The Research Start-Up Fund of Wuxi School of
Medicine, Jiangnan University (grant no. 1286010242190060).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WJZ designed the study and drafted the initial
manuscript. WJZ and WWL collated the data. WWL, GYO, JZL, SJY, HCP
and WCY analyzed the data and wrote the initial draft of the
manuscript. WJZ and HCP critically revised the manuscript for
important intellectual content. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sathornsumetee S, Reardon DA, Desjardins
A, Quinn JA, Vredenburgh JJ and Rich JN: Molecularly targeted
therapy for malignant glioma. Cancer. 110:13–24. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Davis ME: Glioblastoma: Overview of
disease and treatment. Clin J Oncol Nurs. 20 (5 Suppl):S2–S8. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stupp R, Hegi ME, Mason WP, van den Bent
MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B,
Belanger K, et al: Effects of radiotherapy with concomitant and
adjuvant temozolomide versus radiotherapy alone on survival in
glioblastoma in a randomised phase III study: 5-year analysis of
the EORTC-NCIC trial. Lancet Oncol. 10:459–466. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ostrom QT, Bauchet L, Davis FG, Deltour I,
Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA, Turner MC, Walsh
KM, et al: The epidemiology of glioma in adults: A ‘state of the
science’ review. Neuro Oncol. 16:896–913. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Morgan MA and Canman CE: Replication
stress: An achilles' heel of glioma cancer stem-like cells. Cancer
Res. 78:6713–6716. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Weller M, van den Bent M, Tonn JC, Stupp
R, Preusser M, Cohen-Jonathan-Moyal E, Henriksson R, Rhun EL,
Balana C, Chinot O, et al: European association for neuro-oncology
(EANO) guideline on the diagnosis and treatment of adult astrocytic
and oligodendroglial gliomas. Lancet Oncol. 18:e315–e329. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nilsson P, Gedda L, Sjöström A and
Carlsson J: Effects of dextranation on the uptake of peptides in
micrometastases: Studies on binding of EGF in tumor spheroids.
Tumour Biol. 22:229–238. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Thuringer D, Hammann A, Benikhlef N,
Fourmaux E, Bouchot A, Wettstein G, Solary E and Garrido C:
Transactivation of the epidermal growth factor receptor by heat
shock protein 90 via Toll-like receptor 4 contributes to the
migration of glioblastoma cells. J Biol Chem. 286:3418–3428. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
O-charoenrat P, Rhys-Evans P, Court WJ,
Box GM and Eccles SA: Differential modulation of proliferation,
matrix metalloproteinase expression and invasion of human head and
neck squamous carcinoma cells by c-ErbB ligands. Clin Exp
Metastasis. 17:631–639. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Forster JA, Paul AB, Harnden P and Knowles
MA: Expression of NRG1 and its receptors in human bladder cancer.
Br J Cancer. 104:1135–1143. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cheng H, Terai M, Kageyama K, Ozaki S,
McCue PA, Sato T and Aplin AE: Paracrine effect of NRG1 and HGF
drives resistance to MEK inhibitors in metastatic uveal melanoma.
Cancer Res. 75:2737–2748. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yarden Y and Sliwkowski MX: Untangling the
ErbB signalling network. Nat Rev Mol Cell Biol. 2:127–137. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Buonanno A and Fischbach GD: Neuregulin
and ErbB receptor signaling pathways in the nervous system. Curr
Opin Neurobiol. 11:287–296. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ritch PS, Carroll SL and Sontheimer H:
Neuregulin-1 enhances survival of human astrocytic glioma cells.
Glia. 51:217–228. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Senchenko VN, Krasnov GS, Dmitriev AA,
Kudryavtseva AV, Anedchenko EA, Braga EA, Pronina IV, Kondratieva
TT, Ivanov SV, Zabarovsky ER and Lerman MI: Differential expression
of CHL1 gene during development of major human cancers. PLoS One.
6:e156122011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schmid RS and Maness PF: L1 and NCAM
adhesion molecules as signaling coreceptors in neuronal migration
and process outgrowth. Curr Opin Neurobiol. 18:245–250. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Panicker AK, Buhusi M, Thelen K and Maness
PF: Cellular signalling mechanisms of neural cell adhesion
molecules. Front Biosci. 8:d900–d911. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sasaki H, Yoshida K, Ikeda E, Asou H,
Inaba M, Otani M and Kawase T: Expression of the neural cell
adhesion molecule in astrocytic tumors: An inverse correlation with
malignancy. Cancer. 82:1921–1931. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Long MJ, Wu FX, Li P, Liu M, Li X and Tang
H: MicroRNA-10a targets CHL1 and promotes cell growth, migration
and invasion in human cervical cancer cells. Cancer Lett.
324:186–196. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu Y, Yu Y, Schachner M and Zhao W:
Neuregulin 1-β regulates cell adhesion molecule L1 expression in
the cortex and hippocampus of mice. Biochem Biophys Res Commun.
441:7–12. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao WJ and Schachner M: Neuregulin 1
enhances cell adhesion molecule l1 expression in human glioma cells
and promotes their migration as a function of malignancy. J
Neuropathol Exp Neurol. 72:244–255. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella- Branger D, Cavenee WK, Ohgaki H, Wiestler
OD, Kleihues P and Ellison DW: The 2016 world health organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ritch PA, Carroll SL and Sontheimer H:
Neuregulin-1 enhances motility and migration of human astrocytic
glioma cells. J Biol Chem. 278:20971–20978. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Demyanenko GP, Schachner M, Anton E,
Schmid R, Feng G, Sanes J and Maness PF: Close homolog of L1
modulates area-specific neuronal positioning and dendrite
orientation in the cerebral cortex. Neuron. 44:423–437. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sytnyk V, Leshchyns'ka I and Schachner M:
Neural cell adhesion molecules of the immunoglobulin superfamily
regulate synapse formation, maintenance, and function. Trends
Neurosci. 40:295–308. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
He LH, Ma Q, Shi YH, Ge J, Zhao HM, Li SF
and Tong ZS: CHL1 is involved in human breast tumorigenesis and
progression. Biochem Biophys Res Commun. 438:433–438. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang Z, Xie Q, Hu CL, Jiang Q, Shen HF,
Schachner M and Zhao WJ: CHL1 is expressed and functions as a
malignancy promoter in glioma cells. Front Mol Neurosci.
10:3242017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fan HC, Chen CM, Chi CS, Tsai JD, Chiang
KL, Chang YK, Lin SZ and Harn HJ: Targeting telomerase and
ATRX/DAXX inducing tumor senescence and apoptosis in the malignant
glioma. Int J Mol Sci. 20(pii): E2002019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Debacq-Chainiaux F, Erusalimsky JD,
Campisi J and Toussaint O: Protocols to detect
senescence-associated beta-galactosidase (SA-beta gal) activity, a
biomarker of senescent cells in culture and in vivo. Nat Protoc.
4:1798–1806. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Alver TN, Lavelle TJ, Longva AS, Øy GF,
Hovig E and Bøe SL: MITF depletion elevates expression levels of
ERBB3 receptor and its cognate ligand NRG1-beta in melanoma.
Oncotarget. 7:55128–55140. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Asati V, Mahapatra DK and Bharti SK:
PI3K/Akt/mTOR and Ras/Raf/MEK/ERK signaling pathways inhibitors as
anticancer agents: Structural and pharmacological perspectives. Eur
J Med Chem. 109:314–341. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao HF, Wang J, Shao W, Wu CP, Chen ZP,
To ST and Li WP: Recent advances in the use of PI3K inhibitors for
glioblastoma multiforme: Current preclinical and clinical
development. Mol Cancer. 16:1002017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li X, Wu C, Chen N, Gu H, Yen A, Cao L,
Wang E and Wang L: PI3K/Akt/mTOR signaling pathway and targeted
therapy for glioblastoma. Oncotarget. 7:33440–33450.
2016.PubMed/NCBI
|
|
36
|
Rodon J, Dienstmann R, Serra V and
Tabernero J: Development of PI3K inhibitors: Lessons learned from
early clinical trials. Nat Rev Clin Oncol. 10:143–153. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yin F, Zhang JN, Wang SW, Zhou CH, Zhao
MM, Fan WH, Fan M and Liu S: MiR-125a-3p regulates glioma apoptosis
and invasion by regulating Nrg1. PLoS One. 10:e01167592015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li Y, Li Y, Ge P and Ma C: MiR-126
regulates the ERK pathway via targeting KRAS to inhibit the glioma
cell proliferation and invasion. Mol Neurobiol. 54:137–145. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li B, Wang F, Liu N, Shen W and Huang T:
Astragaloside IV inhibits progression of glioma via blocking
MAPK/ERK signaling pathway. Biochem Biophys Res Commun. 491:98–103.
2017. View Article : Google Scholar : PubMed/NCBI
|