Introduction
Colorectal carcinoma (CRC) is the third most common
cancer type worldwide, and its prevalence is rapidly increasing in
the Republic of Korea with an annual increment of ~6.5% in males
and females (1). Despite the
decrease in the associated mortality rate and moderate advances in
treatment, CRC remains the third highest cause of cancer-associated
mortality in South Korea, with a 5-year overall survival (OS) rate
of 59% (2,3).
To date, molecular genetic studies have identified
several dominant somatic or germline genetic alterations associated
with an underlying predisposition to CRCs. There are three common
pathways responsible for sporadic CRCs: Chromosomal instability,
microsatellite instability and CpG island methylator phenotype
pathways (4). Most cases of CRCs
result from the chromosomal instability pathway, which is
characterized by the traditional adenoma-carcinoma sequence,
including early loss of the adenomatous polyposis coli gene,
KRAS point mutation, 18q loss of heterozygosity and late
tumor protein (TP)53 inactivation. KRAS mutation has a role
in the chromosomal instability pathway, mainly in the progression
from early to late advanced adenoma in ~30% of adenomas and 30-50%
of CRCs (5,6).
Ras family proteins are the prototypes of the small
guanosine triphosphatases (GTPases) and regulate multiple
intracellular processes, including growth, differentiation,
immunity and survival. The KRAS oncogene belongs to the Ras
family, which also includes HRAS and NRAS.
KRAS encodes a 21-kDa GTPase protein called K-Ras, which is,
under normal circumstances, temporarily activated as a response to
certain signals, including cytokines, hormones, growth factors and
external stimuli (7). However,
KRAS mutation leads to continuous activation of the
mitogen-activated protein kinase (MAPK) and PI3K/AKT signaling
pathways by the K-Ras protein, which may potentially stimulate
tumorigenesis or tumor progression. KRAS point mutation
frequently occurs at exon 2, particularly in codons 12 and 13.
Clinically, KRAS is a well-established biomarker of
resistance to anti-epidermal growth factor receptor (EGFR) antibody
treatment in advanced CRCs (7).
However, the prognostic significance of KRAS mutation
remains controversial (8,9).
A number of studies have demonstrated an association
between certain molecular subtypes and histology of CRCs; for
instance, a previous study revealed that the majority of sessile
serrated adenomas harbor a B-Raf proto-oncogene serine/threonine
kinase (BRAF) mutation or a CpG island hypermethylation
(9). Microsatellite instability-high
CRCs frequently exhibit poorly differentiated and mucinous features
(10). Regarding KRAS
mutation, an association with well-to moderately differentiated
conventional adenocarcinoma histology has been suggested, but as
yet there is no consensus on this (8,11). Thus,
the present study investigated the prognostic value of
KRAS-mutation in CRCs and its association with histologic
features.
Materials and methods
Patient cohort
Using Systematized Nomenclature of Medicine-Clinical
Terms (SNOMED-CT) (12), 310
patients who had been diagnosed with adenocarcinoma of the colon or
rectum at various stages were selected, and KRAS mutation
testing was performed between January 2011 and December 2014 at the
Konkuk University Medical Center (KUMC; Seoul, Republic of Korea).
Clinicopathological data including age, sex, patient history and
reports of imaging, surgery and pathology, were obtained from the
electronic medical records. Based on the patients' age, they were
divided into 2 groups according to a study by Yang et al
(13): Those under 60 years and
those ≥60 years. Based on the tumor location, the patients were
stratified into the following subgroups: ‘Right-sided’ (from the
cecum to the transverse colon) and ‘left-sided’ (from the
descending colon to the rectum). According to tumor size, the
patients were also divided into 2 groups, <5 cm and ≥5 cm, based
on previous studies (14–16).
Hematoxylin and eosin (H&E) stained slides were
reviewed for 267/310 patients who underwent surgical resection for
CRC. For survival analyses, 22/267 patients were excluded due to
inter-hospital transfer during the follow-up period. The median
follow-up period of the 245 patients was 37.4 months (range,
1.0–60.0 months).
KRAS mutation analysis
All DNA extraction and pyrosequencing were performed
according to methods routinely used at KUMC (17). In all cases, tumor-rich areas
detected on microscopic examination were marked on the
formalin-fixed, paraffin-embedded tissue slides by a pathologist
(HSL). After removing the cover glass, tumor cells were scraped
using a 26-gauge needle and 50–100 µl of DNA extraction buffer
solution [including 50 mM Tris buffer, pH 8.3; 1 mM EDTA, pH 8.0;
5% Tween-20 (Sigma-Aldrich; Merck KGaA, Germany), 200 µg/ml
proteinase K; and 10% resin] was added to the scraped cells. After
incubation for at least 1 h at 56°C, each tube was heated for 20
min at 100°C, followed by centrifugation at 4°C for 10 min at
13,000 × g to pellet the debris. The recovered supernatant was used
for PCR. The amount of genomic DNA was spectrophotometrically
determined using a Qubit assay kit 3.0 (Thermo Fisher Scientific,
Inc.).
PCR primer sequences used for the amplification of
KRAS gene were as follows: Codons 12 and 13,
5′-CTGGTGGAGTATTTGATAGTGTA-3′ (forward) and
5′-biotin-TGGTCCTGCACCAGTAATAT-3′ (reverse); and codon 61,
5′-biotin-TCCAGACTGTGTTTCTCCCTTC-3′ (forward) and
5′-TACTGGTCCCTCATTGCACTGT-3′ (reverse). A total of 10–20 ng/µl of
DNA were added to each 50 µl of PCR solution mixture [0.2 mmol each
of deoxynucleoside triphosphate, 1.5 mmol/l MgCl2, 1X
PCR buffer, 1.5 U of Immolase™ DNA polymerase (Bioline) and 20 pmol
of each primer]. PCR was performed with an initial denaturation for
5 min at 95°C followed by 40 cycles of 30 sec at 95°C, annealing of
the primers for codons 12, 13 and 61 for 30 sec at 55°C, primer
extension for 30 sec at 72°C, and a final incubation for 10 min at
72°C. Electrophoresis of the PCR products was performed in an
agarose gel to confirm amplification. Immobilization of
biotinylated PCR products onto streptavidin-coated beads (GE
Healthcare) using the solution from the PSQ™ 96 sample Preparation
kit (Qiagen) was performed, according to the manufacturer's
instructions. Following 20-fold dilution of 3 µl beads in binding
buffer (37 µl) and distilled water (20 µl) with 10 µl biotinylated
PCR products, incubation for 10 min at room temperature was
performed. The beads were then transferred to a filter probe and
the liquid was removed by vacuum filtration. The DNAs were
separated in PyroMark denaturation solution (Qiagen GmbH) for 2 min
at room temperature. The templates were washed in PyroMark wash
buffer (Qiagen GmbH), transferred to a PSQ 96 single nucleotide
polymorphism (SNP) plate and then annealed with sequencing primers
5′-ATAAACTTGTGGTAGTTGG-3′ (codons 12 and 13) and
5′-CCTCATTGCACTGTAC-3′ (codon 61), in PyroMark annealing buffer
(Qiagen GmbH) at room temperature. Finally, pyrosequencing was
performed using the PyroMark Q96 ID system (Qiagen), with PyroMark
Gold Q96 Reagents (Qiagen GmbH), according to the manufacturer's
instructions.
Histological evaluation
One pathologist (HSL) reviewed all H&E and
immunohistochemistry slides according to the 2010 World Health
Organization (WHO) classification and American Joint Committee on
Cancer (AJCC) staging manual, 8th edition TNM staging (18,19).
Histological features were comprehensively evaluated for tumor
border, differentiation, degree of patterns (cribriform, serrated,
mucinous, signet ring cell, solid, papillary and micropapillary),
degree of inflammatory reactions associated with CRCs [Crohn's-like
lymphoid reaction, neutrophilic infiltration and tumor-infiltrating
lymphocytes (TILs)], dirty necrosis, tumor budding counts,
lymphatic invasion, vascular invasion and perineural invasion.
Specifically, the microscopic tumor border was
divided into two groups according to the fraction of the
infiltrative border area: <50 and ≥50%. Tumor differentiation
was graded into three groups according to the 2010 WHO
classification: Well, moderately and poorly differentiated. Each
histological feature was divided into two or three categories,
depending on the degree of each pattern: Cribriform (absent, mild
or moderate), serrated (absent, mild or moderate), signet ring cell
(absent or present), solid (absent or present), papillary (absent
or present) and micropapillary (absent or present).
Mucinous components were divided into two groups by
the quality of mucin (low or high grade) (20). The Q score for mucinous components
was calculated by multiplying the quality of mucin by the
proportion of the area. Crohn's-like lymphoid reaction was defined
as the degree of peritumoral lymphoid aggregate ≥1 mm (absent to
mild or moderate) (21).
Intraluminal necrotic debris (dirty necrosis) was graded into five
degrees: Absent, low, moderate, high or confluent. Neutrophil
infiltration was graded into four degrees: Absent, small numbers
and scattered in the stroma, focal abscesses within tumor glands or
numerous abscesses disrupting tumor glands (22). The density of TILs at the hot spot
area of the tumor border was evaluated at a magnification of ×20
and subdivided into two groups: <50 and ≥50% (23). The tumor budding count was
determined, and cases were subdivided into low, intermediate and
high groups, according to the recommendations of the International
Tumor Budding Consensus Conference in 2016 (24). The presence of lymphatic, vascular
and perineural invasion was also re-evaluated. For TP53,
expression in at least 50% of the tumor nuclei was regarded as
positive, according to previous studies (25,26).
Statistical analysis
Statistical analysis was performed using SPSS
version 19 (IBM, Corp.). Comparison of clinicopathological factors
between patients with KRAS mutation and wild type
KRAS was performed using Pearson's χ2 test
(n=310). Histological findings between patients with KRAS
mutation and wild type KRAS were also compared using
Pearson's χ2 test (n=267). In survival analyses (n=245),
the OS rate was calculated as the rate of survivors from the onset
of typical CRC symptoms, including bowel habit changes, bloody
stool, continuously localized abdominal pain or anemia until the
date of the last clinical follow-up. The DFS rate was calculated
based on the date of symptom onset until the date of the detection
of local recurrence or metastasis by imaging observation. The Cox
proportional hazards model was used for univariate and multivariate
survival analyses to estimate the hazard ratios (HRs) for patients.
The results are presented as estimated HRs with 95% confidence
intervals (CIs) and Wald test P-values. The parameters used in the
multivariate analysis of 245 patients are as follows: KRAS
mutation, age, sex, grade of regression after neoadjuvant therapy,
tumor location, tumor size, T stage, N stage, M stage, total TNM
stage, TP53 expression, tumor border and differentiation,
and comprehensive histological features including lymphatic,
vascular, and perineural invasions. The parameters used in the
separate multivariate analysis in each ‘KRAS-mutated’ (n=91)
and ‘wild type’ (n=154) subgroup are as follows: Age, sex, tumor
location, tumor size, M stage, tumor border, tumor differentiation,
and comprehensive histological features except lymphovascular and
perineural invasions. The proportional hazards assumption was
assessed using graphical methods and tests based on Schoenfeld
residuals. The Kaplan-Meier method and the log-rank test was used
to estimate the OS curves. A two-tailed P-value was used for all
analyses, with P<0.05 considered to indicate statistical
significance.
Results
Patient characteristics and KRAS
mutation subtypes
The clinicopathological characteristics stratified
by KRAS mutation status are presented in Table I. Of all patients, 195 (62.9%) had
wild type KRAS and 115 (37.1%) had
mutations of KRAS. There were 176 males and 134 females,
with a median age of 62 years (age range, 27–88 years). The samples
were predominantly acquired from the primary tumor sites (n=298,
96.1%). A total of 28 patients (9.0%) received neoadjuvant therapy
and exhibited minimal to near complete regression. A total of 34
patients (11.0%) received adjuvant anti-EGFR therapy (cetuximab) in
combination with irinotecan (n=14, 41.2%), folinic acid +
fluorouracil + irinotecan (FOLFIRI; n=19, 55.9%) or folinic acid +
fluorouracil + oxaliplatin (FOLFOX; n=1, 2.9%). The mean tumor size
was 4.83 cm (range, 1–13 cm) and the number of samples with ≥5 cm
was 140 (45.3%). The initial distribution of TNM stages in the
cohort was as follows: 0 (1.6%), I (9.4%), II (14.2%), III (28.1%)
and IV (46.8%). There were no significant differences in age, sex,
acquisition site, grade of regression after neoadjuvant therapy,
tumor size, T stage, N stage, M stage, and total TNM stage between
the groups with different KRAS mutation status. Generally,
the tumors were in the left colon in both KRAS-mutated and
wild type groups. However, KRAS-mutated CRCs were more
likely to be in the right colon than wild type (P=0.014). In
addition, absence of TP53 expression was apparent in only
wild type CRCs (P=0.038).
 | Table I.Clinicopathological features of 310
patients with primary colorectal cancer stratified by KRAS
mutation. |
Table I.
Clinicopathological features of 310
patients with primary colorectal cancer stratified by KRAS
mutation.
|
|
| KRAS
mutation status |
|
|---|
|
|
|
|
|
|---|
| Characteristic | Total (n=310) | Wild type (n=195,
62.9%) | Mutated (n=115,
37.1%) | P-value |
|---|
| Sex |
|
|
| 0.587 |
|
Female | 134 (43.2) | 82 (42.1) | 52 (45.2) |
|
|
Male | 176 (56.8) | 113 (57.9) | 63 (54.8) |
|
| Age (years) |
|
|
| 0.281 |
|
<60 | 136 (43.9) | 81 (41.5) | 55 (47.8) |
|
|
≥60 | 174 (56.1) | 114 (58.5) | 60 (52.2) |
|
| Acquisition
site |
|
|
| 0.738 |
|
Primary | 298 (96.1) | 188 (96.4) | 110 (95.7) |
|
|
Metastatic | 12 (3.9) | 7 (3.6) | 5 (4.3) |
|
| Grade of regression
after neoadjuvant therapy |
|
|
| 0.778 |
| Not
received | 282 (91.0) | 178 (91.3) | 104 (90.4) |
|
|
Minimal | 16 (5.2) | 11 (5.6) | 5 (4.3) |
|
|
Moderate | 10 (3.2) | 5 (2.6) | 5 (4.3) |
|
| Near
complete | 2 (0.6) | 1 (0.5) | 1 (0.9) |
|
| Anti-EGFR
therapy |
|
|
| <0.001 |
| Not
received | 276 (89.0) | 161 (82.6) | 115 (100.0) |
|
| Cetuximab +
Irinotecan | 14 (4.5) | 14 (7.2) | 0 (0.0) |
|
| Cetuximab +
FOLFIRI | 19 (6.1) | 19 (9.7) | 0 (0.0) |
|
| Cetuximab +
FOLFOX | 1 (0.3) | 1 (0.5) | 0 (0.0) |
|
| Tumor location |
|
|
| 0.014 |
|
Right-sided | 78 (25.2) | 40 (20.5) | 38 (33.0) |
|
|
Left-sided | 232 (74.8) | 155 (79.5) | 77 (67.0) |
|
| Tumor size
(cm) |
|
|
| 0.463 |
|
<5 | 169 (54.7) | 103 (53.1) | 66 (57.4) |
|
| ≥5 | 140 (45.3) | 91 (46.9) | 49 (42.6) |
|
| T stage |
|
|
| 0.713 |
|
Tis | 5 (1.6) | 3 (1.5) | 2 (1.7) |
|
| T1 | 20 (6.5) | 13 (6.7) | 7 (6.1) |
|
| T2 | 48 (15.5) | 32 (16.4) | 16 (13.9) |
|
| T3 | 177 (57.1) | 114 (58.5) | 63 (54.8) |
|
| T4 | 60 (19.4) | 33 (16.9) | 27 (23.5) |
|
| N stage |
|
|
| 0.511 |
| N0 | 111 (35.8) | 71 (36.4) | 40 (34.8) |
|
| N1 | 96 (31.0) | 56 (28.7) | 40 (34.8) |
|
| N2 | 103 (33.2) | 68 (34.9) | 35 (30.4) |
|
| M stage |
|
|
| 0.645 |
| M0 | 167 (53.9) | 107 (54.9) | 60 (52.2) |
|
| M1 | 143 (46.1) | 88 (45.1) | 55 (47.8) |
|
| TNM stage |
|
|
| 0.692 |
| 0 | 5 (1.6) | 3 (1.5) | 2 (1.7) |
|
| I | 29 (9.4) | 18 (9.2) | 11 (9.6) |
|
| II | 44 (14.2) | 25 (12.8) | 19 (16.5) |
|
|
III | 87 (28.1) | 60 (30.8) | 27 (23.5) |
|
| IV | 145 (46.8) | 89 (45.6) | 56 (48.7) |
|
| TP53
expression |
|
|
| 0.038 |
|
Negative (<50%) | 221 (78.6) | 140 (79.5) | 81 (77.1) |
|
|
Positive (≥50%) | 8 (2.8) | 8 (4.5) | 0 (0.0) |
|
| Not
determined | 52 (18.5) | 28 (15.9) | 24 (22.9) |
|
The distribution of KRAS mutational changes
is presented in Table II. Most of
the mutations occurred in exon 2 (114/115, 99.1%), and among these,
mutations in codon 12 were more common compared with those in codon
13 (76.3% vs. 23.7%, respectively). The three most common amino
acid changes were G12D (n=44, 38.3%), G12V (n=26, 22.6%) and G13D
(n=24, 20.9%).
 | Table II.Distribution of KRAS mutation
variants in colorectal carcinoma (n=115). |
Table II.
Distribution of KRAS mutation
variants in colorectal carcinoma (n=115).
| A, Exon 2
(n=114) |
|---|
|
|---|
| Codon/position/base
change | Amino acid
change | n (%) |
|---|
| Codon 12
(n=87) |
|
Position 34 (n=12) |
|
|
|
G>T | G12C | 8 (7.0) |
|
G>A | G12S | 3 (2.6) |
|
G>C | G12R | 1 (0.9) |
|
Position 35 (n=75) |
|
|
|
G>A | G12D | 44 (38.3) |
|
G>T | G12V | 26 (22.6) |
|
G>C | G12A | 5 (4.3) |
| Codon 13
(n=27) |
|
|
|
Position 37 (n=3) |
|
|
|
G>T | G13C | 2 (1.7) |
|
G>C | G13R | 1 (0.9) |
|
Position 38 (n=24) |
|
|
|
G>A | G13D | 24 (20.9) |
|
| B, Exon 3
(n=1) |
|
|
Codon/position/base change | Amino acid
change | n (%) |
|
| Codon 61 |
|
|
|
Position 182 |
|
|
|
A>G | Q61R | 1 (0.9) |
Association of KRAS mutation with
survival
The results of univariate and multivariate survival
analyses in 245 patients are presented in Table SI (OS) and Table SII (DFS), respectively. The factors
for the multivariate analysis included KRAS mutation, age,
sex, tumor location, tumor size, TNM stage and comprehensive
histological features. Overall, KRAS mutation was an
unfavorable prognostic factor in terms of OS, according to the
univariate (P=0.023; HR, 1.593; 95% CI, 1.065–2.382) and
multivariate analyses (P=0.001; HR, 2.49; 95% CI, 1.427–4.343)
(Table SI). The cumulative OS rate
of patients with KRAS mutation using log-rank test was lower
compared with that of patients with wild type KRAS (P=0.021,
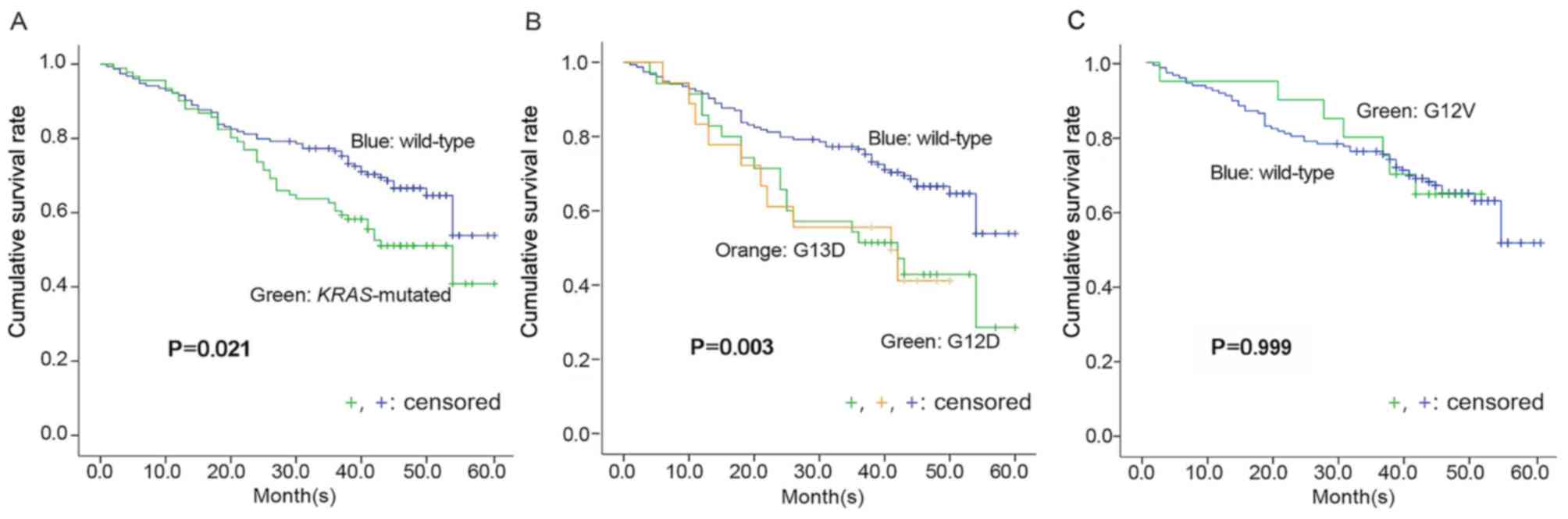
40.9 vs. 53.8%) (Fig. 1A). However,
the KRAS mutation status was not associated with DFS in
univariate (P=0.611; HR, 1.097; 95% CI, 0.769–1.564) and
multivariate (P=0.365; HR, 1.221; 95% CI, 0.793–1.878) analyses
(Table SII). Of the common mutation
types, patients with G12D and G13D mutations had significantly
lower OS rates compared with those with wild type KRAS
(P=0.003; Fig. 1B). However, there
was no significant difference between the OS curves for G12V and
wild type KRAS (P=0.999; Fig.
1C).
Histological features according to
KRAS mutation status
The cross-tabulation analysis results of the
comprehensive histological examination are summarized in Table III. For accurate morphological
analysis, only surgically resected specimens (n=267) were included.
In general, similar histological findings were revealed in patients
with mutations and those with wild type KRAS; however,
samples from those with mutations exhibited more prominent serrated
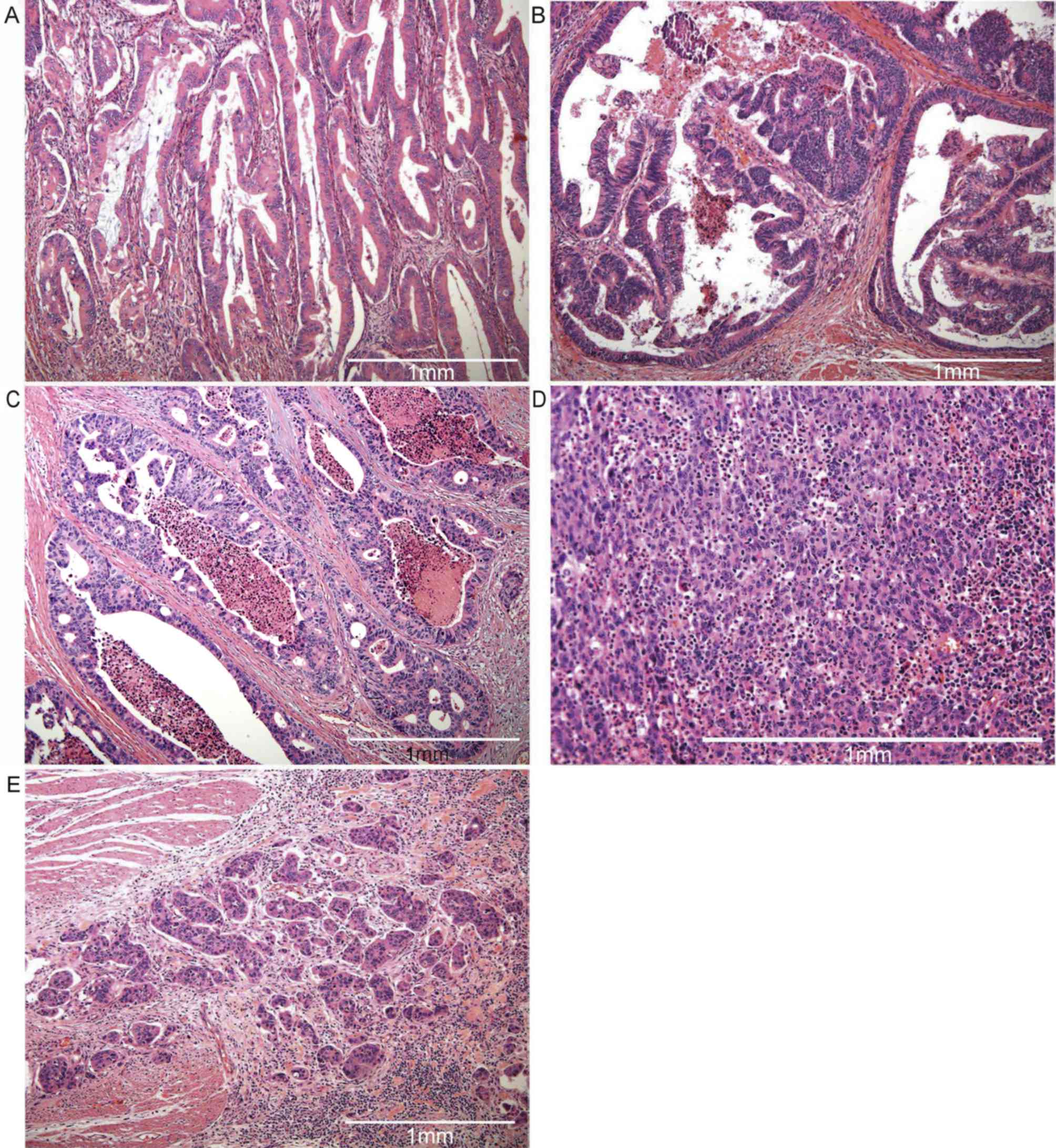
(Fig. 2A) and/or papillary (Fig. 2B) patterns (P=0.009 and P=0.014,
respectively).
 | Table III.Comparison of histologic findings
according to KRAS mutation status. |
Table III.
Comparison of histologic findings
according to KRAS mutation status.
|
|
| KRAS
mutation status |
|
|---|
|
|
|
|
|
|---|
| Morphologic
characteristic | Total (n=267) | Wild type | Mutated | P-value |
|---|
| Infiltrative tumor
border (%) |
|
|
| 0.932 |
|
<50 | 48 (18.0) | 31 (18.1) | 17 (17.7) |
|
|
≥50 | 219 (82.0) | 140 (81.9) | 79 (82.3) |
|
| Degree of
differentiation |
|
|
| 0.208 |
|
Well | 35 (13.1) | 20 (11.7) | 15 (15.6) |
|
|
Moderate | 208 (77.9) | 132 (77.2) | 76 (79.2) |
|
|
Poor | 24 (9.0) | 19 (11.1) | 5 (5.2) |
|
| Cribriform
pattern |
|
|
| 0.734 |
|
Absent | 35 (13.1) | 22 (12.9) | 13 (13.5) |
|
|
Mild | 172 (64.4) | 108 (63.2) | 64 (66.7) |
|
|
Moderate | 60 (22.5) | 41 (24.0) | 19 (19.8) |
|
| Serrated
pattern |
|
|
| 0.009 |
|
Absent | 104 (39.0) | 75 (43.9) | 29 (30.2) |
|
|
Mild | 142 (53.2) | 88 (51.5) | 54 (56.3) |
|
|
Moderate | 21 (7.9) | 8 (4.7) | 13 (13.5) |
|
| Quality of mucin
(n=66) |
|
|
| 0.190 |
|
Low | 50 (75.8) | 25 (69.4) | 25 (83.3) |
|
|
High | 16 (24.2) | 11 (30.6) | 5 (16.7) |
|
| Q score of mucin
(n=66) |
|
|
| 0.327 |
|
<60 | 42 (63.6) | 21 (58.3) | 21 (70.0) |
|
|
≥60 | 24 (36.4) | 15 (41.7) | 9 (30.0) |
|
| Signet ring
cells |
|
|
| 0.245 |
|
Absent | 253 (94.8) | 160 (93.6) | 93 (96.9) |
|
|
Present | 14 (5.2) | 11 (6.4) | 3 (3.1) |
|
| Solid component
(%) |
|
|
| 0.195 |
|
<50 | 208 (77.9) | 129 (75.4) | 79 (82.3) |
|
|
≥50 | 59 (22.1) | 42 (24.6) | 17 (17.7) |
|
| Papillary
component |
|
|
| 0.014 |
|
Absent | 149 (55.8) | 105 (61.4) | 44 (45.8) |
|
|
Present | 118 (44.2) | 66 (38.6) | 52 (54.2) |
|
| Micropapillary
component |
|
|
| 0.763 |
|
Absent | 220 (82.4) | 140 (81.9) | 80 (83.3) |
|
|
Present | 47 (17.6) | 31 (18.1) | 16 (16.7) |
|
| Crohn's-like
lymphoid reaction |
|
|
| 0.923 |
| Absent
to mild | 177 (66.3) | 113 (66.1) | 64 (66.7) |
|
|
Moderate | 90 (33.7) | 58 (33.9) | 32 (33.3) |
|
| Dirty necrosis |
|
|
| 0.345 |
|
Absent | 17 (6.4) | 11 (6.4) | 6 (6.3) |
|
|
Low | 51 (19.1) | 32 (18.7) | 19 (19.8) |
|
|
Moderate | 81 (30.3) | 51 (29.8) | 30 (31.3) |
|
|
High | 48 (18.0) | 26 (15.2) | 22 (22.9) |
|
|
Confluent | 70 (26.2) | 51 (29.8) | 19 (19.8) |
|
| Neutrophilic
infiltration |
|
|
| 0.055 |
|
Absent | 58 (21.7) | 45 (26.3) | 13 (13.5) |
|
| Low and
scattered | 88 (33.0) | 56 (32.7) | 32 (33.3) |
|
| Focal
abscesses | 65 (24.3) | 35 (20.5) | 30 (31.3) |
|
|
Numerous abscesses | 56 (21.0) | 35 (20.5) | 21 (21.9) |
|
| Tumor-infiltrating
lymphocytes (%) |
|
|
| 0.133 |
|
<50 | 194 (72.7) | 119 (69.6) | 75 (78.1) |
|
|
≥50 | 73 (27.3) | 52 (30.4) | 21 (21.9) |
|
| Tumor budding
grade |
|
|
| 0.872 |
|
Low | 150 (56.2) | 97 (56.7) | 53 (55.2) |
|
|
Intermediate | 73 (27.3) | 45 (26.3) | 28 (29.2) |
|
|
High | 44 (16.5) | 29 (17.0) | 15 (15.6) |
|
Influence of clinicopathological
features on OS according to KRAS mutation status
Further univariate and multivariate survival
analyses for OS were performed on the ‘KRAS-mutated’ and
‘wild type’ CRC subgroups.
On the basis of the univariate analysis of
‘KRAS-mutated’ subgroup (Table
SIII), tumor size ≥5 cm (P=0.030), high initial TNM stage
(P<0.001), presence of signet ring cells (P=0.021), absent to
mild Crohn's-like lymphoid reaction (P=0.020), absence of dirty
necrosis (P=0.021) and paucity of neutrophilic infiltration
(P=0.002) were associated with lower OS rate. However, in
multivariate analysis (Table IV),
those who were aged ≥60 years (P=0.023; HR, 3.058; 95% CI,
1.169–7.995), male (P=0.034; HR, 2.747; 95% CI, 1.079–6.995), M1
stage at diagnosis (P=0.029; HR, 5.608; 95% CI, 1.197–26.279) and
particularly the presence of solid component (representative
histology image, see Fig. 2C) were
associated with a lower OS rate (P=0.032; HR, 4.040; 95% CI,
1.127–14.488).
 | Table IV.Results of the multivariate analysis
of the influence of various factors on overall survival according
to KRAS mutation status. |
Table IV.
Results of the multivariate analysis
of the influence of various factors on overall survival according
to KRAS mutation status.
|
|
KRAS-mutated | Wild type |
|---|
|
|
|
|
|---|
| Characteristic | P-value | HR (95% CI) | P-value | HR (95% CI) |
|---|
| Age, years (≥60 vs.
<60) | 0.023 | 3.058
(1.169–7.995) | 0.111 | 1.914
(0.862–4.250) |
| Sex (Male vs.
Female) | 0.034 | 2.747
(1.079–6.995) | 0.123 | 0.544
(0.251–1.180) |
| Tumor location
(Left vs. Right) | 0.326 | 0.589
(0.205–1.694) | 0.106 | 0.450
(0.171–1.185) |
| Tumor size, cm (≥5
vs. <5) | 0.800 | 1.118
(0.471–2.655) | 0.695 | 0.855
(0.391–1.869) |
| M stage (M1 vs.
M0) | 0.029 | 5.608
(1.197–26.279) | <0.001 | 10.176
(3.488–29.686) |
| Infiltrative tumor
border, % (≥50 vs. <50) | 0.658 | 0.729
(0.180–2.956) | 0.575 | 1.880
(0.207–17.088) |
| Tumor
differentiation (Poor vs. Moderate/Well) | 0.644 | 0.660
(0.113–3.838) | 0.061 | 3.874
(1.106–8.574) |
| Cribriform
pattern | 0.606 |
| 0.059 |
|
| Mild
vs. Absent | 0.564 | 0.677
(0.180–2.549) | 0.906 | 0.945
(0.372–2.405) |
|
Moderate vs. Absent | 0.812 | 1.240
(0.211–7.298) | 0.040 | 0.186
(0.037–0.926) |
| Serrated
pattern | 0.242 |
| 0.402 |
|
| Mild
vs. Absent | 0.093 | 0.442
(0.171–1.145) | 0.392 | 0.675
(0.275–1.660) |
|
Moderate vs. Absent | 0.790 | 0.829
(0.208–3.297) | 0.201 | 0.358
(0.074–1.727) |
| Signet ring cells
(Present vs. Absent) | 0.284 | 4.886
(0.268–89.090) | 0.224 | 2.478
(0.574–10.708) |
| Solid component, %
(≥50 vs. <50) | 0.032 | 4.040
(1.127–14.488) | 0.086 | 0.424
(0.159–1.129) |
| Papillary component
(Present vs. Absent) | 0.852 | 1.096
(0.416–2.891) | 0.536 | 0.741
(0.286–1.918) |
| Micropapillary
component (Present vs. Absent) | 0.562 | 1.502
(0.379–5.956) | 0.018 | 2.908
(1.205–7.017) |
| Quality of
mucin | 0.795 |
| 0.251 |
|
|
Low-grade vs. Absent | 0.501 | 0.659
(0.195–2.222) | 0.563 | 1.422
(0.431–4.693) |
|
High-grade vs. Absent | 0.947 | 0.001
(0.000–0.000) | 0.252 | 0.352
(0.059–2.097) |
| Q score of mucin
(≥60 vs. <60) | 0.953 | 0.001
(0.000–0.000) | 0.357 | 2.237
(0.403–12.416) |
| Crohn's-like
lymphoid reaction (Moderate vs. Absent-Mild) | 0.107 | 0.358
(0.102–1.251) | 0.987 | 1.008
(0.395–2.574) |
| Dirty necrosis | 0.205 |
| 0.314 |
|
| Low vs.
Absent | 0.307 | 2.161
(0.493–9.470) | 0.151 | 0.468
(0.166–1.318) |
|
Moderate vs. Absent | 0.115 | 4.583
(0.689–30.476) | 0.151 | 0.374
(0.097–1.434) |
| High
vs. Absent | 0.061 | 5.055
(0.927–27.560) | 0.574 | 0.569
(0.080–4.063) |
|
Confluent vs. Absent | 0.656 | 1.684
(0.170–16.668) | 0.964 | 0.965
(0.209–4.451) |
| Neutrophilic
infiltration | 0.136 |
| 0.411 |
|
| Low vs.
Absent | 0.220 | 0.394
(0.089–1.744) | 0.680 | 1.306
(0.368–4.626) |
| Focal
vs. Absent | 0.753 | 0.797
(0.195–3.266) | 0.160 | 2.325
(0.717–7.540) |
|
Numerous vs. Absent | 0.048 | 0.153
(0.024–0.980) | 0.208 | 2.141
(0.655–7.003) |
| Tumor-infiltrating
lymphocytes, % (≥50 vs. <50) | 0.759 | 1.185
(0.402–3.492) | 0.942 | 1.036
(0.404–2.656) |
| Tumor budding
grade | 0.060 |
| 0.222 |
|
|
Intermediate vs. Low | 0.018 | 3.519
(1.243–9.964) | 0.108 | 2.219
(0.839–5.865) |
| High
vs. Low | 0.530 | 1.948
(0.243–15.583) | 0.194 | 2.347
(0.649–8.491) |
By contrast, in the ‘wild type’ subgroup,
right-sided tumor location (P=0.021), high initial TNM stage
(P<0.001), infiltrative tumor border ≥50% (P=0.046),
poorly-differentiated tumor (P=0.006), moderate cribriform pattern
(P=0.006), presence of signet ring cells (P=0.002), absence of
dirty necrosis (P=0.002), absence of neutrophilic infiltration
(P=0.041) and high tumor budding grade (P=0.005) were the
independent prognostic factors for worse OS rate in univariate
analysis (Table SIII). However, in
the multivariate analysis (Table
IV), initial M1 stage (P<0.001) and the presence of a
micropapillary component (representative histology image, see
Fig. 2D) were associated with a
lower OS rate (P=0.018; HR, 2.908; 95% CI, 1.205–7.017).
Influence of clinicopathological
features on DFS according to KRAS mutation status
Further univariate and multivariate survival
analyses for DFS were performed on the ‘KRAS-mutated’ and
‘wild type’ CRC subgroups.
On the basis of the univariate analysis in the
‘KRAS-mutated’ subgroup (Table
SIV), high initial TNM stage (P<0.001), mild or absence of
Crohn's-like lymphoid reaction (P=0.033) and paucity of
neutrophilic infiltration (P=0.004) were associated with lower DFS
rate. However, only initial M1 stage (P<0.001) exhibited
statistical significance following multivariate analysis (Table SV).
On the basis of univariate analysis in the ‘wild
type’ subgroup (Table SIV), high
initial TNM stage (P<0.001), presence of signet ring cells
(P=0.020), paucity of neutrophilic infiltration (P=0.027) and
TIL<50% (P=0.018) were independent prognostic factors for DFS.
However, only initial M1 stage (P<0.001) was associated with
lower DFS in multivariate analysis (Table SV).
Influence of clinicopathological
factors on OS
The results of univariate and multivariate analyses
of OS are presented in Table SI.
Right-sided tumor location (P=0.016), tumor size ≥5 cm (P=0.025),
high initial TNM stage (P<0.001), TP53 positivity
(P=0.005), high-grade tumor differentiation (P=0.006), a moderate
cribriform pattern (P=0.047), a signet ring cell pattern
(P<0.001), absence of Crohn's-like lymphoid reaction (P=0.009),
absence of dirty necrosis (P<0.001), paucity of neutrophilic
infiltration (P=0.003), a high tumor budding grade (P=0.050) and
presence of lymphatic or vascular or perineural invasion (P=0.001,
P<0.001, and P<0.001, respectively) were unfavorable
prognostic factors regarding OS, according to univariate analysis.
However, according to the multivariate analysis, age ≥60 (P=0.001),
right-sided tumor location (P=0.036), tumor size ≥5 cm, a high
initial TNM stage (P=0.001), a high tumor budding grade (P=0.021)
and vascular invasion (P=0.049) were associated with poor OS.
Influence of clinicopathological
factors on DFS
Local recurrence or distant metastasis occurred in
128 (47.8%) patients during the follow-up period, but there was no
statistical significance (30.692 vs. 35.013 months, respectively).
The cumulative DFS was 44.9% for the KRAS-mutated group and
49.1% for the wild type group (P=0.518). The results of univariate
and multivariate analyses between clinicopathological factors and
DFS are summarized in Table SII.
Tumor size ≥5 cm (P=0.025) and high initial TNM stage (P<0.001),
a signet ring cell pattern (P=0.006), absence of Crohn's-like
lymphoid reaction (P=0.038), less neutrophilic infiltration
(P=0.001), TIL <50% (P=0.013) and presence of lymphatic or
vascular or perineural invasions (P<0.001, P=0.001 and P=0.009,
respectively) were associated with a lower DFS rate based on
univariate analysis. However, only initial M1 stage (P<0.001)
reached significance in the multivariate analysis.
Discussion
In the present study, the prevalence of KRAS
mutation and common amino acid changes determined were consistent
with the results of previous studies (27,28). The
results of the present study support those of previous studies,
with a more frequent right-sided tumor location in patients with
mutations and rare simultaneous TP53 and KRAS
mutation (29,30). Of note, KRAS mutation was a
significant prognostic marker of OS with a 2.5-fold increased HR.
There has been controversy over the prognostic value of KRAS
mutation, but a recent review strengthens the evidence of a
negative clinical effect of the mutation in metastatic CRC
(P<0.001; HR, 1.674; 95% CI, 1.341–2.089) (8,31). In
addition, the cumulative OS rate of patients with KRAS
mutations was lower compared with that reported previously and this
difference may be due to low patient numbers or shorter
observational periods compared with those in the present study
(32–34).
Furthermore, G12D and G13D mutations resulted in a
worse OS rate compared with wild type KRAS (P=0.035). While
mutations of codons 12 or 13 have been widely studied in CRCs,
their impact on clinical action has been debated (35). Thus far, the prevailing view is that
codon 12 mutation results in poor clinical outcome, contrary to the
case with codon 13 (36,37). There is more evidence of decreased
sensitivity or reduced survival in patients with G12D and G13D
mutations (38,39). Different results were reported for
G12V, which has been indicated to result in lower DFS and OS rates
(32,37). However, further verification is
required, since, unlike the present study, most studies involved
only metastatic CRCs.
The prognostic role of KRAS mutation in DFS
has been debated. In particular, most studies, including those on
non-metastatic CRCs, concluded that KRAS mutations are
associated with poor DFS (40–43).
However, studies including metastatic CRCs indicated no association
between KRAS mutations and DFS (44–46).
Inoue et al (47) also
revealed that the KRAS genotype had no effect on DFS of
patients at stage IV, but G13D was a poor prognostic factor for DFS
of patients at stage I–III. Similarly, the present study revealed
no significant prognostic value of KRAS mutation for DFS,
including metastatic CRCs (stage 0-IV). However, stage 0-III CRCs
exhibited a trend of association between the KRAS-mutated genotype
and lower DFS (P=0.059), in accordance with the results of previous
studies (40–43).
The present study was performed on a specific ethnic
group as the cohort. Most of the studies from South Korea have
indicated that KRAS is not associated with either OS or DFS
(48,49) except for that by Lee et al
(42), according to which DFS was
shorter in stage II and III patients treated with FOLFOX. However,
the present results are in accordance with those obtained by
certain other studies in East Asian countries, which indicated an
association between KRAS mutation and a lower OS rate
(32,47). By contrast, KRAS was not
associated with prognosis in most studies from in Southwest Asia
and South America (34,50–52).
However, in Caucasians, the results were controversial. Studies
performed in Italy and Spain suggested shorter OS and DFS of
patients with KRAS mutation (53,54).
However, studies performed in Sweden and Australia indicated no
association between KRAS mutation and OS (8,55).
Furthermore, a study from Austria reported that KRAS-mutated
patients had better OS, except for those with G12V mutation
(56).
Of note, patients with KRAS mutation
presented with more serrated and/or papillary features compared
with those with wild type KRAS. A previous study also
demonstrated that KRAS is involved in the traditional
serrated pathway, in addition to the conventional pathway of the
pathogenesis of CRCs (57). However,
in the mutation group, the serrated feature itself was not
associated with prognosis (P=0.242) and this result was similar to
that in the whole population (P=0.329). The biological behavior of
CRCs with marked serrated features has been debated; however, their
prognostic impact appears to be defined by molecular traits rather
than by morphology (58,59). Taken together, KRAS mutation
may be involved in the morphogenesis of serration, but the
prognostic value of the serrated pattern in patients with mutations
remains uncertain.
Similarly, certain studies reported on CRCs with
papillary or villous features and designated them as papillary,
adenoma-like or villous adenocarcinoma (60–62).
These tumors were less likely to be associated with lymph node
metastasis, absence of aberrant p53 expression and frequent
KRAS mutation (62,63). However, the papillary architecture
was not associated with survival in patients with mutations and the
whole population (P=0.852 and P=0.135, respectively). Overall,
KRAS mutation has a moderate association with papillary
architecture but the prognostic effect of the predominant papillary
architecture in CRC requires further verification.
The present study also demonstrated the association
of solid architecture with a lower OS rate in the group with
KRAS mutation, whereas micropapillary features were
associated with a lower OS rate in the wild type group. A possible
explanation is that the solid pattern, by definition, includes
poorly differentiated tumors, resulting in a more accurate estimate
of high-grade differentiated tumors than the original diagnoses,
which are directed toward moderately differentiated tumors. For the
micropapillary feature, this has been associated with frequent
lymph node metastasis in numerous solid organ tumors, including
CRCs (64,65). However, the results of the present
study support the fact that KRAS mutation has a more
significant impact on prognosis compared with micropapillary
histology.
Most of the results of the survival analysis for
clinicopathological factors of the present study are in accordance
with those of previous studies (19,66,67). In
addition, in previous studies, a better OS has been indicated in
stage IV/KRAS wild type and anti-EGFR-treated subgroups
(68,69). A higher 3-year OS rate suggests that
anti-EGFR treatment may have a positive effect on short-term
survival.
However, there are certain differences between the
present results and those of previous studies. First, an
association of dirty necrosis or tumor necrosis with better OS is
not reported in the literature. While there have only been a few
studies that have directly investigated the role of dirty necrosis
on survival in CRC (70,71), a previous study by Pollheimer et
al (70) reasoned that tumor
necrosis reflects a hypoxic environment due to rapid proliferation
of the tumor and is therefore associated with a poor prognosis.
Väyrynen et al (71) also
reported that tumor necrosis was associated with high T stage,
vascular invasion and short DFS time in CRC, but the degree of
necrosis was not proportional to the Ki-67 proliferation index.
Overall, dirty necrosis is not merely an indicator of the tumor
growth rate but may also reflect intraluminal growth rather than
invasiveness. The second point is regarding neutrophilic
infiltration. In the present study, the paucity of neutrophilic
infiltration was associated with poor OS. Certain studies have
reported neutrophil infiltration as a favorable prognostic factor,
whilst other studies have proposed that the induction of immune
escape by intratumoral neutrophils results in stimulation of tumor
growth (72,73). The results of the present study
support an activated anti-tumor immune response induced by
intratumoral neutrophils rather than immune evasion.
There are certain limitations to this type of
retrospective study using previously processed material for
diagnostic purposes. First, only a small number of patients (2.6%,
8/310) were able to undergo BRAF testing, as the procedure
was not covered by their health insurance. BRAF, like
KRAS, is also known to cause constitutive activation of the
MAPK pathway and BRAF mutations have been associated with
adverse clinical outcomes in advanced CRC (74–76). Won
et al (44) even concluded
that BRAF mutations, rather than KRAS mutations, were
significant prognostic factors in Korean patients with CRC.
Accordingly, simultaneous testing for KRAS and BRAF is
required in future studies in CRC. In addition, the detection kit
used in the present study only included common codon changes.
Therefore, further studies on the histological results and their
prognostic value of rare KRAS codon variants are
necessary.
In summary, the present study demonstrated a
moderate association between KRAS-mutated CRCs and specific
histology, and, to a certain degree, an association between
histology and prognosis, according to KRAS mutation status.
Due to the different prognostic value of KRAS mutations in
patients with different ethnicities, the present study holds
scientific value as the patient cohort consists of a specific
ethnic group. Given that the results of the present study vary from
previous findings performed in South Korea, based on the prognostic
value of KRAS mutation, which have indicated that
KRAS is not associated with either OS or DFS (48,49),
this should be further clarified by meta-analysis.
Supplementary Material
Supporting Data
Acknowledgements
The authors greatly appreciate the valuable work
carried out by Ms Seo Young Oh at the Department of Pathology, KUMC
(Seoul, Korea) for performing the KRAS mutation test, and
her interpretation of the KRAS test results.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
HSH designed and supervised the study. DYH
contributed to data collection and data analysis. HSL contributed
to data collection, histological and statistical analyses,
constructed the figures and tables and drafted the initial
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
This retrospective study was reviewed and a waiver
of informed consent was provided by the Institutional Review Board
of KUMC (Seoul, Korea; ethics approval no. KUH1210056).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal carcinoma
|
|
KUMC
|
Konkuk University Medical Center
|
|
TIL
|
tumor-infiltrating lymphocytes
|
|
OS
|
overall survival
|
|
DFS
|
disease-free survival
|
|
HR
|
hazard ratio
|
References
|
1
|
Shin A, Kim KZ, Jung KW, Park S, Won YJ,
Kim J, Kim DY and Oh JH: Increasing trend of colorectal cancer
incidence in Korea, 1999–2009. Cancer Res Treat. 44:219–226. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jung KW, Won YJ, Kong HJ and Lee ES:
Prediction of cancer incidence and mortality in Korea, 2018. Cancer
Res Treat. 50:317–323. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
De Angelis R, Sant M, Coleman MP,
Francisci S, Baili P, Pierannunzio D, Trama A, Visser O, Brenner H,
Ardanaz E, et al: Cancer survival in Europe 1999–2007 by country
and age: Results of EUROCARE-5-a population-based study. Lancet
Oncol. 15:23–34. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mundade R, Imperiale TF, Prabhu L, Loehrer
PJ and Lu T: Genetic pathways, prevention, and treatment of
sporadic colorectal cancer. Oncoscience. 1:400–406. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Castagnola P and Giaretti W: Mutant KRAS,
chromosomal instability and prognosis in colorectal cancer. Biochim
Biophys Acta. 1756:115–125. 2005.PubMed/NCBI
|
|
6
|
Suehiro Y, Wong CW, Chirieac LR, Kondo Y,
Shen L, Webb CR, Chan YW, Chan AS, Chan TL, Wu TT, et al:
Epigenetic-genetic interactions in the APC/WNT, RAS/RAF, and P53
pathways in colorectal carcinoma. Clin Cancer Res. 14:2560–2569.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Malumbres M and Barbacid M: RAS oncogenes:
The first 30 years. Nat Rev Cancer. 3:459–465. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rosty C, Young JP, Walsh MD, Clendenning
M, Walters RJ, Pearson S, Pavluk E, Nagler B, Pakenas D, Jass JR,
et al: Colorectal carcinomas with KRAS mutation are associated with
distinctive morphological and molecular features. Mod Pathol.
26:825–834. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tanaka M, Omura K, Watanabe Y, Oda Y and
Nakanishi I: Prognostic factors of colorectal cancer: K-ras
mutation, overexpression of the p53 protein, and cell proliferative
activity. J Surg Oncol. 57:57–64. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kakar S, Aksoy S, Burgart LJ and Smyrk TC:
Mucinous carcinoma of the colon: Correlation of loss of mismatch
repair enzymes with clinicopathologic features and survival. Mod
Pathol. 17:696–700. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rimbert J, Tachon G, Junca A, Villalva C,
Karayan-Tapon L and Tougeron D: Association between
clinicopathological characteristics and RAS mutation in colorectal
cancer. Mod Pathol. 31:517–526. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Millar J: The Need for a Global
Language-SNOMED CT introduction. Stud Health Technol Inform.
225:683–685. 2016.PubMed/NCBI
|
|
13
|
Yang D, Lai X, Xu F, Li Y, Jiang W and Ma
D: Prognosis and clinical characteristics of colorectal cancer
patients with KRAS gene mutation: A 5-year follow-up study. Int J
Clin Exp Pathol. 12:409–418. 2019.PubMed/NCBI
|
|
14
|
Lim DR, Kuk JK, Kim T and Shin EJ:
Comparison of oncological outcomes of right-sided colon cancer
versus left-sided colon cancer after curative resection: Which side
is better outcome? Medicine (Baltimore). 96:e82412017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen CH, Hsieh MC, Hsiao PK, Lin EK, Lu YJ
and Wu SY: A critical reappraisal for the value of tumor size as a
prognostic variable in rectal adenocarcinoma. J Cancer.
8:1927–1934. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhai ZW and Gu J: Influence of tumor size
on the prognosis in patients with colon cancer. Zhonghua Wei Chang
Wai Ke Za Zhi. 15:495–498. 2012.(In Chinese). PubMed/NCBI
|
|
17
|
Oh SY, Han JY, Lee SR and Lee HT: Improved
DNA extraction method for molecular diagnosis from smaller numbers
of cells. Korean J Clin Lab Sci. 46:99–105. 2014. View Article : Google Scholar
|
|
18
|
Bosman FT, Carneiro F, Hruban RH and
Theise ND: WHO classification of tumours of the digestive system.
Fourth Edition. IARC. 2010.
|
|
19
|
Weiser MR: AJCC 8th edition: Colorectal
cancer. Ann Surg Oncol. 25:1454–1455. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Onodera M, Nishigami T, Torii I, Sato A,
Tao LH, Kataoka TR, Yoshikawa R and Tsujimura T: Comparison between
colorectal low- and high-grade mucinous adenocarcinoma with MUC1
and MUC5AC. World J Gastrointest Oncol. 1:69–73. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ueno H, Hashiguchi Y, Shimazaki H, Shinto
E, Kajiwara Y, Nakanishi K, Kato K, Maekawa K, Miyai K, Nakamura T,
et al: Objective criteria for crohn-like lymphoid reaction in
colorectal cancer. Am J Clin Pathol. 139:434–441. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shia J, Schultz N, Kuk D, Vakiani E,
Middha S, Segal NH, Hechtman JF, Berger MF, Stadler ZK, Weiser MR,
et al: Morphological characterization of colorectal cancers in The
Cancer Genome Atlas reveals distinct morphology-molecular
associations: Clinical and biological implications. Mod Pathol.
30:599–609. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Iseki Y, Shibutani M, Maeda K, Nagahara H,
Fukuoka T, Matsutani S, Kashiwagi S, Tanaka H, Hirakawa K and Ohira
M: A new method for evaluating tumor-infiltrating lymphocytes
(TILs) in colorectal cancer using hematoxylin and eosin
(H-E)-stained tumor sections. PLoS One. 13:e01927442018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lugli A, Kirsch R, Ajioka Y, Bosman F,
Cathomas G, Dawson H, El Zimaity H, Flèjou JF, Hansen TP, Hartmann
A, et al: Recommendations for reporting tumor budding in colorectal
cancer based on the International Tumor Budding Consensus
Conference (ITBCC) 2016. Mod Pathol. 30:1299–1311. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kruschewski M, Mueller K, Lipka S,
Budczies J, Noske A, Buhr HJ and Elezkurtaj S: The prognostic
impact of p53 expression on sporadic colorectal cancer is dependent
on p21 status. Cancers (Basel). 3:1274–1284. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ogino S, Kawasaki T, Kirkner GJ, Ogawa A,
Dorfman I, Loda M and Fuchs CS: Down-regulation of p21
(CDKN1A/CIP1) is inversely associated with microsatellite
instability and CpG island methylator phenotype (CIMP) in
colorectal cancer. J Pathol. 210:147–154. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guo F, Gong H, Zhao H, Chen J, Zhang Y,
Zhang L, Shi X, Zhang A, Jin H, Zhang J and He Y: Mutation status
and prognostic values of KRAS, NRAS, BRAF and PIK3CA in 353 Chinese
colorectal cancer patients. Sci Rep. 8:60762018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lee WS, Lee JN, Baek JH and Park YH: RAS
status in Korean patients with stage III and IV colorectal cancer.
Clin Transl Oncol. 17:751–756. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Byun JH, Ahn JB, Kim SY, Kang JH, Zang DY,
Kang SY, Kang MJ, Shim BY, Baek SK, Kim BS, et al: The impact of
primary tumor location in patients with metastatic colorectal
cancer: A Korean Cancer Study Group CO12-04 study. Korean J Intern
Med. 34:165–177. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Calistri D, Rengucci C, Seymour I,
Leonardi E, Truini M, Malacarne D, Castagnola P and Giaretti W:
KRAS, p53 and BRAF gene mutations and aneuploidy in sporadic
colorectal cancer progression. Cell Oncol. 28:161–166.
2006.PubMed/NCBI
|
|
31
|
Tosi F, Magni E, Amatu A, Mauri G,
Bencardino K, Truini M, Veronese S, De Carlis L, Ferrari G,
Nichelatti M, et al: Effect of KRAS and BRAF mutations on survival
of metastatic colorectal cancer after liver resection: A systematic
review and meta-analysis. Clin Colorectal Cancer. 16:e153–e163.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bai B, Shan L, Xie B, Huang X, Mao W, Wang
X, Wang D and Zhu H: Mutations in KRAS codon 12 predict poor
survival in Chinese patients with metastatic colorectal cancer.
Oncol Lett. 15:3161–3166. 2018.PubMed/NCBI
|
|
33
|
Lee JH, Ahn J, Park WS, Choe EK, Kim E,
Shin R, Heo SC, Jung S, Kim K, Chai YJ and Chae H: Colorectal
cancer prognosis is not associated with BRAF and KRAS Mutations-A
STROBE compliant study. J Clin Med. 8(pii): E1112019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Payandeh M, Shazad B, Sadeghi M and
Shahbazi M: Correlation between RAS test results and prognosis of
metastatic colorectal cancer patients: A report from Western Iran.
Asian Pac J Cancer Prev. 17:1729–1732. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Modest DP, Brodowicz T, Stintzing S, Jung
A, Neumann J, Laubender RP, Ocvirk J, Kurteva G, Papai Z,
Knittelfelder R, et al: Impact of the specific mutation in KRAS
codon 12 mutated tumors on treatment efficacy in patients with
metastatic colorectal cancer receiving cetuximab-based first-line
therapy: A pooled analysis of three trials. Oncology. 83:241–247.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Renaud S, Guerrera F, Seitlinger J,
Costardi L, Schaeffer M, Romain B, Mossetti C, Claire-Voegeli A,
Filosso PL, Legrain M, et al: KRAS exon 2 codon 13 mutation is
associated with a better prognosis than codon 12 mutation following
lung metastasectomy in colorectal cancer. Oncotarget. 8:2514–2524.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jones RP, Sutton PA, Evans JP, Clifford R,
McAvoy A, Lewis J, Rousseau A, Mountford R, McWhirter D and Malik
HZ: Specific mutations in KRAS codon 12 are associated with worse
overall survival in patients with advanced and recurrent colorectal
cancer. Br J Cancer. 116:923–929. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kaczirek K, Ciuleanu TE, Vrbanec D, Marton
E, Messinger D, Liegl-Atzwanger B, Wrba F, Knittelfelder R, Lindner
E, Zielinski CC, et al: FOLFOX4 plus cetuximab for patients with
previously untreated metastatic colorectal cancer according to
tumor RAS and BRAF mutation status: Updated analysis of the
CECOG/CORE 1.2.002 study. Clin Colorectal Cancer. 14:91–98. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yoon HH, Tougeron D, Shi Q, Alberts SR,
Mahoney MR, Nelson GD, Nair SG, Thibodeau SN, Goldberg RM, Sargent
DJ, et al: KRAS codon 12 and 13 mutations in relation to
disease-free survival in BRAF-wild-type stage III colon cancers
from an adjuvant chemotherapy trial (N0147 alliance). Clin Cancer
Res. 20:3033–3043. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kadowaki S, Kakuta M, Takahashi S,
Takahashi A, Arai Y, Nishimura Y, Yatsuoka T, Ooki A, Yamaguchi K,
Matsuo K, et al: Prognostic value of KRAS and BRAF mutations in
curatively resected colorectal cancer. World J Gastroenterol.
21:1275–1283. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Alberts SR, Sargent DJ, Nair S, Mahoney
MR, Mooney M, Thibodeau SN, Smyrk TC, Sinicrope FA, Chan E, Gill S,
et al: Effect of oxaliplatin, fluorouracil, and leucovorin with or
without cetuximab on survival among patients with resected stage
III colon cancer: A randomized trial. JAMA. 307:1383–1393. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lee DW, Kim KJ, Han SW, Lee HJ, Rhee YY,
Bae JM, Cho NY, Lee KH, Kim TY, Oh DY, et al: KRAS mutation is
associated with worse prognosis in stage III or high-risk stage II
colon cancer patients treated with adjuvant FOLFOX. Ann Surg Oncol.
22:187–194. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Blons H, Emile JF, Le Malicot K, Juliè C,
Zaanan A, Tabernero J, Mini E, Folprecht G, Van Laethem JL, Thaler
J, et al: Prognostic value of KRAS mutations in stage III colon
cancer: Post hoc analysis of the PETACC8 phase III trial dataset.
Ann Oncol. 25:2378–2385. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Won DD, Lee JI, Lee IK, Oh ST, Jung ES and
Lee SH: The prognostic significance of KRAS and BRAF mutation
status in Korean colorectal cancer patients. BMC Cancer.
17:4032017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gao XH, Yu GY, Hong YG, Lian W, Chouhan H,
Xu Y, Liu LJ, Bai CG and Zhang W: Clinical significance of multiple
gene detection with a 22-gene panel in formalin-fixed
paraffin-embedded specimens of 207 colorectal cancer patients. Int
J Clin Oncol. 24:141–152. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Natsume S, Yamaguchi T, Takao M, Iijima T,
Wakaume R, Takahashi K, Matsumoto H, Nakano D, Horiguchi SI,
Koizumi K and Miyaki M: Clinicopathological and molecular
differences between right-sided and left-sided colorectal cancer in
Japanese patients. Jpn J Clin Oncol. 48:609–618. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Inoue Y, Saigusa S, Iwata T, Okugawa Y,
Toiyama Y, Tanaka K, Uchida K, Mohri Y and Kusunoki M: The
prognostic value of KRAS mutations in patients with colorectal
cancer. Oncol Rep. 28:1579–1584. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chang MH, Lee IK, Si Y, Lee KS, Woo IS and
Byun JH: Clinical impact of K-ras mutation in colorectal cancer
patients treated with adjuvant FOLFOX. Cancer Chemother Pharmacol.
68:317–323. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kim HS, Heo JS, Lee J, Lee JY, Lee MY, Lim
SH, Lee WY, Kim SH, Park YA, Cho YB, et al: The impact of KRAS
mutations on prognosis in surgically resected colorectal cancer
patients with liver and lung metastases: A retrospective analysis.
BMC Cancer. 16:1202016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Buim ME, Fanelli MF, Souza VS, Romero J,
Abdallah EA, Mello CA, Alves V, Ocea LM, Mingues NB, Barbosa PN, et
al: Detection of KRAS mutations in circulating tumor cells from
patients with metastatic colorectal cancer. Cancer Biol Ther.
16:1289–1295. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cabrera-Mendoza F, Gainza-Lagunes S,
Castañeda-Andrade I and Castro-Zarate A: Clinical relevance of the
K-ras oncogene in colorectal cancer: Experience in a Mexican
population. Rev Gastroenterol Mex. 79:166–170. 2014.(In Spanish).
PubMed/NCBI
|
|
52
|
Zekri J, Rizvi A, Al-Maghrabi J and Bin
Sadiq B: K-ras in colorectal cancer tumors from saudi patients:
Frequency, Clinco-pathological Association and clinical outcome.
Open Colorectal Cancer J. 5:22–27. 2012. View Article : Google Scholar
|
|
53
|
Sastre J, Vidaurreta M, Gómez A, Rivera F,
Massutí B, López MR, Abad A, Gallen M, Benavides M, Aranda E, et
al: Prognostic value of the combination of circulating tumor cells
plus KRAS in patients with metastatic colorectal cancer treated
with chemotherapy plus bevacizumab. Clin Colorectal Cancer.
12:280–286. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Serenari M, Alvarez FA, Ardiles V, de
Santibañes M, Pekolj J and de Santibañes E: The ALPPS approach for
colorectal liver metastases: Impact of KRAS mutation status in
survival. Dig Surg. 35:303–310. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Graf W, Cashin PH, Ghanipour L, Enblad M,
Botling J, Terman A and Birgisson H: Prognostic impact of BRAF and
KRAS mutation in patients with colorectal and appendiceal
peritoneal metastases scheduled for CRS and HIPEC. Ann Surg Oncol.
27:293–300. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Winder T, Mündlein A, Rhomberg S,
Dirschmid K, Hartmann BL, Knauer M, Drexel H, Wenzl E, De Vries A
and Lang A: Different types of K-Ras mutations are conversely
associated with overall survival in patients with colorectal
cancer. Oncol Rep. 21:1283–1287. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
East JE, Atkin WS, Bateman AC, Clark SK,
Dolwani S, Ket SN, Leedham SJ, Phull PS, Rutter MD, Shepherd NA, et
al: British society of gastroenterology position statement on
serrated polyps in the colon and rectum. Gut. 66:1181–1196. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lee CT, Huang YC, Hung LY, Chow NH, Su PF,
Ho CL, Tsai HW, Chen YL, Lin SC, Lin BW, et al: Serrated
adenocarcinoma morphology in colorectal mucinous adenocarcinoma is
associated with improved patient survival. Oncotarget.
8:35165–35175. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Patai AV, Molnár B, Tulassay Z and Sipos
F: Serrated pathway: Alternative route to colorectal cancer. World
J Gastroenterol. 19:607–615. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Loy TS and Kaplan PA: Villous
adenocarcinoma of the colon and rectum: A clinicopathologic study
of 36 cases. Am J Surg Pathol. 28:1460–1465. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Palazzo JP, Edmonston TB, Chaille-Arnold
LM and Burkholder S: Invasive papillary adenocarcinoma of the
colon. Hum Pathol. 33:372–375. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Gonzalez RS, Cates JM, Washington MK,
Beauchamp RD, Coffey RJ and Shi C: Adenoma-like adenocarcinoma: A
subtype of colorectal carcinoma with good prognosis, deceptive
appearance on biopsy and frequent KRAS mutation. Histopathology.
68:183–190. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Fecteau RE, Lutterbaugh J, Markowitz SD,
Willis J and Guda K: GNAS mutations identify a set of right-sided,
RAS mutant, villous colon cancers. PLoS One. 9:e879662014.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Kim MJ, Hong SM, Jang SJ, Yu E, Kim JS,
Kim KR, Gong G and Ro JY: Invasive colorectal micropapillary
carcinoma: An aggressive variant of adenocarcinoma. Hum Pathol.
37:809–815. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Pettinato G, Manivel CJ, Panico L, Sparano
L and Petrella G: Invasive micropapillary carcinoma of the breast:
Clinicopathologic study of 62 cases of a poorly recognized variant
with highly aggressive behavior. Am J Clin Pathol. 121:857–866.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Hendifar A, Yang D, Lenz F, Lurje G, Pohl
A, Lenz C, Ning Y, Zhang W and Lenz HJ: Gender disparities in
metastatic colorectal cancer survival. Clin Cancer Res.
15:6391–6397. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Majek O, Gondos A, Jansen L, Emrich K,
Holleczek B, Katalinic A, Nennecke A, Eberle A and Brenner H; GEKID
Cancer Survival Working Group, : Sex differences in colorectal
cancer survival: Population-based analysis of 164,996 colorectal
cancer patients in Germany. PLoS One. 8:e680772013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Soulierès D, Greer W, Magliocco AM,
Huntsman D, Young S, Tsao MS and Kamel-Reid S: KRAS mutation
testing in the treatment of metastatic colorectal cancer with
anti-EGFR therapies. Curr Oncol. 17 (Suppl 1):S31–S40.
2010.PubMed/NCBI
|
|
69
|
Chuko J, Yeh MK, Chen BJ and Hu KY:
Efficacy of cetuximab on wild-type and mutant KRAS in colorectal
cancer: Systematic review and meta-analysis. J Med Sci. 30:189–198.
2010.
|
|
70
|
Pollheimer MJ, Kornprat P, Lindtner RA,
Harbaum L, Schlemmer A, Rehak P and Langner C: Tumor necrosis is a
new promising prognostic factor in colorectal cancer. Hum Pathol.
41:1749–1757. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Väyrynen SA, Väyrynen JP, Klintrup K,
Mäkelä J, Karttunen TJ, Tuomisto A and Mäkinen MJ: Clinical impact
and network of determinants of tumour necrosis in colorectal
cancer. Br J Cancer. 114:1334–1342. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wikberg ML, Ling A, Li X, Öberg A, Edin S
and Palmqvist R: Neutrophil infiltration is a favorable prognostic
factor in early stages of colon cancer. Hum Pathol. 68:193–202.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Rao HL, Chen JW, Li M, Xiao YB, Fu J, Zeng
YX, Cai MY and Xie D: Increased intratumoral neutrophil in
colorectal carcinomas correlates closely with malignant phenotype
and predicts patients' adverse prognosis. PLoS One. 7:e308062012.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Sanz-Garcia E, Argiles G, Elez E and
Tabernero J: BRAF mutant colorectal cancer: Prognosis, treatment,
and new perspectives. Ann Oncol. 28:2648–2657. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Chen D, Huang JF, Liu K, Zhang LQ, Yang Z,
Chuai ZR, Wang YX, Shi DC, Huang Q and Fu WL: BRAFV600E mutation
and its association with clinicopathological features of colorectal
cancer: A systematic review and meta-analysis. PLoS One.
9:e906072014. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Davies H, Bignell GR, Cox C, Stephens P,
Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W,
et al: Mutations of the BRAF gene in human cancer. Nature.
417:949–954. 2002. View Article : Google Scholar : PubMed/NCBI
|