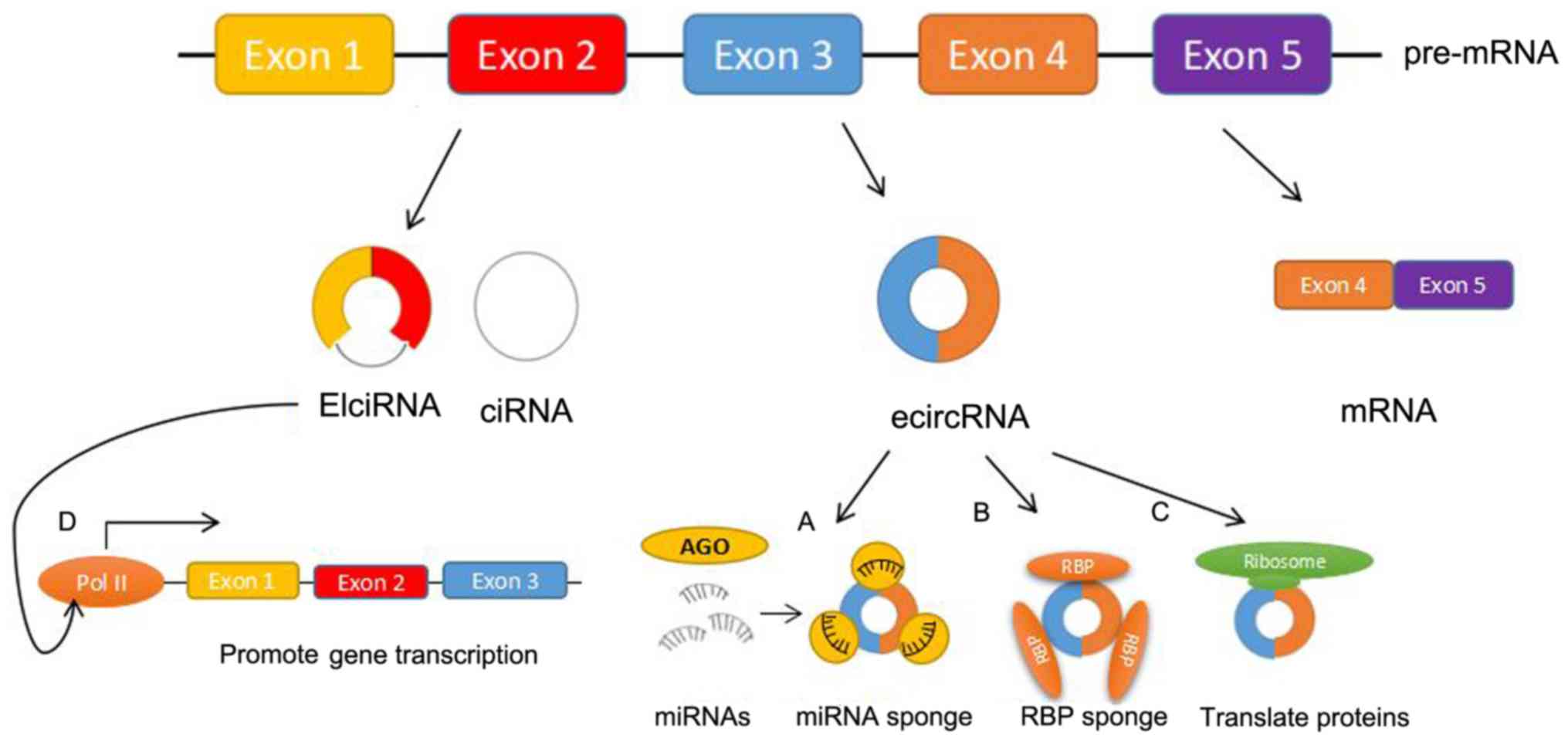
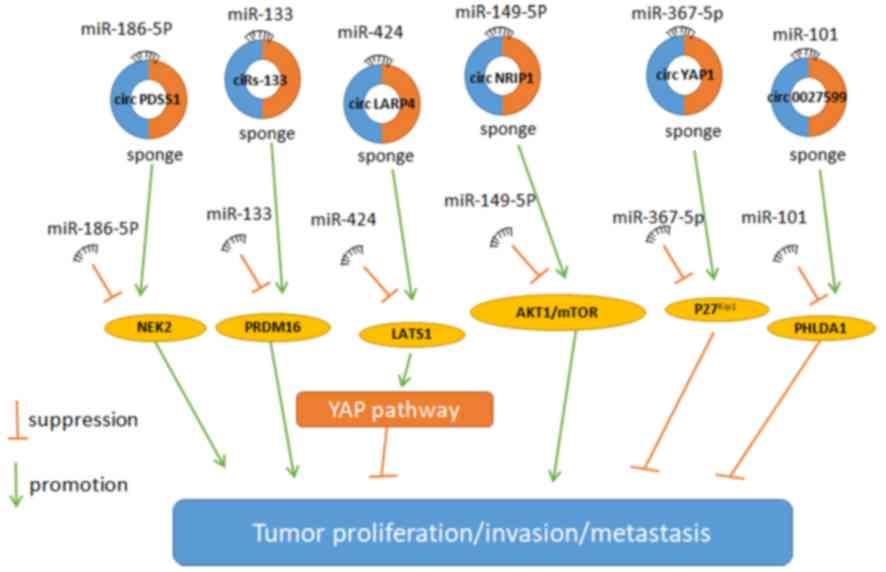
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhu L, Qin J, Wang J, Guo T, Wang Z and
Yang J: Early gastric cancer: Current advances of endoscopic
diagnosis and treatment. Gastroenterol Res Pract. 2016:96380412016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huang RJ, Choi AY, Truong CD, Yeh MM and
Hwang JH: Diagnosis and management of gastric intestinal
metaplasia: Current status and future directions. Gut Liver.
15:596–603. 2019. View
Article : Google Scholar
|
|
4
|
Hentze MW and Preiss T: Circular RNAs:
Splicing's enigma variations. EMBO J. 32:923–925. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao
L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, et al: The
landscape of circular RNA in cancer. Cell. 176:869–881. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wilusz JE and Sharp PA: Molecular biology.
A circuitous route to noncoding RNA. Science. 340:440–441. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Broadbent KM, Broadbent JC, Ribacke U,
Wirth D, Rinn JL and Sabeti PC: Strand-Specific RNA sequencing in
Plasmodium falciparum malaria identifies developmentally regulated
long non-coding RNA and circular RNA. BMC Genomics. 16:4542015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lu T, Cui L, Zhou Y, Zhu C, Fan D, Gong H,
Zhao Q, Zhou C, Zhao Y, Lu D, et al: Transcriptome-Wide
investigation of circular RNAs in rice. RNA. 21:2076–2087. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lu Z, Filonov GS, Noto JJ, Schmidt CA,
Hatkevich TL, Wen Y, Jaffrey SR and Matera AG: Metazoan tRNA
introns generate stable circular RNAs in vivo. RNA. 21:1554–1565.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schmidt CA, Noto JJ, Filonov GS and Matera
AG: A method for expressing and imaging abundant, stable, circular
RNAs in vivo using tRNA splicing. Methods Enzymol. 572:215–236.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jin M, Shi C, Yang C, Liu J and Huang G:
Upregulated CircRNA ARHGAP10 Predicts an unfavorable prognosis in
NSCLC through regulation of the miR-150-5p/GLUT-1 axis. Mol Ther
Nucleic Acids. 18:219–231. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ye F, Gao G, Zou Y, Zheng S, Zhang L, Ou
X, Xie X and Tang H: CircFBXW7 inhibits malignant progression by
sponging miR-197-3p and encoding a 185-aa protein in
triple-negative breast cancer. Mol Ther Nucleic Acids. 18:88–98.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu K, Zhan H, Peng Y, Yang L, Gao Q, Jia
H, Dai Z, Tang Z, Fan J and Zhou J: Plasma hsa_circ_0027089 is a
diagnostic biomarker for hepatitis B virus-related hepatocellular
carcinoma. Carcinogenesis. 19:1542019.
|
|
16
|
Chen J, Chen T, Zhu Y, Li Y, Zhang Y, Wang
Y, Li X, Xie X, Wang J, Huang M, et al: CircPTN sponges
miR-145-5p/miR-330-5p to promote proliferation and stemness in
glioma. J Exp Clin Cancer Res. 38:3982019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu G, Chen Y, Fu M, Zang X, Cang M, Niu Y,
Zhang W, Zhang Y, Mao Z, Shao M, et al: Circular RNA CCDC66
promotes gastric cancer progression by regulating c-Myc and TGF-β
signaling pathways. J Cancer. 11:2759–2768. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Song H, Xu Y, Xu T, Fan R, Jiang T, Cao M,
Shi L and Song J: CircPIP5K1A activates KRT80 and PI3K/AKT pathway
to promote gastric cancer development through sponging miR-671-5p.
Biomed Pharmacother. 126:1099412020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang L, Chang X, Zhai T, Yu J, Wang W, Du
A and Liu N: A novel circular RNA, circ-ATAD1, contributes to
gastric cancer cell progression by targeting miR-140-3p/YY1/PCIF1
signaling axis. Biochem Biophys Res Commun. 14:841–849. 2020.
View Article : Google Scholar
|
|
20
|
Hu K, Qin X, Shao Y, Zhou Y, Ye G and Xu
S: Circular RNA MTO1 suppresses tumorigenesis of gastric carcinoma
by sponging miR-3200-5p and targeting PEBP1. Mol Cell Probes.
56:1015622020. View Article : Google Scholar
|
|
21
|
Ren S, Liu J, Feng Y, Li Z, He L, Li L,
Cao X, Wang Z and Zhang Y: Knockdown of circDENND4C inhibits
glycolysis, migration and invasion by up-regulating miR-200b/c in
breast cancer under hypoxia. J Exp Clin Cancer Res. 38:3882019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu Q, Zhang W, Wu Z, Liu H, Hu H, Shi H,
Li S and Zhang X: Construction of a circular
RNA-microRNA-messengerRNA regulatory network in stomach
adenocarcinoma. J Cell Biochem. 12:1317–1331. 2019.
|
|
23
|
Bi J, Liu H, Dong W, Xie W, He Q, Cai Z,
Huang J and Lin T: Circular RNA circ-ZKSCAN1 inhibits bladder
cancer progression through miR-1178-3p/p21 axis and acts as a
prognostic factor of recurrence. Mol Cancer. 18:1332019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang H, Wei L, Qin T, Yang N, Li Z and Xu
Z: Circular RNA ciRS-7 triggers the migration and invasion of
esophageal squamous cell carcinoma via miR-7/KLF4 and NF-κB
signals. Cancer Biol Ther. 20:73–80. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chung DJ, Wang CJ, Yeh CW and Tseng TH:
Inhibition of the proliferation and invasion of C6 glioma cells by
tricin via the upregulation of focal-adhesion-kinase-targeting
MicroRNA-7. J Agric Food Chem. 66:6708–6716. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pan M, Li M, You C, Zhao F, Guo M, Xu H,
Li L, Wang L and Dou J: Inhibition of breast cancer growth via
miR-7 suppressing ALDH1A3 activity concomitant with decreasing
breast cancer stem cell subpopulation. J Cell Physiol.
235:1405–1416. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ye T, Yang M, Huang D, Wang X, Xue B, Tian
N, Xu X, Bao L, Hu H, Lv T and Huang Y: MicroRNA-7 as a potential
therapeutic target for aberrant NF-κB-driven distant metastasis of
gastric cancer. J Exp Clin Cancer Res. 38:552019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ling Y, Cao C, Li S, Qiu M, Shen G, Chen
Z, Yao F and Chen W: TRIP6, as a target of miR-7, regulates the
proliferation and metastasis of colorectal cancer cells. Biochem
Biophys Res Commun. 514:231–238. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang
C, Liu D, Wang M, Wang L, Zeng F and Jiang G: CircHIPK3 sponges
miR-558 to suppress heparanase expression in bladder cancer cells.
EMBO Rep. 18:1646–1659. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y,
Yang S, Zeng Z, Liao W, Ding YQ and Liang L: Emerging roles of
circRNA_001569 targeting miR-145 in the proliferation and invasion
of colorectal cancer. Oncotarget. 7:26680–26691. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Meng Q, Li S, Liu Y, Zhang S, Jin J, Zhang
Y, Guo C, Liu B and Sun Y: Circular RNA circSCAF11 accelerates the
glioma tumorigenesis through the miR-421/SP1/VEGFA axis. Mol Ther
Nucleic Acids. 17:669–677. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yan Y, Fu G, Ye Y and Ming L: Exosomes
participate in the carcinogenesis and the malignant behavior of
gastric cancer. Scand J Gastroenterol. 52:499–504. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Thomas LF and Sætrom P: Circular RNAs are
depleted of polymorphisms at microRNA binding sites.
Bioinformatics. 30:2243–2246. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhao ZJ and Shen J: Circular RNA
participates in the carcinogenesis and the malignant behavior of
cancer. RNA Biol. 14:514–521. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lu J, Wang YH, Huang XY, Xie JW, Wang JB,
Lin JX, Chen QY, Cao LL, Huang CM, Zheng CH and Li P: Circ-CEP85L
suppresses the proliferation and invasion of gastric cancer by
regulating NFKBIA expression via miR-942-5p. J Cell Physiol.
5:102020.
|
|
37
|
Li H, Wei X, Yang J, Dong D, Hao D, Huang
Y, Lan X, Plath M, Lei C, Ma Y, et al: CircFGFR4 promotes
differentiation of myoblasts via binding miR-107 to relieve its
inhibition of Wnt3a. Mol Ther Nucleic Acids. 11:272–283. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: Circrna biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zheng J, Liu X, Xue Y, Gong W, Ma J, Xi Z,
Que Z and Liu Y: TTBK2 circular RNA promotes glioma malignancy by
regulating miR-217/HNF1β/Derlin-1 pathway. J Hematol Oncol.
10:522017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang ZG, Awan FM, Du WW, Zeng Y, Lyu J,
Wu, Gupta S, Yang W and Yang BB: The Circular RNA interacts with
stat3, increasing its nuclear translocation and wound repair by
modulating Dnmt3a and miR-17 function. Mol Ther. 25:2062–2074.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu KS, Pan F, Mao XD, Liu C and Chen J:
Biological functions of circular RNAs and their roles in occurrence
of reproduction and gynecological diseases. Am J Transl Res.
11:1–15. 2019.PubMed/NCBI
|
|
42
|
Zhang Y, Liang W, Zhang P, Chen J, Qian H,
Zhang X and Xu W: Circular RNAs: Emerging cancer biomarkers and
targets. J Exp Clin Cancer Res. 36:1522017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of CircRNAs. Mol
Cell. 66:9–21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N(6)-methyladenosine. Cell Res. 27:626–641.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z
and Sharpless NE: Expression of linear and novel circular forms of
an INK4/ARF-associated non-coding RNA correlates with
atherosclerosis risk. PLoS Genet. 6:e10012332010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Abdelmohsen K, Panda AC, Munk R,
Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM,
Martindale JL and Gorospe M: Identification of HuR target circular
RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA
Biol. 14:361–369. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang Y and Wang Z: Efficient backsplicing
produces translatablecircular mRNAs. RNA. 21:172–179. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-Intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li F, Ma K, Sun M and Shi S:
Identification of the tumor-suppressive function of circular RNA
ITCH in glioma cells through sponging miR-214 and promoting linear
ITCH expression. Am J Transl Res. 10:1373–1386. 2018.PubMed/NCBI
|
|
50
|
Ouyang Y, Li Y, Huang Y, Li X, Zhu Y, Long
Y, Wang Y, Guo X and Gong K: CircRNA circPDSS1 promotes the gastric
cancer progression by sponging miR-186-5p and modulating NEK2. J
Cell Physiol. 234:10458–10469. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Fry AM, O'Regan L, Sabir SR and Bayliss R:
Cell cycle regulation by the NEK family of protein kinases. J Cell
Sci. 125:4423–4433. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen J, Li Y, Zheng Q, Bao C, He J, Chen
B, Lyu D, Zheng B, Xu Y, Long Z, et al: Circular RNA profile
identifies circPVT1 as a proliferative factor and prognostic marker
in gastric cancer. Cancer Lett. 388:208–219. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liu H, Liu Y, Bian Z, Zhang J, Zhang R,
Chen X, Huang Y, Wang Y and Zhu J: Circular RNA YAP1 inhibits the
proliferation and invasion of gastric cancer cells by regulating
the miR-367-5p/p27 (Kip1) axis. Mol Cancer. 17:1512018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhang Y, Liu H, Li W, Yu J, Li J, Shen Z,
Ye G, Qi X and Li G: CircRNA_100269 is downregulated in gastric
cancer and suppresses tumor cell growth by targeting miR-630. Aging
(Albany NY). 9:1585–1594. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang L, Shen J and Jiang YL:
Circ_0027599/PHDLA1 suppresses gastric cancer progression by
sponging miR-101-3p.1. Cell Biosci. 8:582018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW,
Carmichael GG and Chen LL: Long noncoding RNAs with snoRNA ends.
Mol Cell. 48:219–230. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Han B, Chao J and Yao H: Circular RNA and
its mechanisms in disease: From the bench to the clinic. Pharmacol
Ther. 187:31–44. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Sun HD, Xu ZP, Sun ZQ, Zhu B, Wang Q, Zhou
J, Jin H, Zhao A, Tang WW and Cao XF: Down-Regulation of circPVRL3
promotes the proliferation and migration of gastric cancer cells.
Sci Rep. 8:101112018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei
P, Liu H, Xu J, Xiao F, Zhou H, et al: A peptide encoded by
circular form of LINC-PINT suppresses oncogenic transcriptional
elongation in glioblastoma. Nat Commun. 9:44752018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Huang H, Weng H and Chen J: m6A
Modification in coding and non-coding RNAs: Roles and therapeutic
implications in cancer. Cancer Cell. 37:270–288. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng
X, Xiong F, Guo C, Wu X, Li Y, et al: Emerging role of
tumor-related functional peptides encoded by lncRNA and circRNA.
Mol Cancer. 19:222020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Fang J, Hong H, Xue X, Zhu X, Jiang L, Qin
M, Liang H and Gao L: A novel circular RNA, circFAT1(e2), inhibits
gastric cancer progression by targeting miR-548g in the cytoplasm
and interacting with YBX1 in the nucleus. Cancer Lett. 442:222–232.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Warburg O, Wind F and Negelein E: The
metabolism of tumors in the body. J Gen Physiol. 8:519–530. 1927.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Kimmelman AC and White E: Autophagy and
tumor metabolism. Cell Metab. 25:1037–1043. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhang X, Wang S, Wang H, Cao J, Huang X,
Chen Z, Xu P, Sun G, Xu J, Lv J and Xu Z: Circular RNA circNRIP1
acts as a microRNA-149-5p sponge to promote gastric cancer
progression via the AKT1/mTOR pathway. Mol Cancer. 18:202019.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Heras-Sandoval D, Perez-Rojas JM,
Hernandez-Damian J and Pedraza-Chaverri J: The role of
PI3K/AKT/mTOR pathway in the modulation of autophagy and the
clearance of protein aggregates in neurodegeneration. Cell Signal.
26:2694–2701. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wu ZH, Lin C, Liu CC, Jiang WW, Huang MZ,
Liu X and Guo WJ: MiR-616-3p promotes angiogenesis and EMT in
gastric cancer via the PTEN/AKT/mTOR pathway. Biochem Biophys Res
Commun. 501:1068–1073. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhang H, Zhu L, Bai M, Liu Y, Zhan Y, Deng
T, Yang H, Sun W, Wang X, Zhu K, et al: Exosomal circRNA derived
from gastric tumor promotes white adipose browning by targeting the
miR-133/PRDM16 pathway. Int J Cancer. 144:2501–2515. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wei J, Wei W, Xu H, Wang Z, Gao W, Wang T,
Zheng Q, Shu Y and De W: Circular RNA hsa_circRNA_102958 may serve
as a diagnostic marker for gastric cancer. Cancer Biomark.
27:139–145. 2019. View Article : Google Scholar
|
|
70
|
Shao Y, Ye M, Li Q, Sun W, Ye G, Zhang X,
Yang Y, Xiao B and Guo J: LncRNA-RMRP promotes carcinogenesis by
acting as a miR-206 sponge and is used as a novel biomarker for
gastric cancer. Oncotarget. 7:37812–37824. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Park YM, Kim JK, Baik SJ, Park JJ, Youn YH
and Park H: Clinical risk assessment for gastric cancer in
asymptomatic population after a health check-up: An individualized
consideration of the risk factors. Medicine (Baltimore).
95:e53512016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Shao Y, Li J, Lu R, Li T, Yang Y, Xiao B
and Guo J: Global circular RNA expression profile of human gastric
cancer and its clinical significance. Cancer Med. 6:1173–1180.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Duffy MJ, Lamerz R, Haglund C, Nicolini A,
Kalousová M, Holubec L and Sturgeon C: Tumor markers in colorectal
cancer, gastric cancer and gastrointestinal stromal cancers:
European group on tumor markers 2014 guidelines update. Int J
Cancer. 134:2513–2522. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhao Q, Chen S, Li T, Xiao B and Zhang X:
Clinical values of circular RNA 0000181 in the screening of gastric
cancer. J Clin Lab Anal. 32:e223332018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Lu J, Zhang PY, Xie JW, Wang JB, Lin JX,
Chen QY, Cao LL, Huang CM, Li P and Zheng CH: Hsa_circ_0000467
promotes cancer progression and serves as a diagnostic and
prognostic biomarker for gastric cancer. J Clin Lab Anal.
33:e227262019. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Tang W, Fu K, Sun H, Rong D, Wang H and
Cao H: CircRNA microarray profiling identifies a novel circulating
biomarker for detection of gastric cancer. Mol Cancer. 17:1372018.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Yang W, Gu J, Wang X, Wang Y, Feng M, Zhou
D, Guo J and Zhou M: Inhibition of circular RNA CDR1as increases
chemosensitivity of 5-FU-resistant BC cells through up-regulating
miR-7. J Cell Mol Med. 23:3166–3177. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Pan H, Li T, Jiang Y, Pan C, Ding Y, Huang
Z, Yu H and Kong D: Overexpression of circular RNA ciRS-7 abrogates
the tumor suppressive effect of miR-7 on gastric cancer via
PTEN/PI3K/AKT signaling pathway. J Cell Biochem. 119:440–446. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhang H, Wang X, Huang H, Wang Y, Zhang F
and Wang S: Hsa_circ_0067997 promotes the progression of gastric
cancer by inhibition of miR-515-5p and activation of X
chromosome-linked inhibitor of apoptosis (XIAP). Artif Cells
Nanomed Biotechnol. 47:308–318. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Lencz T, Guha S, Liu C, Rosenfeld J,
Mukherjee S, De Rosse P, John M, Cheng L, Zhang C, Badner JA, et
al: Genome-Wide association study implicates NDST3 in schizophrenia
and bipolar disorder. Nat Commun. 4:27392013. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Sun J, Li J, Zhang W, Zhang J, Sun S, Li
G, Song H and Wan D: MicroRNA-509-3p inhibits cancer cell
proliferation and migration via upregulation of XIAP in gastric
cancer cells. Oncol Res. 25:455–461. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Dang Y, Ouyang X, Zhang F, Wang K, Lin Y,
Sun B, Wang Y, Wang L and Huang Q: Circular RNAs expression
profiles in human gastric cancer. Sci Rep. 7:90602017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao
F, Huang N, Yang X, Zhao K, Zhou H, et al: Novel role of FBXW7
circular RNA in repressing glioma tumorigenesis. J Natl Cancer
Inst. 110:32018. View Article : Google Scholar
|
|
84
|
Zhou C, Molinie B, Daneshvar K, Pondick
JV, Wang J, Van Wittenberghe N, Xing Y, Giallourakis CC and Mullen
AC: Genome-Wide maps of m6A circRNAs identify widespread and
cell-type-specific methylation patterns that are distinct from
mRNAs. Cell Rep. 20:2262–2276. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Li WH, Song YC, Zhang H, Zhou ZJ, Xie X,
Zeng QN, Guo K, Wang T, Xia P and Chang DM: Decreased expression of
Hsa_circ_00001649 in gastric cancer and its clinical significance.
Dis Markers. 2017:45876982017. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Li J, Zhen L, Zhang Y, Zhao L, Liu H, Cai
D, Chen H, Yu J, Qi X and Li G: Circ-104916 is downregulated in
gastric cancer and suppresses migration and invasion of gastric
cancer cells. Onco Targets Ther. 10:3521–3529. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Huang M, He YR, Liang LC, Huang Q and Zhu
ZQ: Circular RNA hsa_circ_0000745 may serve as a diagnostic marker
for gastric cancer. World J Gastroenterol. 23:6330–6338. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Li P, Chen H, Chen S, Mo X, Li T, Xiao B,
Yu R and Guo J: Circular RNA 0000096 affects cell growth and
migration in gastric cancer. Br J Cancer. 116:626–633. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Li T, Shao Y, Fu L, Xie Y, Zhu L, Sun W,
Yu R, Xiao B and Guo J: Plasma circular RNA profiling of patients
with gastric cancer and their droplet digital RT-PCR detection. J
Mol Med (Berl). 96:85–96. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Fang Y, Ma M, Wang J, Liu X and Wang Y:
Circular RNAs play an important role in late-stage gastric cancer:
Circular RNA expression profiles and bioinformatics analyses.
Tumour Biol. 39:13933838142017. View Article : Google Scholar
|
|
92
|
Li P, Chen S, Chen H, Mo X, Li T, Shao Y,
Xiao B and Guo J: Using circular RNA as a novel type of biomarker
in the screening of gastric cancer. Clin Chim Acta. 444:132–136.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Lu R, Shao Y, Ye G, Xiao B and Guo J: Low
expression of hsa_circ_0006633 in human gastric cancer and its
clinical significances. Tumour Biol. 39:13933854892017. View Article : Google Scholar
|
|
94
|
Sun H, Tang W, Rong D, Jin H, Fu K, Zhang
W, Liu Z, Cao H and Cao X: Hsa_circ_0000520, a potential new
circular RNA biomarker, is involved in gastric carcinoma. Cancer
Biomark. 21:299–306. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Xie Y, Shao Y, Sun W, Ye G, Zhang X, Xiao
B and Guo J: Downregulated expression of hsa_circ_0074362 in
gastric cancer and its potential diagnostic values. Biomark Med.
12:11–20. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Rong D, Dong C, Fu K, Wang H, Tang W and
Cao H: Upregulation of circ_0066444 promotes the proliferation,
invasion, and migration of gastric cancer cells. Onco Targets Ther.
11:2753–2761. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Zhong S, Wang J, Hou J, Zhang Q, Xu H, Hu
J, Zhao J and Feng J: Circular RNA hsa_circ_0000993 inhibits
metastasis of gastric cancer cells. Epigenomics. 10:1301–1313.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Wei H, Pan L, Tao D and Li R: Circular RNA
circZFR contributes to papillary thyroid cancer cell proliferation
and invasion by sponging miR-1261 and facilitating C8orf4
expression. Biochem Biophys Res Commun. 503:56–61. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Tian M, Chen R, Li T and Xiao B: Reduced
expression of circRNA hsa_circ_0003159 in gastric cancer and its
clinical significance. J Clin Lab Anal. 32:32018. View Article : Google Scholar
|
|
100
|
Chen S, Li T, Zhao Q, Xiao B and Guo J:
Using circular RNA hsa_circ_0000190 as a new biomarker in the
diagnosis of gastric cancer. Clin Chim Acta. 466:167–171. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Zhou LH, Yang YC, Zhang RY, Wang P, Pang
MH and Liang LQ: CircRNA_0023642 promotes migration and invasion of
gastric cancer cells by regulating EMT. Eur Rev Med Pharmacol Sci.
22:2297–2303. 2018.PubMed/NCBI
|
|
102
|
Sun H, Xi P, Sun Z, Wang Q, Zhu B, Zhou J,
Jin H, Zheng W, Tang W, Cao H and Cao X: Circ-SFMBT2 promotes the
proliferation of gastric cancer cells through sponging miR-182-5p
to enhance CREB1 expression. Cancer Manag Res. 10:5725–5734. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Shen F, Liu P, Xu Z, Li N, Yi Z, Tie X,
Zhang Y and Gao L: CircRNA_001569 promotes cell proliferation
through absorbing miR-145 in gastric cancer. J Biochem. 165:27–36.
2019. View Article : Google Scholar : PubMed/NCBI
|