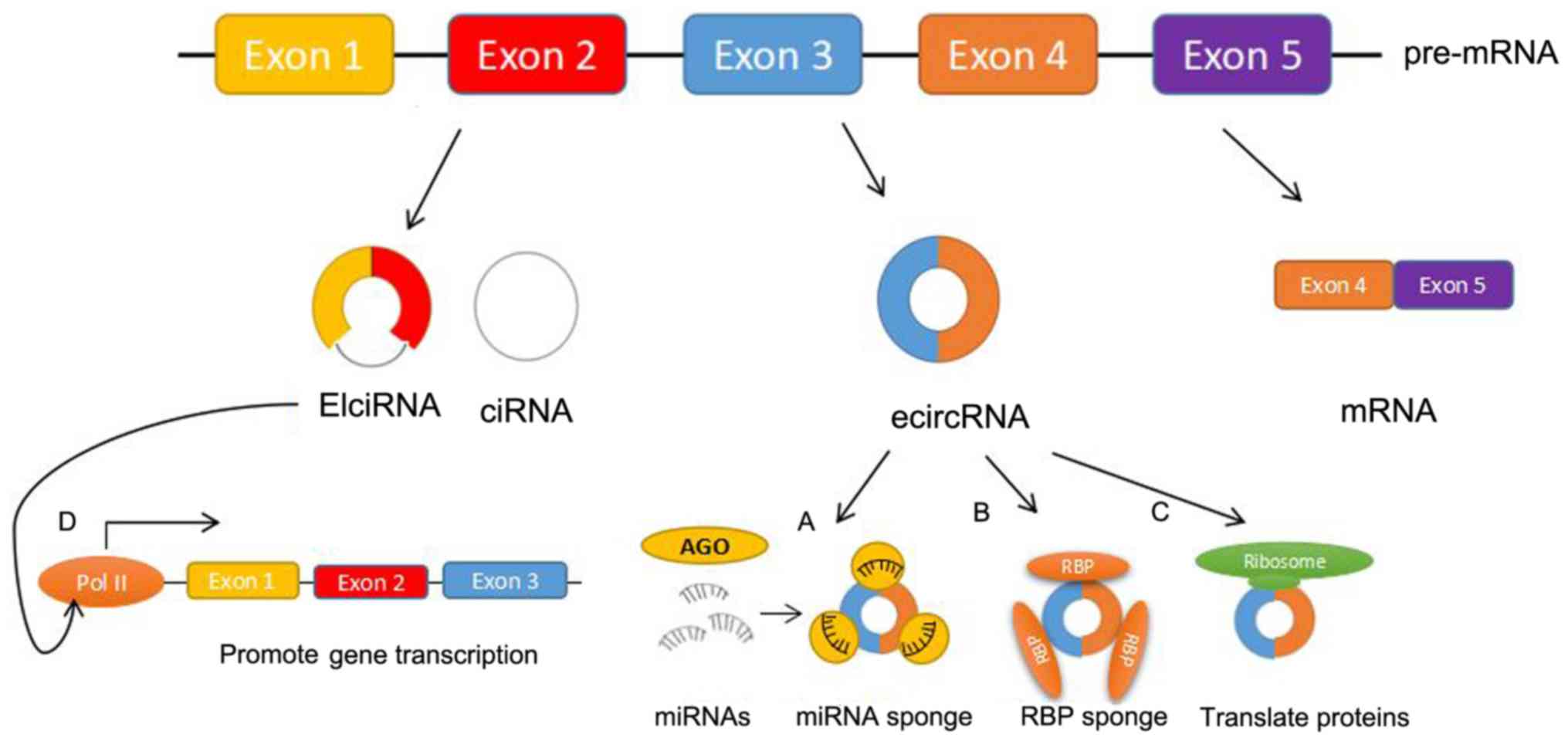
Gastric cancer is the fifth most common cancer and
third leading cause of cancer mortality in the world, according to
the International Agency for Research on Cancer in 2018 (1). The morbidity of gastric cancer affected
>1 million in 2018, accounting for 5.7% of new cancer cases
globally. Worldwide, gastric cancer mortality is ~783,000,
accounting for 8.2% of the total global deaths caused by cancer in
2018 (1). Diagnosis of gastric
cancer relies on endoscopic examination and analyses of serum
biomarkers in the clinical practice (2). However, endoscopy cannot be used for
large-scale screening, and detection by serum biomarkers lacks
sensitivity and specificity (3). To
date, there is no simple and efficient diagnostic method that
allows for the early diagnosis of gastric cancer to improve its
prognosis.
Circular RNAs (circRNAs) are a group of
single-strand closed-loop RNAs that are derived from back-splicing
and are widely found in serum and saliva (4,5).
circRNAs serve a regulatory role at the transcriptional and/or
post-transcriptional level, but some can only function at the
transcriptional level. Previous research has found that biological
functions of circRNAs include microRNA (miRNA) sponging, regulation
of transcription and translation, and interaction with RNA-binding
proteins (4,6–8).
Additionally, the dysregulation of circRNA expression is suggested
to play an important role in the pathogenesis and diagnosis of
gastric cancer.
Although little is known about the role of circRNAs
in the development of gastric cancer at the molecular level,
circRNAs secreted into the extracellular environment can be
identified in blood, stool and other body fluids (9–12). Thus,
circRNAs might be used as new potential biomarkers for the
diagnosis and therapeutic targeting of gastric cancer.
circRNAs are abnormally expressed in various cancer
types, including gastric cancer (13–16),
such as circCCDC66, circPIP5K1A, circ-ATAD1 are higher expressed,
but circMTO1 expression is lower in gastric cancer (17–20).
circRNAs may cause cancer by interacting with tumor-associated
miRNAs, proteins and genes, and by participating in
pathophysiological activities, including proliferation, invasion
and apoptosis (21–23). CDR1 antisense RNA (CDR1as) is an
abundantly expressed circRNA in the mammalian brain with >70
miRNA (miR)-7 binding sites (24).
CDR1as/miR-7 is a classical tumor-associated circRNA-miRNA sponge
system. CDR1as inhibits miR-7 activity by competing with miR-7,
which ultimately upregulates the expression of oncogenes, such as
epidermal growth factor receptor, NF-κB, and eventually promotes
the development of cancer (25).
Overexpression of CDR1as inhibits the proliferation
and invasion of glioma, breast cancer, gastric cancer and
colorectal cancer (26–29). By contrast, upregulation of other
circRNAs, such as circHIPK3, hsa_circ_001569 and circSCAF11,
promotes the development of bladder and colorectal cancers, as well
as glioma (30–32). In addition, abundant exosome circRNAs
in the serum differentiate gastric cancer by transmitting a signal
to local and remote cells, such as infiltrating immune cells,
stromal cells and endothelial cells. These cells are vital
components of the tumor microenvironment and enhance tumor growth,
angiogenesis and metastasis (33).
Such circRNAs may be useful for early and specific diagnosis of
tumors.
circRNAs are involved in numerous physiological
processes by interacting with a variety of RBPs to form RNA-protein
complexes, regulating gene transcription and influencing the
formation of circRNAs (Fig. 1B).
RBPs affect the expression of downstream target genes and miRNA
sponges by competing with miRNAs (6,7). RBPs
also act as trans-factors to regulate circRNA generation.
Modulation of splicing factor muscle blind (MBL) expression affects
the biosynthesis of circMBL through specific binding to MBL binding
sites within introns of circMBL (38). circMBL regulates parental gene
expression by binding to its own pre-mRNA flanking introns binding
sites at the transcriptional level. circRNAs have a binding site
for the enzyme and its substrate, thus it can directly bind to
protein or act as a scaffold between two or more proteins. It has
been reported that circ-Amotl1 interacts with proto-oncogene c-Myc,
STAT3, pyruvate dehydrogenase kinase 1and AKT1 and affects the
expression of its target genes by promoting nuclear translocation
of these proteins (39,40). In addition, RBPs are also involved in
the post-transcriptional regulation of miRNA expression and
interact with miRNA regulators by cooperative and competitive
inhibition (41). Pumilioprotein, a
cooperative RBP, promotes miRNA binding to target mRNA, which
inhibits protein translation by binding to a specific short
sequence in the 3′UTR of target mRNA, leading to altered spatial
conformation of mRNA (42).
circRNAs derived from exons are located in the
cytoplasm and have an open reading frame (ORF) to drive translation
(Fig. 1C) (43). In addition, some circRNA protein
translation is initiated after being modified by
N6-methyladenosine (m6A), such as circRNAs
initiates the protein translation by recruit YTHDF3 and EIf4G2
through the m6A modification site (44). circANRIL regulates the expression of
INK4/ARF by affecting the binding of ANRIL to polycomb-group
complex and inhibiting the transcription of the coding gene INK4
and its variable reading frame genes (45). circPABPN1 reduces the translational
efficiency of PABPN1 mRNA by competitively binding the RBP human
antigen R (HuR) and prevents HuR binding to PABPN1 mRNA (46). In addition, a small amount of
circRNAGFP participates in the translation process as a template
(47).
circRNA sequences are highly similar to their
homogenous linear mRNA. circRNAs act as positive regulators of the
activity of RNA polymerase II (RNA pol II), affecting expression of
parental genes and transcription (Fig.
1D). circEIF3J and circPAIP2, both exon-intron circRNAs
(EIciRNAs), interact with U1 small nuclear ribonucleoprotein
(snRNP) through RNA-RNA interactions to form EIciRNA-U1 snRNP
complexes. This complex interacts with RNA pol II in the promoter
region of host genes to cis-regulate the expression of its
host genes (48). Additionally,
circ-ITCH has common miRNA binding sites with the 3′ untranslated
region of itchy E3 ubiquitin protein ligase (ITCH). circ-ITCH
interacts with miR-7, miR-17 and miR-214 to promote the expression
of ITCH (49).
circRNAs promote malignant cell development,
proliferation and metastasis by sponging miRNAs resulting in the
dysregulated expression of miRNAs and the disruption of target
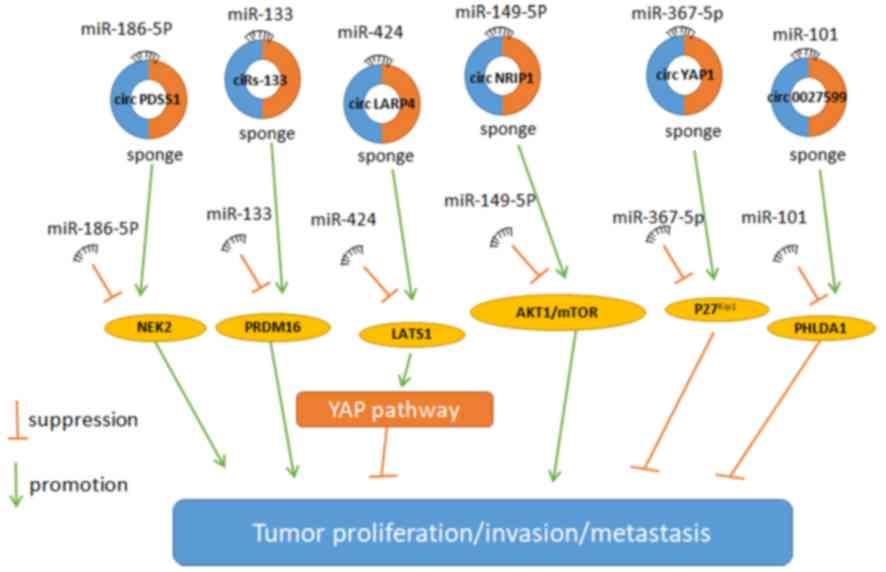
genes in gastric cancer. Upregulated circPDSS1 expression promotes
the expression of NIMA-related kinase 2 (NEK2) in gastric cancer
through a miR-186-5p sponge in vivo and in vitro
(Fig. 2) (50). Subsequently NEK2, which is associated
with cell centrosome and cell cycle in gastric cancer, enhances
cell proliferation and cell cycle by regulating cell mitosis in
this type of cancer (51). circPVT1,
a potential oncogenic gene, inhibits the activity of miR-125 and
promotes the proliferation of gastric cancer through c-Myc
(52). In addition, circLARP4 is
inversely associated with tumor size and lymph node metastasis in
gastric cancer by regulating the target gene large tumor suppressor
kinase 1 and Yes-associated protein (YAP) of miR-424, resulting in
the inhibition of DNA synthesis, cell proliferation and invasion
(Fig. 2) (53). circYAP1 inhibits cellular
proliferation, invasion and cell cycle by suppressing cyclin
dependent kinase inhibitor 1B. Downregulated circYAP1 in gastric
cancer is linked to poor prognosis and low 5 year survival rate
(Fig. 2) (53).
circ_100269 suppresses the p53 signaling pathway by
sponging miR-630 to inhibit proliferation and invasion of gastric
cancer (54). In addition,
circ_0027599 sponges miR-101 to inhibit the migration and invasion
of gastric cancer by targeting pleckstrin homology-like domain
family A member 1 gene (Fig. 2)
(55). These differential
expressions of circRNAs in the different aforementioned cancers
play crucial but different roles in determining cell fate, inducing
tumor-suppressive or oncogenic effects by acting as a sponge to
multiple different miRNAs, forming a circRNA-miRNA-mRNA axis
(50–55). Thus, circRNAs have the ability to
participate in many physiological and pathological processes,
suggesting that the regulation of circRNAs could offer a potential
therapeutic window for gastric cancer.
circRNAs not only function as miRNA sponges to
regulate miRNA expression, but also serve important roles in
regulating protein production (56).
circRNAs affect the proliferation and metastasis potential of
malignant cells, including gastric cancer cells, by directly
interacting with proteins or binding to the miRNA-acting factor to
regulate specific downstream target genes (57).
Downregulated circPVRL3 also promotes the
proliferation of gastric cancer by promoting protein encoding
(58). It might play a role in the
assembling of RBP complexes by binding to AGO2, FUS RNA-binding
protein, lin-28 homolog A, polypyrimidine tract binding protein and
eukaryotic translation initiation factor 4A3. In addition,
circPVRL3 contains internal ribosome entry site, ORF and
m6A modifications, these modifications initiate the
protein translation by recruiting ribosomes (59,60). The
translational products may exert specific biological effects or
interfere with protein-protein interactions to affect tumor
progression (44,61). Another study reported that
circFAT1(e2), located in cytoplasm and nucleus of gastric cancer
cells, is downregulated in gastric cancer (62). circFAT1(e2) directly binds to Y-box
binding protein-1 (YBX1) for DNA- and RNA-binding in the nucleus.
Upregulated circFAT1(e2) inhibits the growth of gastric cancer by
suppressing the expression of three targeted genes of YBX1
(epidermal growth factor receptor, MET proto-oncogene and cell
division cycle 25A).
Results from these studies suggest that the
stability of circRNA-protein interaction might act as ‘scaffolding
molecules’ in gastric cancer. Consequently, circRNAs acting as
scaffolding molecules for protein complexes and network functional
modules can modulate protein-protein interactions. circRNAs might
work as sequence targeting elements, affecting the function of
downstream target genes by simultaneously binding to RBPs.
Cell glycolytic activity, including in aerobic
conditions, is more active in cancer cells compared with normal
cells, process known as the Warburg effect (63). In addition, tumor cells exhibit
abnormal fatty acid and amino acid metabolism (64). Recent studies demonstrated that
circRNAs are closely related to tumor metabolism and these are able
to regulate gastric cancer metabolism through miRNA (65–67).
Upregulated circNRIP1 expression promotes the
proliferation, invasion and migration of gastric cancer through
activating AKT1/mTOR cell pathway by sponging miR-149-5p (Fig. 2) (65). circNRIP1 promotes anabolic activities
and prevents metabolic decomposition by maintaining energy
homeostasis, suggesting that circNRIP1 plays a crucial role in the
regulation of tumor metabolism (66,67).
In gastric cancer, ciRS-133 regulates substance
metabolism and organic metabolism through PR domain-containing
16(PRDM16), enhancing the Warburg effect. ciRS-133 accelerates
oxygen consumption and glucose consumption of brown fat cells,
promoting white adipose tissue (WAT) burning through enhancing the
expression of PRDM16 by adsorbing miR-133 in gastric cancer cells
(Fig. 2) (68). Consequently, such effects are known
to cause weight loss and systemic inflammation in patients with
poor prognosis. Moreover, knockdown of ciRS-133 reduces cancer
cachexia, decreasing oxygen consumption and glucose expenditure in
mouse animal models (68). Thus,
circRNAs contribute to cancer-associated cachexia and metabolic
disorder by sponging miRNA to regulate WAT browning.
Several studies have demonstrated that circRNAs are
differentially expressed in gastric cancer, as summarized in
Table I. circRNAs are widely present
in blood and gastric juice, and may have potential value for
diagnosis (9–12). Therefore, circRNAs may work as novel
biomarker-based screening tools for gastric cancer diagnosis and
evaluation.
In gastric cancer, circ_102958 is closely associated
to tumor, node, metastasis (TNM) cancer staging, but inversely
associated to age, gender, tumor diameter and differentiation
degree in lymph node metastasis (69). In addition, receiver operating
characteristic curve analysis indicated that circ_102958 has a high
diagnostic value. Together, these features suggested that
circ_102958 may be a biomarker for early diagnosis of gastric
cancer.
Owing to the lack of clinical symptoms and detection
methods at the early stages of gastric cancer, the analysis of
gastric juice could be advantageous for the detection of this type
of cancer (70). For instance,
circ_0014717 is downregulated in gastric juice of chronic atrophic
gastritis. Individuals with chronic atrophic gastritis are at high
risk of gastric cancer (71). This
implies that circ_0014717 may be used for early screening of
gastric cancer (72). In addition,
circ_0000181 is downregulated in the plasma of patients with
gastric cancer and is positively associated with the
differentiation of gastric cancer cells and the level of
carcinoembryonic antigen (CEA), which is a well-known gastric
cancer biomarker (73). In addition,
the specificity and sensitivity of hsa_circ_0000181 is higher than
CEA and carbohydrate antigen19-9 in screening gastric cancer,
suggesting that hsa_circ_0000181 may be a reliable plasma-based
biomarker (74).
circRNAs can potentially be used as a biomarker for
the prognosis and the evaluation of treatment efficacy in gastric
cancer. It has been demonstrated that circ_0000467 is highly
expressed in tissue and plasma samples of patients with gastric
cancer associated with a poor prognosis. Cox multivariate analysis
showed that circ_0000467 might be an ideal independent prognostic
factor (75). In addition, a
previous study reported that the determination of prognostic
factors in gastric cancer using circ_0000467 along with TNM stage
is more accurate compared to that using TNM stage alone (75).
The expression of circ_KIAA1244 in plasma of
patients with gastric cancer is lower than that in healthy patients
without gastric cancer, which is inversely associated with TNM
stage and lymph node metastasis, as well as a shorter overall
survival. This is supported by univariate and multivariate
analysis, which demonstrated that the expression of circ_KIAA1244
may be used as an independent prognostic indicator of the overall
survival in patients with gastric cancer (76).
More recently, with the development of molecular
biology and in-depth pathogenesis studies, the mechanistic function
of circRNAs in metabolic and signal transduction pathways has also
been investigated for gastric cancer. ciRS-7 possesses about 70
binding sites for miR-7 and has significant functions in several
types of tumors, including gastric cancer, where it is highly
expressed (77). ciRS-7 is able to
enhance proliferation and migration of gastric cancer cells by
upregulating PTEN/PI3K/AKT signaling pathway through sponging of
miR-7 (78). Overexpression of
ciRS-7 is also shown to promote the growth of xenograft tumors
in vivo exhibiting a promising therapeutic value for gastric
cancer (78). Therefore, developing
inhibitors that can efficiently target ciRS-7 may be an important
approach for therapy.
Highly expressed circ_0067997 promotes tumor growth
in gastric cancer by sponging miR-515-5p, which results in the
regulation of x-linked inhibitor of apoptosis (XIAP) expression
(79,80). Suppressing circ_0067997 by siRNA
blocks a variety of apoptosis-related pathways and reduces cell
viability, colony formation, proliferation and invasion of gastric
cancer cell lines in vitro. circ_0067997 siRNAs treated
MGC-803 cells inhibits the growth of tumors in vivo compared
with si-NC treated cells, also suggesting a potential therapeutic
target (81).
Finally, using Gene Ontology functional term and
Kyoto Encyclopedia of Genes and Genomes signaling pathway analyses,
gastric cancer was found to be linked to circRNAs in several
important physiological processes, such as extracellular matrix
degradation, binding to cell adhesion molecules and reaction with
transforming growth factor β (82).
Data from these studies demonstrated that circRNAs
may contribute to the occurrence and development of gastric cancer,
and its application in tumor therapy presents a valuable prospect.
circRNAs may also be used as a therapeutic target for inhibiting
tumor proliferation and metastasis.
Increasing evidence demonstrates that the expression
of circRNAs affect the development and progression of gastric
cancer and may exhibit great potential in diagnosis, prognosis and
treatment of gastric cancer. Research within the mechanistic
function of circRNAs is an emerging scientific field with an
enormous potential. The differential expression of circRNAs in
gastric cancer compared to non-cancer samples enables the use of
these circRNAs as potential biomarkers (41,83,84).
circRNAs are have a stable structure and are widely expressed in
various body fluids, including blood, saliva and gastric fluid
(9–12). In addition, RNA sequencing and
reverse transcription-quantitative PCR have made the detection of
circRNAs more convenient and accurate than proteins as detection
time is shorter, smaller amount of sample is required and the
sensitivity is higher. Numerous circRNAs are highly enriched in the
blood compared with the corresponding linear RNAs. Moreover, the
expression of circRNAs in cancer tissue is higher than in
precancerous tissue derived exosomes (85), which is consistent with another study
showing that the levels of circ-KIAA1244 in plasma and plasma
exosomes are similar (76).
Therefore, circRNAs in extracellular exosomes represent the ideal
candidates as reliable biomarkers for prognosis and diagnosis in
gastric cancer. However, at present, circRNAs can only be used in
combination with other biomarkers for cancer diagnosis, and the
clinical application of circRNAs still requires further
investigation and optimization. Although there are many studies
about circRNAs, there no current standardized methodology for
detecting circRNAs. Also, to the best of our knowledge there is no
detailed study about the application of circRNAs as biomarkers for
malignant cells, particularly regarding cost and the specificity
and sensitivity for detecting cancer cells.
The biological functions of several circRNAs may be
utilized to explore the underlying mechanism of circRNAs during the
development of gastric cancer. Despite many specific circRNAs
reportedly expressed in gastric cancer, the features of circRNAs,
including their biogenesis, degradation, location and function,
remain to be explored. Therefore, comprehensive studies are
required to determine the function and molecular mechanisms of
circRNAs in gastric cancer. Moreover, further understanding of the
relationship between circRNAs and gastric cancer may reveal the
complex regulatory networks of carcinogenesis and accelerate the
clinical application of circRNAs in prognosis, diagnosis and
therapy of gastric cancer.
In the present review, the biological functions and
molecular mechanisms of circRNAs were examined, and the clinical
values of circRNAs with an emphasis on gastric cancer were
summarized. Although numerous circRNAs have been identified, only a
small amount of functional circRNAs has been elucidated, and most
studies on circRNAs have focused on their regulation of cancer
development through interactions with miRNAs and RBPs. In the
future, the mechanistic study of its roles in genomic and mRNA
transcription needs to be further explored. As circRNAs are known
to be stable in different body fluids, including blood, circRNAs
may act as ideal biomarkers and therapeutic targets for gastric
cancer.
Not applicable.
This study was supported by grants from Shanghai
Tongren Hospital (grant no. 2019shtrxx09), The Project of Shanghai
Municipal Commission of Health and Family Planning (grant no.
20174Y0231), The National Natural Science Foundation of China
(grant no. 81672335), Shanghai Jiaotong University, Medical
Professionals Cross Fund (grant no. YG2016ZD10) and SJTU Research
Project Grants 2019, University of Sydney, Australia.
Not applicable.
CY and GQ conceived the study and wrote the
manuscript. LS generated the tables and diagrams. LM and SB revised
the manuscript for important intellectual content. All authors have
read and approved the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhu L, Qin J, Wang J, Guo T, Wang Z and
Yang J: Early gastric cancer: Current advances of endoscopic
diagnosis and treatment. Gastroenterol Res Pract. 2016:96380412016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huang RJ, Choi AY, Truong CD, Yeh MM and
Hwang JH: Diagnosis and management of gastric intestinal
metaplasia: Current status and future directions. Gut Liver.
15:596–603. 2019. View
Article : Google Scholar
|
|
4
|
Hentze MW and Preiss T: Circular RNAs:
Splicing's enigma variations. EMBO J. 32:923–925. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao
L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, et al: The
landscape of circular RNA in cancer. Cell. 176:869–881. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wilusz JE and Sharp PA: Molecular biology.
A circuitous route to noncoding RNA. Science. 340:440–441. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Broadbent KM, Broadbent JC, Ribacke U,
Wirth D, Rinn JL and Sabeti PC: Strand-Specific RNA sequencing in
Plasmodium falciparum malaria identifies developmentally regulated
long non-coding RNA and circular RNA. BMC Genomics. 16:4542015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lu T, Cui L, Zhou Y, Zhu C, Fan D, Gong H,
Zhao Q, Zhou C, Zhao Y, Lu D, et al: Transcriptome-Wide
investigation of circular RNAs in rice. RNA. 21:2076–2087. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lu Z, Filonov GS, Noto JJ, Schmidt CA,
Hatkevich TL, Wen Y, Jaffrey SR and Matera AG: Metazoan tRNA
introns generate stable circular RNAs in vivo. RNA. 21:1554–1565.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schmidt CA, Noto JJ, Filonov GS and Matera
AG: A method for expressing and imaging abundant, stable, circular
RNAs in vivo using tRNA splicing. Methods Enzymol. 572:215–236.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jin M, Shi C, Yang C, Liu J and Huang G:
Upregulated CircRNA ARHGAP10 Predicts an unfavorable prognosis in
NSCLC through regulation of the miR-150-5p/GLUT-1 axis. Mol Ther
Nucleic Acids. 18:219–231. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ye F, Gao G, Zou Y, Zheng S, Zhang L, Ou
X, Xie X and Tang H: CircFBXW7 inhibits malignant progression by
sponging miR-197-3p and encoding a 185-aa protein in
triple-negative breast cancer. Mol Ther Nucleic Acids. 18:88–98.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu K, Zhan H, Peng Y, Yang L, Gao Q, Jia
H, Dai Z, Tang Z, Fan J and Zhou J: Plasma hsa_circ_0027089 is a
diagnostic biomarker for hepatitis B virus-related hepatocellular
carcinoma. Carcinogenesis. 19:1542019.
|
|
16
|
Chen J, Chen T, Zhu Y, Li Y, Zhang Y, Wang
Y, Li X, Xie X, Wang J, Huang M, et al: CircPTN sponges
miR-145-5p/miR-330-5p to promote proliferation and stemness in
glioma. J Exp Clin Cancer Res. 38:3982019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu G, Chen Y, Fu M, Zang X, Cang M, Niu Y,
Zhang W, Zhang Y, Mao Z, Shao M, et al: Circular RNA CCDC66
promotes gastric cancer progression by regulating c-Myc and TGF-β
signaling pathways. J Cancer. 11:2759–2768. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Song H, Xu Y, Xu T, Fan R, Jiang T, Cao M,
Shi L and Song J: CircPIP5K1A activates KRT80 and PI3K/AKT pathway
to promote gastric cancer development through sponging miR-671-5p.
Biomed Pharmacother. 126:1099412020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang L, Chang X, Zhai T, Yu J, Wang W, Du
A and Liu N: A novel circular RNA, circ-ATAD1, contributes to
gastric cancer cell progression by targeting miR-140-3p/YY1/PCIF1
signaling axis. Biochem Biophys Res Commun. 14:841–849. 2020.
View Article : Google Scholar
|
|
20
|
Hu K, Qin X, Shao Y, Zhou Y, Ye G and Xu
S: Circular RNA MTO1 suppresses tumorigenesis of gastric carcinoma
by sponging miR-3200-5p and targeting PEBP1. Mol Cell Probes.
56:1015622020. View Article : Google Scholar
|
|
21
|
Ren S, Liu J, Feng Y, Li Z, He L, Li L,
Cao X, Wang Z and Zhang Y: Knockdown of circDENND4C inhibits
glycolysis, migration and invasion by up-regulating miR-200b/c in
breast cancer under hypoxia. J Exp Clin Cancer Res. 38:3882019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu Q, Zhang W, Wu Z, Liu H, Hu H, Shi H,
Li S and Zhang X: Construction of a circular
RNA-microRNA-messengerRNA regulatory network in stomach
adenocarcinoma. J Cell Biochem. 12:1317–1331. 2019.
|
|
23
|
Bi J, Liu H, Dong W, Xie W, He Q, Cai Z,
Huang J and Lin T: Circular RNA circ-ZKSCAN1 inhibits bladder
cancer progression through miR-1178-3p/p21 axis and acts as a
prognostic factor of recurrence. Mol Cancer. 18:1332019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang H, Wei L, Qin T, Yang N, Li Z and Xu
Z: Circular RNA ciRS-7 triggers the migration and invasion of
esophageal squamous cell carcinoma via miR-7/KLF4 and NF-κB
signals. Cancer Biol Ther. 20:73–80. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chung DJ, Wang CJ, Yeh CW and Tseng TH:
Inhibition of the proliferation and invasion of C6 glioma cells by
tricin via the upregulation of focal-adhesion-kinase-targeting
MicroRNA-7. J Agric Food Chem. 66:6708–6716. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pan M, Li M, You C, Zhao F, Guo M, Xu H,
Li L, Wang L and Dou J: Inhibition of breast cancer growth via
miR-7 suppressing ALDH1A3 activity concomitant with decreasing
breast cancer stem cell subpopulation. J Cell Physiol.
235:1405–1416. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ye T, Yang M, Huang D, Wang X, Xue B, Tian
N, Xu X, Bao L, Hu H, Lv T and Huang Y: MicroRNA-7 as a potential
therapeutic target for aberrant NF-κB-driven distant metastasis of
gastric cancer. J Exp Clin Cancer Res. 38:552019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ling Y, Cao C, Li S, Qiu M, Shen G, Chen
Z, Yao F and Chen W: TRIP6, as a target of miR-7, regulates the
proliferation and metastasis of colorectal cancer cells. Biochem
Biophys Res Commun. 514:231–238. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang
C, Liu D, Wang M, Wang L, Zeng F and Jiang G: CircHIPK3 sponges
miR-558 to suppress heparanase expression in bladder cancer cells.
EMBO Rep. 18:1646–1659. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y,
Yang S, Zeng Z, Liao W, Ding YQ and Liang L: Emerging roles of
circRNA_001569 targeting miR-145 in the proliferation and invasion
of colorectal cancer. Oncotarget. 7:26680–26691. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Meng Q, Li S, Liu Y, Zhang S, Jin J, Zhang
Y, Guo C, Liu B and Sun Y: Circular RNA circSCAF11 accelerates the
glioma tumorigenesis through the miR-421/SP1/VEGFA axis. Mol Ther
Nucleic Acids. 17:669–677. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yan Y, Fu G, Ye Y and Ming L: Exosomes
participate in the carcinogenesis and the malignant behavior of
gastric cancer. Scand J Gastroenterol. 52:499–504. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Thomas LF and Sætrom P: Circular RNAs are
depleted of polymorphisms at microRNA binding sites.
Bioinformatics. 30:2243–2246. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhao ZJ and Shen J: Circular RNA
participates in the carcinogenesis and the malignant behavior of
cancer. RNA Biol. 14:514–521. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lu J, Wang YH, Huang XY, Xie JW, Wang JB,
Lin JX, Chen QY, Cao LL, Huang CM, Zheng CH and Li P: Circ-CEP85L
suppresses the proliferation and invasion of gastric cancer by
regulating NFKBIA expression via miR-942-5p. J Cell Physiol.
5:102020.
|
|
37
|
Li H, Wei X, Yang J, Dong D, Hao D, Huang
Y, Lan X, Plath M, Lei C, Ma Y, et al: CircFGFR4 promotes
differentiation of myoblasts via binding miR-107 to relieve its
inhibition of Wnt3a. Mol Ther Nucleic Acids. 11:272–283. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: Circrna biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zheng J, Liu X, Xue Y, Gong W, Ma J, Xi Z,
Que Z and Liu Y: TTBK2 circular RNA promotes glioma malignancy by
regulating miR-217/HNF1β/Derlin-1 pathway. J Hematol Oncol.
10:522017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang ZG, Awan FM, Du WW, Zeng Y, Lyu J,
Wu, Gupta S, Yang W and Yang BB: The Circular RNA interacts with
stat3, increasing its nuclear translocation and wound repair by
modulating Dnmt3a and miR-17 function. Mol Ther. 25:2062–2074.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu KS, Pan F, Mao XD, Liu C and Chen J:
Biological functions of circular RNAs and their roles in occurrence
of reproduction and gynecological diseases. Am J Transl Res.
11:1–15. 2019.PubMed/NCBI
|
|
42
|
Zhang Y, Liang W, Zhang P, Chen J, Qian H,
Zhang X and Xu W: Circular RNAs: Emerging cancer biomarkers and
targets. J Exp Clin Cancer Res. 36:1522017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of CircRNAs. Mol
Cell. 66:9–21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N(6)-methyladenosine. Cell Res. 27:626–641.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z
and Sharpless NE: Expression of linear and novel circular forms of
an INK4/ARF-associated non-coding RNA correlates with
atherosclerosis risk. PLoS Genet. 6:e10012332010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Abdelmohsen K, Panda AC, Munk R,
Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM,
Martindale JL and Gorospe M: Identification of HuR target circular
RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA
Biol. 14:361–369. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang Y and Wang Z: Efficient backsplicing
produces translatablecircular mRNAs. RNA. 21:172–179. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-Intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li F, Ma K, Sun M and Shi S:
Identification of the tumor-suppressive function of circular RNA
ITCH in glioma cells through sponging miR-214 and promoting linear
ITCH expression. Am J Transl Res. 10:1373–1386. 2018.PubMed/NCBI
|
|
50
|
Ouyang Y, Li Y, Huang Y, Li X, Zhu Y, Long
Y, Wang Y, Guo X and Gong K: CircRNA circPDSS1 promotes the gastric
cancer progression by sponging miR-186-5p and modulating NEK2. J
Cell Physiol. 234:10458–10469. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Fry AM, O'Regan L, Sabir SR and Bayliss R:
Cell cycle regulation by the NEK family of protein kinases. J Cell
Sci. 125:4423–4433. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen J, Li Y, Zheng Q, Bao C, He J, Chen
B, Lyu D, Zheng B, Xu Y, Long Z, et al: Circular RNA profile
identifies circPVT1 as a proliferative factor and prognostic marker
in gastric cancer. Cancer Lett. 388:208–219. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liu H, Liu Y, Bian Z, Zhang J, Zhang R,
Chen X, Huang Y, Wang Y and Zhu J: Circular RNA YAP1 inhibits the
proliferation and invasion of gastric cancer cells by regulating
the miR-367-5p/p27 (Kip1) axis. Mol Cancer. 17:1512018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhang Y, Liu H, Li W, Yu J, Li J, Shen Z,
Ye G, Qi X and Li G: CircRNA_100269 is downregulated in gastric
cancer and suppresses tumor cell growth by targeting miR-630. Aging
(Albany NY). 9:1585–1594. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang L, Shen J and Jiang YL:
Circ_0027599/PHDLA1 suppresses gastric cancer progression by
sponging miR-101-3p.1. Cell Biosci. 8:582018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW,
Carmichael GG and Chen LL: Long noncoding RNAs with snoRNA ends.
Mol Cell. 48:219–230. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Han B, Chao J and Yao H: Circular RNA and
its mechanisms in disease: From the bench to the clinic. Pharmacol
Ther. 187:31–44. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Sun HD, Xu ZP, Sun ZQ, Zhu B, Wang Q, Zhou
J, Jin H, Zhao A, Tang WW and Cao XF: Down-Regulation of circPVRL3
promotes the proliferation and migration of gastric cancer cells.
Sci Rep. 8:101112018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei
P, Liu H, Xu J, Xiao F, Zhou H, et al: A peptide encoded by
circular form of LINC-PINT suppresses oncogenic transcriptional
elongation in glioblastoma. Nat Commun. 9:44752018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Huang H, Weng H and Chen J: m6A
Modification in coding and non-coding RNAs: Roles and therapeutic
implications in cancer. Cancer Cell. 37:270–288. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng
X, Xiong F, Guo C, Wu X, Li Y, et al: Emerging role of
tumor-related functional peptides encoded by lncRNA and circRNA.
Mol Cancer. 19:222020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Fang J, Hong H, Xue X, Zhu X, Jiang L, Qin
M, Liang H and Gao L: A novel circular RNA, circFAT1(e2), inhibits
gastric cancer progression by targeting miR-548g in the cytoplasm
and interacting with YBX1 in the nucleus. Cancer Lett. 442:222–232.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Warburg O, Wind F and Negelein E: The
metabolism of tumors in the body. J Gen Physiol. 8:519–530. 1927.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Kimmelman AC and White E: Autophagy and
tumor metabolism. Cell Metab. 25:1037–1043. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhang X, Wang S, Wang H, Cao J, Huang X,
Chen Z, Xu P, Sun G, Xu J, Lv J and Xu Z: Circular RNA circNRIP1
acts as a microRNA-149-5p sponge to promote gastric cancer
progression via the AKT1/mTOR pathway. Mol Cancer. 18:202019.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Heras-Sandoval D, Perez-Rojas JM,
Hernandez-Damian J and Pedraza-Chaverri J: The role of
PI3K/AKT/mTOR pathway in the modulation of autophagy and the
clearance of protein aggregates in neurodegeneration. Cell Signal.
26:2694–2701. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wu ZH, Lin C, Liu CC, Jiang WW, Huang MZ,
Liu X and Guo WJ: MiR-616-3p promotes angiogenesis and EMT in
gastric cancer via the PTEN/AKT/mTOR pathway. Biochem Biophys Res
Commun. 501:1068–1073. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhang H, Zhu L, Bai M, Liu Y, Zhan Y, Deng
T, Yang H, Sun W, Wang X, Zhu K, et al: Exosomal circRNA derived
from gastric tumor promotes white adipose browning by targeting the
miR-133/PRDM16 pathway. Int J Cancer. 144:2501–2515. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wei J, Wei W, Xu H, Wang Z, Gao W, Wang T,
Zheng Q, Shu Y and De W: Circular RNA hsa_circRNA_102958 may serve
as a diagnostic marker for gastric cancer. Cancer Biomark.
27:139–145. 2019. View Article : Google Scholar
|
|
70
|
Shao Y, Ye M, Li Q, Sun W, Ye G, Zhang X,
Yang Y, Xiao B and Guo J: LncRNA-RMRP promotes carcinogenesis by
acting as a miR-206 sponge and is used as a novel biomarker for
gastric cancer. Oncotarget. 7:37812–37824. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Park YM, Kim JK, Baik SJ, Park JJ, Youn YH
and Park H: Clinical risk assessment for gastric cancer in
asymptomatic population after a health check-up: An individualized
consideration of the risk factors. Medicine (Baltimore).
95:e53512016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Shao Y, Li J, Lu R, Li T, Yang Y, Xiao B
and Guo J: Global circular RNA expression profile of human gastric
cancer and its clinical significance. Cancer Med. 6:1173–1180.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Duffy MJ, Lamerz R, Haglund C, Nicolini A,
Kalousová M, Holubec L and Sturgeon C: Tumor markers in colorectal
cancer, gastric cancer and gastrointestinal stromal cancers:
European group on tumor markers 2014 guidelines update. Int J
Cancer. 134:2513–2522. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhao Q, Chen S, Li T, Xiao B and Zhang X:
Clinical values of circular RNA 0000181 in the screening of gastric
cancer. J Clin Lab Anal. 32:e223332018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Lu J, Zhang PY, Xie JW, Wang JB, Lin JX,
Chen QY, Cao LL, Huang CM, Li P and Zheng CH: Hsa_circ_0000467
promotes cancer progression and serves as a diagnostic and
prognostic biomarker for gastric cancer. J Clin Lab Anal.
33:e227262019. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Tang W, Fu K, Sun H, Rong D, Wang H and
Cao H: CircRNA microarray profiling identifies a novel circulating
biomarker for detection of gastric cancer. Mol Cancer. 17:1372018.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Yang W, Gu J, Wang X, Wang Y, Feng M, Zhou
D, Guo J and Zhou M: Inhibition of circular RNA CDR1as increases
chemosensitivity of 5-FU-resistant BC cells through up-regulating
miR-7. J Cell Mol Med. 23:3166–3177. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Pan H, Li T, Jiang Y, Pan C, Ding Y, Huang
Z, Yu H and Kong D: Overexpression of circular RNA ciRS-7 abrogates
the tumor suppressive effect of miR-7 on gastric cancer via
PTEN/PI3K/AKT signaling pathway. J Cell Biochem. 119:440–446. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhang H, Wang X, Huang H, Wang Y, Zhang F
and Wang S: Hsa_circ_0067997 promotes the progression of gastric
cancer by inhibition of miR-515-5p and activation of X
chromosome-linked inhibitor of apoptosis (XIAP). Artif Cells
Nanomed Biotechnol. 47:308–318. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Lencz T, Guha S, Liu C, Rosenfeld J,
Mukherjee S, De Rosse P, John M, Cheng L, Zhang C, Badner JA, et
al: Genome-Wide association study implicates NDST3 in schizophrenia
and bipolar disorder. Nat Commun. 4:27392013. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Sun J, Li J, Zhang W, Zhang J, Sun S, Li
G, Song H and Wan D: MicroRNA-509-3p inhibits cancer cell
proliferation and migration via upregulation of XIAP in gastric
cancer cells. Oncol Res. 25:455–461. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Dang Y, Ouyang X, Zhang F, Wang K, Lin Y,
Sun B, Wang Y, Wang L and Huang Q: Circular RNAs expression
profiles in human gastric cancer. Sci Rep. 7:90602017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao
F, Huang N, Yang X, Zhao K, Zhou H, et al: Novel role of FBXW7
circular RNA in repressing glioma tumorigenesis. J Natl Cancer
Inst. 110:32018. View Article : Google Scholar
|
|
84
|
Zhou C, Molinie B, Daneshvar K, Pondick
JV, Wang J, Van Wittenberghe N, Xing Y, Giallourakis CC and Mullen
AC: Genome-Wide maps of m6A circRNAs identify widespread and
cell-type-specific methylation patterns that are distinct from
mRNAs. Cell Rep. 20:2262–2276. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Li WH, Song YC, Zhang H, Zhou ZJ, Xie X,
Zeng QN, Guo K, Wang T, Xia P and Chang DM: Decreased expression of
Hsa_circ_00001649 in gastric cancer and its clinical significance.
Dis Markers. 2017:45876982017. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Li J, Zhen L, Zhang Y, Zhao L, Liu H, Cai
D, Chen H, Yu J, Qi X and Li G: Circ-104916 is downregulated in
gastric cancer and suppresses migration and invasion of gastric
cancer cells. Onco Targets Ther. 10:3521–3529. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Huang M, He YR, Liang LC, Huang Q and Zhu
ZQ: Circular RNA hsa_circ_0000745 may serve as a diagnostic marker
for gastric cancer. World J Gastroenterol. 23:6330–6338. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Li P, Chen H, Chen S, Mo X, Li T, Xiao B,
Yu R and Guo J: Circular RNA 0000096 affects cell growth and
migration in gastric cancer. Br J Cancer. 116:626–633. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Li T, Shao Y, Fu L, Xie Y, Zhu L, Sun W,
Yu R, Xiao B and Guo J: Plasma circular RNA profiling of patients
with gastric cancer and their droplet digital RT-PCR detection. J
Mol Med (Berl). 96:85–96. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Fang Y, Ma M, Wang J, Liu X and Wang Y:
Circular RNAs play an important role in late-stage gastric cancer:
Circular RNA expression profiles and bioinformatics analyses.
Tumour Biol. 39:13933838142017. View Article : Google Scholar
|
|
92
|
Li P, Chen S, Chen H, Mo X, Li T, Shao Y,
Xiao B and Guo J: Using circular RNA as a novel type of biomarker
in the screening of gastric cancer. Clin Chim Acta. 444:132–136.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Lu R, Shao Y, Ye G, Xiao B and Guo J: Low
expression of hsa_circ_0006633 in human gastric cancer and its
clinical significances. Tumour Biol. 39:13933854892017. View Article : Google Scholar
|
|
94
|
Sun H, Tang W, Rong D, Jin H, Fu K, Zhang
W, Liu Z, Cao H and Cao X: Hsa_circ_0000520, a potential new
circular RNA biomarker, is involved in gastric carcinoma. Cancer
Biomark. 21:299–306. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Xie Y, Shao Y, Sun W, Ye G, Zhang X, Xiao
B and Guo J: Downregulated expression of hsa_circ_0074362 in
gastric cancer and its potential diagnostic values. Biomark Med.
12:11–20. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Rong D, Dong C, Fu K, Wang H, Tang W and
Cao H: Upregulation of circ_0066444 promotes the proliferation,
invasion, and migration of gastric cancer cells. Onco Targets Ther.
11:2753–2761. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Zhong S, Wang J, Hou J, Zhang Q, Xu H, Hu
J, Zhao J and Feng J: Circular RNA hsa_circ_0000993 inhibits
metastasis of gastric cancer cells. Epigenomics. 10:1301–1313.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Wei H, Pan L, Tao D and Li R: Circular RNA
circZFR contributes to papillary thyroid cancer cell proliferation
and invasion by sponging miR-1261 and facilitating C8orf4
expression. Biochem Biophys Res Commun. 503:56–61. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Tian M, Chen R, Li T and Xiao B: Reduced
expression of circRNA hsa_circ_0003159 in gastric cancer and its
clinical significance. J Clin Lab Anal. 32:32018. View Article : Google Scholar
|
|
100
|
Chen S, Li T, Zhao Q, Xiao B and Guo J:
Using circular RNA hsa_circ_0000190 as a new biomarker in the
diagnosis of gastric cancer. Clin Chim Acta. 466:167–171. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Zhou LH, Yang YC, Zhang RY, Wang P, Pang
MH and Liang LQ: CircRNA_0023642 promotes migration and invasion of
gastric cancer cells by regulating EMT. Eur Rev Med Pharmacol Sci.
22:2297–2303. 2018.PubMed/NCBI
|
|
102
|
Sun H, Xi P, Sun Z, Wang Q, Zhu B, Zhou J,
Jin H, Zheng W, Tang W, Cao H and Cao X: Circ-SFMBT2 promotes the
proliferation of gastric cancer cells through sponging miR-182-5p
to enhance CREB1 expression. Cancer Manag Res. 10:5725–5734. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Shen F, Liu P, Xu Z, Li N, Yi Z, Tie X,
Zhang Y and Gao L: CircRNA_001569 promotes cell proliferation
through absorbing miR-145 in gastric cancer. J Biochem. 165:27–36.
2019. View Article : Google Scholar : PubMed/NCBI
|