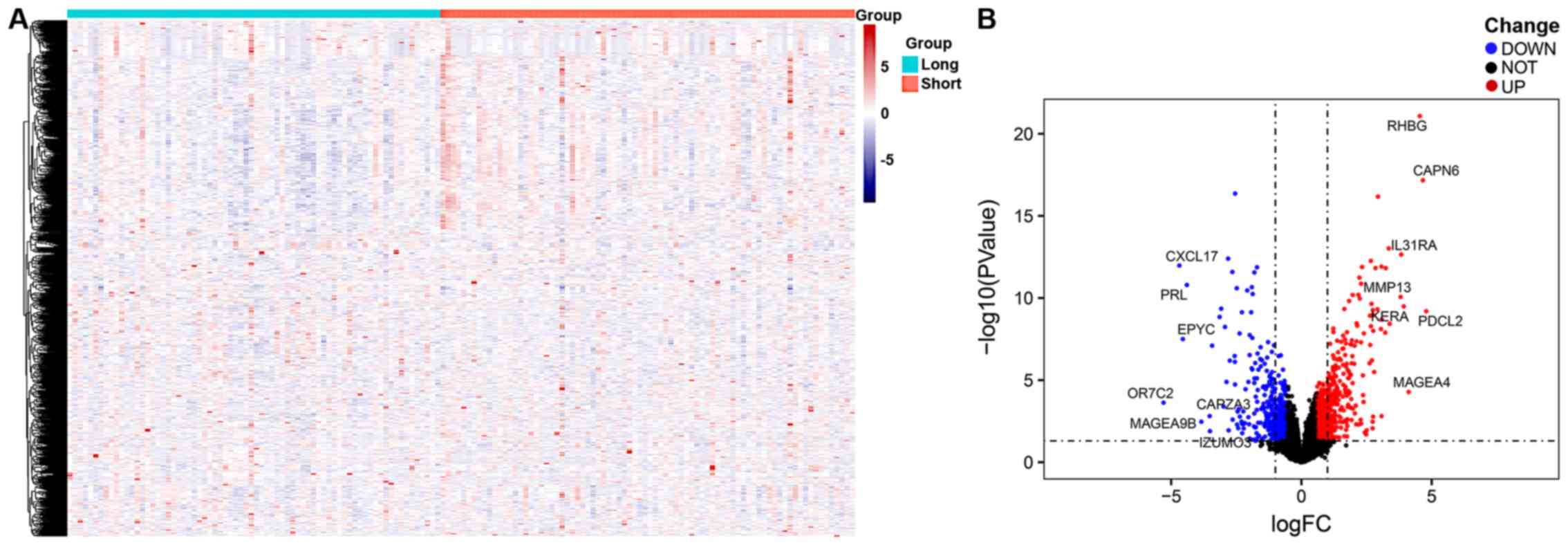
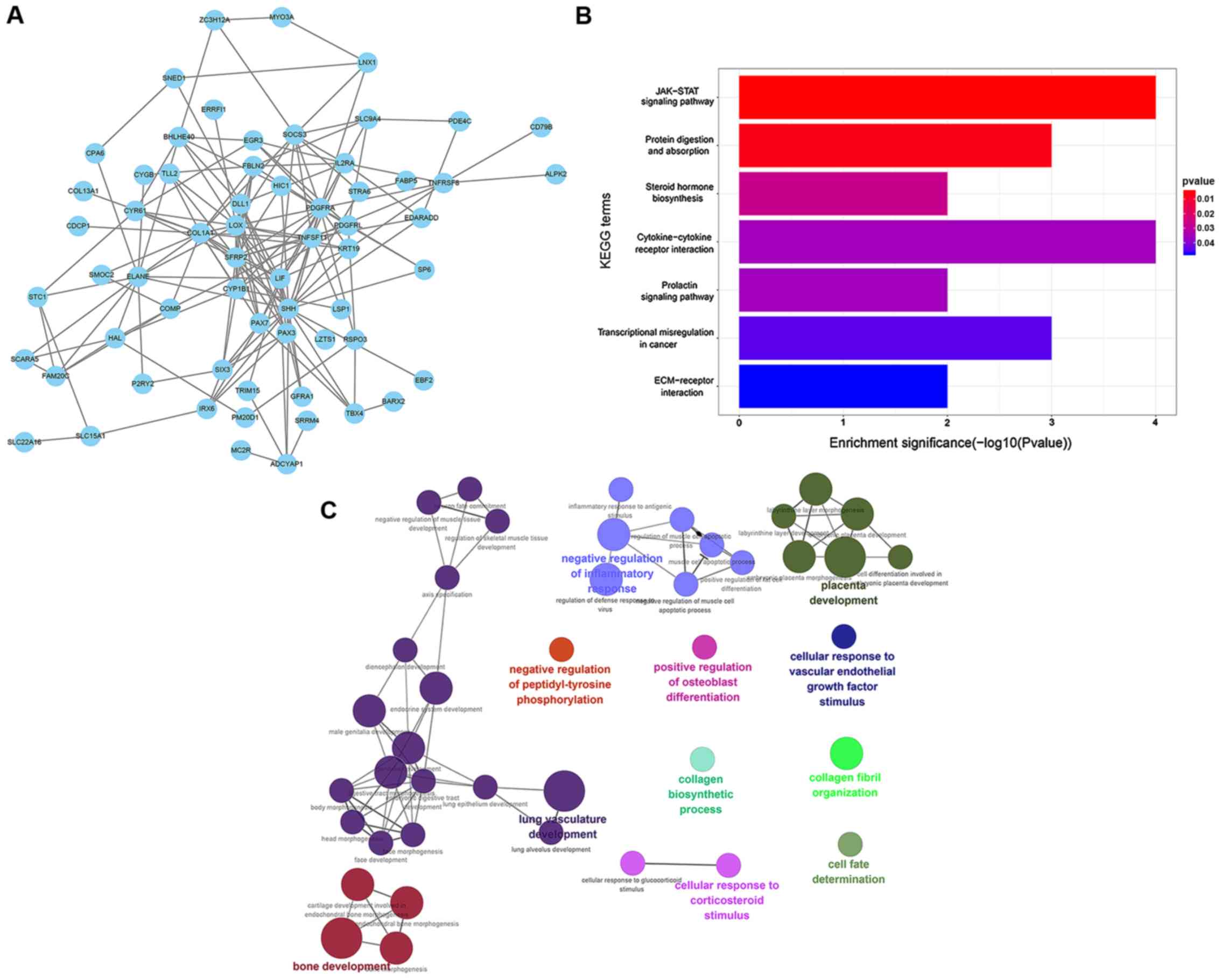
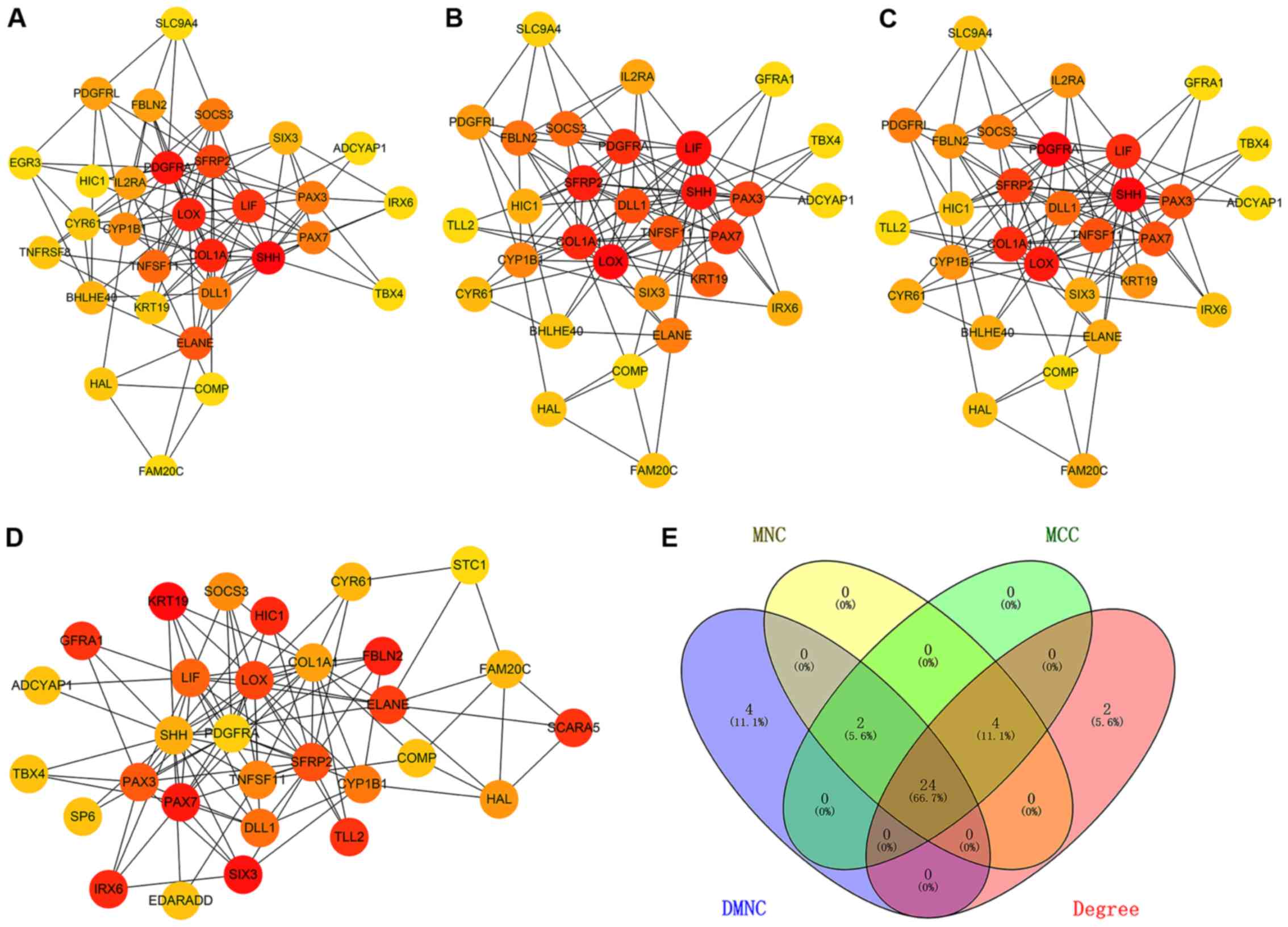
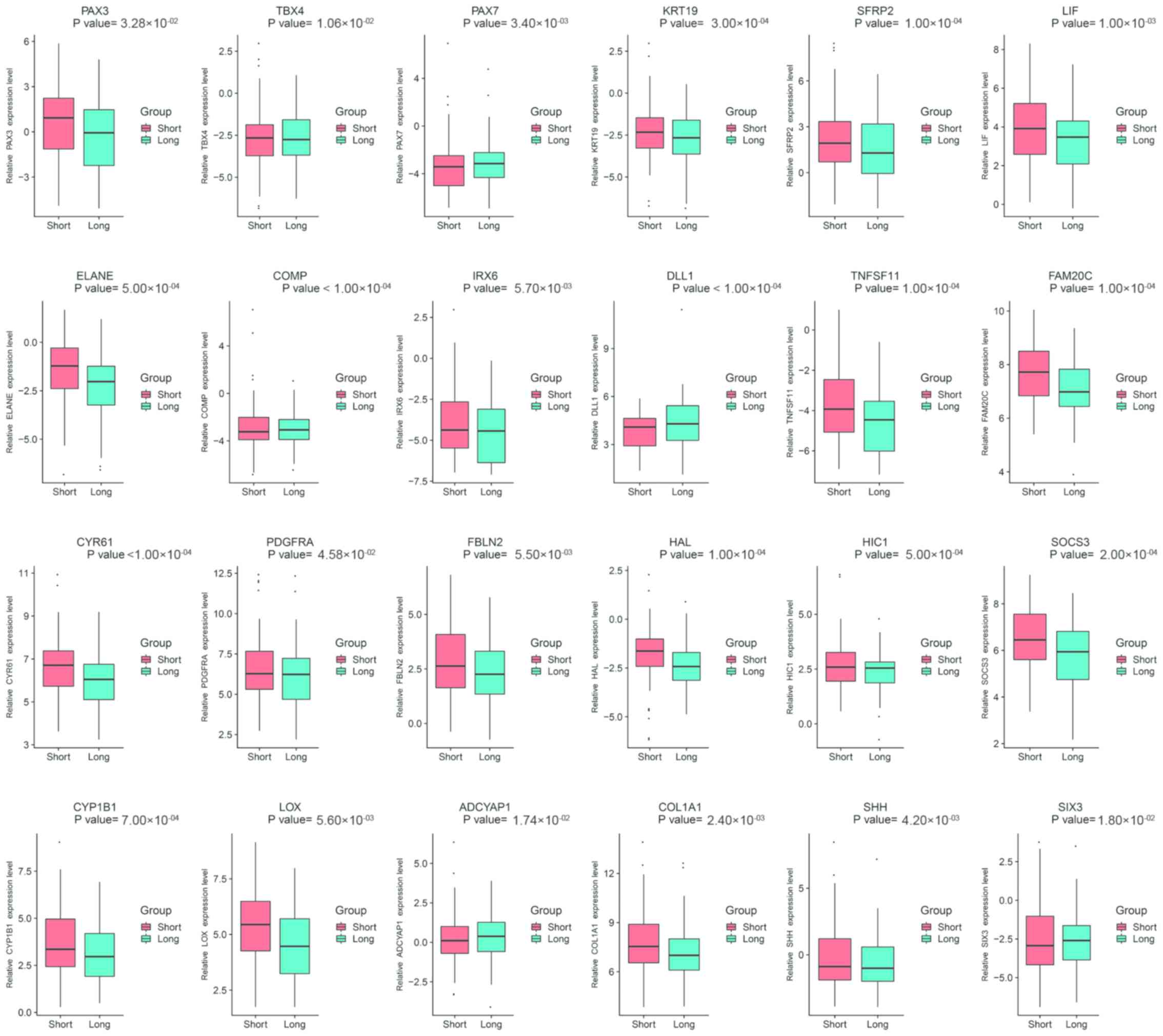
|
1
|
Jackson CM, Choi J and Lim M: Mechanisms
of immunotherapy resistance: Lessons from glioblastoma. Nat
Immunol. 20:1100–1109. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lu VM, Jue TR, McDonald KL and Rovin RA:
The survival effect of repeat surgery at glioblastoma recurrence
and its trend: A systematic review and meta-analysis. World
Neurosurg. 115:453–459. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brandao M, Simon T, Critchley G and Giamas
G: Astrocytes, the rising stars of the glioblastoma
microenvironment. Glia. 67:779–790. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Saadatpour L, Fadaee E, Fadaei S, Mansour
RN, Mohammadi M, Mousavi SM, Goodarzi M, Verdi J and Mirzaei H:
Glioblastoma: Exosome and microRNA as novel diagnosis biomarkers.
Cancer Gene Ther. 23:415–418. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brandes AA, Franceschi E, Paccapelo A,
Tallini G, Biase DD, Ghimenton C, Danieli D, Zunarelli E, Lanza G,
Silini EM, et al: Role of MGMT methylation status at time of
diagnosis and recurrence for patients with glioblastoma: Clinical
implications. Oncologist. 22:432–437. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jhanwar-Uniyal M, Amin AG, Cooper JB, Das
K, Schmidt MH and Murali R: Discrete signaling mechanisms of mTORC1
and mTORC2: Connected yet apart in cellular and molecular aspects.
Adv Biol Regul. 64:39–48. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jhanwar-Uniyal M, Wainwright JV, Mohan AL,
Tobias ME, Murali R, Gandhi CD and Schmidt MH: Diverse signaling
mechanisms of mTOR complexes: MTORC1 and mTORC2 in forming a
formidable relationship. Adv Biol Regul. 72:51–62. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Klughammer J, Kiesel B, Roetzer T,
Fortelny N, Nemc A, Nenning KH, Furtner J, Sheffield NC, Datlinger
P, Peter N, et al: The DNA methylation landscape of glioblastoma
disease progression shows extensive heterogeneity in time and
space. Nat Med. 24:1611–1624. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tan SK, Pastori C, Penas C, Komotar RJ,
Ivan ME, Wahlestedt C and Ayad NG: Serum long noncoding RNA HOTAIR
as a novel diagnostic and prognostic biomarker in glioblastoma
multiforme. Mol Cancer. 17:742018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li X and Diao H: Circular RNA circ_0001946
acts as a competing endogenous RNA to inhibit glioblastoma
progression by modulating miR-671-5p and CDR1. J Cell Physiol.
234:13807–13819. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao H, Shen J, Hodges TR, Song R, Fuller
GN and Heimberger AB: Serum microRNA profiling in patients with
glioblastoma: A survival analysis. Mol Cancer. 16:592017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shajani-Yi Z, de Abreu FB, Peterson JD and
Tsongalis GJ: Frequency of somatic TP53 mutations in combination
with known pathogenic mutations in colon adenocarcinoma, non-small
cell lung carcinoma, and gliomas as identified by next-generation
sequencing. Neoplasia. 20:256–262. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen X, Zhang M, Gan H, Wang H, Lee JH,
Fang D, Kitange GJ, He L, Hu Z, Parney IF, et al: A novel enhancer
regulates MGMT expression and promotes temozolomide resistance in
glioblastoma. Nat Commun. 9:29492018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kessler T, Sahm F, Sadik A, Stichel D,
Hertenstein A, Reifenberger G, Zacher A, Sabel M, Tabatabai G,
Steinbach J, et al: Molecular differences in IDH wildtype
glioblastoma according to MGMT promoter methylation. Neuro Oncol.
20:367–379. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cloughesy T, Finocchiaro G, Belda-Iniesta
C, Recht L, Brandes AA, Pineda E, Mikkelsen T, Chinot OL, Balana C,
Macdonald DR, et al: Randomized, double-blind, placebo-controlled,
multicenter phase II study of onartuzumab plus bevacizumab versus
placebo plus bevacizumab in patients with recurrent glioblastoma:
Efficacy, safety, and hepatocyte growth factor and
O6-Methylguanine-DNA methyltransferase biomarker analyses. J Clin
Oncol. 35:343–351. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Arantes LM, de Carvalho AC, Melendez ME,
Carvalho AL and Goloni-Bertollo EM: Methylation as a biomarker for
head and neck cancer. Oral Oncol. 50:587–592. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Anastasiadi D, Esteve-Codina A and
Piferrer F: Consistent inverse correlation between DNA methylation
of the first intron and gene expression across tissues and species.
Epigenetics Chromatin. 11:372018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Portela A and Esteller M: Epigenetic
modifications and human disease. Nat Biotechnol. 28:1057–1068.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li R, Qu H, Wang S, Wei J, Zhang L, Ma R,
Lu J, Zhu J, Zhong WD and Jia Z: GDCRNATools: An R/bioconductor
package for integrative analysis of lncRNA, miRNA and mRNA data in
GDC. Bioinformatics. 34:2515–2517. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Robinson MD and Oshlack A: A scaling
normalization method for differential expression analysis of
RNA-seq data. Genome Biol. 11:R252010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Weinhold L, Wahl S, Pechlivanis S,
Hoffmann P and Schmid M: A statistical model for the analysis of
beta values in DNA methylation studies. BMC Bioinformatics.
17:4802016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Robinson MD, McCarthy DJ and Smyth GK:
EdgeR: A bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tseng GC: Penalized and weighted K-means
for clustering with scattered objects and prior information in
high-throughput biological data. Bioinformatics. 23:2247–2255.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Morris TJ, Butcher LM, Feber A,
Teschendorff AE, Chakravarthy AR, Wojdacz TK and Beck S: ChAMP: 450
k chip analysis methylation pipeline. Bioinformatics. 30:428–430.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen YA, Lemire M, Choufani S, Butcher DT,
Grafodatskaya D, Zanke BW, Gallinger S, Hudson TJ and Weksberg R:
Discovery of cross-reactive probes and polymorphic CpGs in the
illumina infinium humanmethylation450 microarray. Epigenetics.
8:203–209. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Teschendorff AE, Marabita F, Lechner M,
Bartlett T, Tegner J, Cabrero DG and Beck S: A beta-mixture
quantile normalization method for correcting probe design bias in
illumina infinium 450 k DNA methylation data. Bioinformatics.
29:189–196. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45(D1): D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bindea G, Mlecnik B, Hackl H, Charoentong
P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z and
Galon J: ClueGO: A cytoscape plug-in to decipher functionally
grouped gene ontology and pathway annotation networks.
Bioinformatics. 25:1091–1093. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Woehrer A, Bauchet L and Barnholtz-Sloan
JS: Glioblastoma survival: Has it improved? Evidence from
population-based studies. Curr Opin Neurol. 27:666–674.
2014.PubMed/NCBI
|
|
34
|
Ramos AR, Elong Edimo W and Erneux C:
Phosphoinositide 5-phosphatase activities control cell motility in
glioblastoma: Two phosphoinositides PI(4,5)P2 and PI(3,4)P2 are
involved. Adv Biol Regul. 67:40–48. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Etcheverry A, Aubry M, de Tayrac M,
Vauleon E, Boniface R, Guenot F, Saikali S, Hamlat A, Riffaud L,
Menei P, et al: DNA methylation in glioblastoma: Impact on gene
expression and clinical outcome. BMC Genomics. 11:7012010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Smith AA, Huang YT, Eliot M, Houseman EA,
Marsit CJ, Wiencke JK and Kelsey KT: A novel approach to the
discovery of survival biomarkers in glioblastoma using a joint
analysis of DNA methylation and gene expression. Epigenetics.
9:873–883. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Senft C, Priester M, Polacin M, Schröder
K, Seifert V, Kögel D and Weissenberger J: Inhibition of the
JAK-2/STAT3 signaling pathway impedes the migratory and invasive
potential of human glioblastoma cells. J Neurooncol. 101:393–403.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mukthavaram R, Ouyang X, Saklecha R, Jiang
P, Nomura N, Pingle SC, Guo F, Makale M and Kesari S: Effect of the
JAK2/STAT3 inhibitor SAR317461 on human glioblastoma tumorspheres.
J Transl Med. 13:2692015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Park AK, Kim P, Ballester LY, Esquenazi Y
and Zhao Z: Subtype-specific signaling pathways and genomic
aberrations associated with prognosis of glioblastoma. Neuro Oncol.
21:59–70. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nawaz Z, Patil V, Paul Y, Hegde AS,
Arivazhagan A, Santosh V and Somasundaram K: PI3 kinase pathway
regulated miRNome in glioblastoma: Identification of miR-326 as a
tumour suppressor miRNA. Mol Cancer. 15:742016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Agrawal R, Garg A, Benny Malgulwar P,
Sharma V, Sarkar C and Kulshreshtha R: P53 and miR-210 regulated
NeuroD2, a neuronal basic helix-loop-helix transcription factor, is
downregulated in glioblastoma patients and functions as a tumor
suppressor under hypoxic microenvironment. Int J Cancer.
142:1817–1828. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Park NI, Guilhamon P, Desai K, McAdam RF,
Langille E, O'Connor M, Lan X, Whetstone H, Coutinho FJ, Vanner RJ,
et al: ASCL1 reorganizes chromatin to direct neuronal fate and
suppress tumorigenicity of glioblastoma stem cells. Cell Stem Cell.
21:209–224. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
John S, Sivakumar KC and Mishra R:
Extracellular proton concentrations impacts ln229 glioblastoma
tumor cell fate via differential modulation of surface lipids.
Front Oncol. 7:202017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mammoto T, Jiang A, Jiang E, Panigrahy D,
Kieran MW and Mammoto A: Role of collagen matrix in tumor
angiogenesis and glioblastoma multiforme progression. Am J Pathol.
183:1293–1305. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu J, Li W, Liu S, Zheng X, Shi L, Zhang
W and Yang H: Knockdown of collagen triple helix repeat containing
1 (CTHRC1) inhibits epithelial-mesenchymal transition and cellular
migration in glioblastoma cells. Oncol Res. 25:225–232. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ng SW, Mitchell A, Kennedy JA, Chen WC,
McLeod J, Ibrahimova N, Arruda A, Popescu A, Gupta V, Schimme AD,
et al: A 17-gene stemness score for rapid determination of risk in
acute leukaemia. Nature. 540:433–437. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lin B, Xue Y, Qi C, Chen X and Mao W:
Expression of transmembrane protein 41A is associated with
metastasis via the modulation of E-cadherin in radically resected
gastric cancer. Mol Med Rep. 18:2963–2972. 2018.PubMed/NCBI
|
|
48
|
Balbous A, Cortes U, Guilloteau K,
Villalva C, Flamant S, Gaillard A, Milin S, Wager M, Sorel N,
Guilhot J, et al: A mesenchymal glioma stem cell profile is related
to clinical outcome. Oncogenesis. 3:e912014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xie D, Yin D, Tong X, O'Kelly J, Mori A,
Miller C, Black K, Gui D, Said JW and Koeffler HP: Cyr61 is
overexpressed in gliomas and involved in integrin-linked
kinase-mediated akt and beta-catenin-TCF/Lef signaling pathways.
Cancer Res. 64:1987–1996. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jeansonne D, Pacifici M, Lassak A, Reiss
K, Russo G, Zabaleta J and Peruzzi F: Differential effects of
microRNAs on glioblastoma growth and migration. Genes (Basel).
4:46–64. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Peñuelas S, Anido J, Prieto-Sánchez RM,
Folch G, Barba I, Cuartas I, Dorado DG, Poca MA, Sahuquillo J,
Baselga J and Seoane J: TGF-beta increases glioma-initiating cell
self-renewal through the induction of LIF in human glioblastoma.
Cancer Cell. 15:315–327. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chi KC, Tsai WC, Wu CL, Lin TY and Hueng
DY: An adult drosophila glioma model for studying pathometabolic
pathways of gliomagenesis. Mol Neurobiol. 56:4589–4599. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Xia L, Huang Q, Nie D, Shi J, Gong M, Wu
B, Gong P, Zha L, Zuo H, Ju S, et al: PAX3 is overexpressed in
human glioblastomas and critically regulates the tumorigenicity of
glioma cells. Brain Res. 1521:68–78. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhu H, Wang H, Huang Q, Liu Q, Guo Y, Lu
J, Li X, Xue C and Han Q: Transcriptional repression of p53 by PAX3
contributes to gliomagenesis and differentiation of glioma stem
cells. Front Mol Neurosci. 11:1872018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Götze S, Wolter M, Reifenberger G, Müller
O and Sievers S: Frequent promoter hypermethylation of Wnt pathway
inhibitor genes in malignant astrocytic gliomas. Int J Cancer.
126:2584–2593. 2010.PubMed/NCBI
|
|
56
|
Stricker SH, Feber A, Engström PG, Carén
H, Kurian KM, Takashima Y, Watts C, Way M, Dirks P, Bertone P, et
al: Widespread resetting of DNA methylation in
glioblastoma-initiating cells suppresses malignant cellular
behavior in a lineage-dependent manner. Genes Dev. 27:654–669.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Martini M, Pallini R, Luongo G, Cenci T,
Lucantoni C and Larocca LM: Prognostic relevance of SOCS3
hypermethylation in patients with glioblastoma multiforme. Int J
Cancer. 123:2955–2960. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wirsching HG, Galanis E and Weller M:
Glioblastoma. Handb Clin Neurol. 134:381–397. 2016. View Article : Google Scholar : PubMed/NCBI
|