Introduction
Based on an analysis of data from the World Health
Organization, breast cancer is the most common type of cancer and
affects ~2.1 million women each year (1). In 2018, it is estimated that ~634,000
women worldwide died of breast cancer, accounting for ~15% of all
cancer-associated deaths among women (1). While early-stage breast cancer has been
treated with relatively high success rates, advanced stage breast
cancer remains a challenging disease to manage due to limitations
of currently available therapies (2). To decrease the number of deaths, the
development of new potent anticancer drugs is necessary. However,
most chemotherapeutics used to treat cancer show cytotoxicity to
both normal cells and cancer cells (3). Therefore, new drugs are required with
reduced cytotoxicity towards normal cells and increased selectivity
for cancer cells (3).
There has been a growing interest in recent years in
the study of the anti-cancer activity of mesoionic compounds
(4), which contain a five-carbon
heterocyclic aromatic ring possessing a possessing sextet
configuration of electrons (5). The
heterocyclic ring is associated with a positive charge with a
counterbalancing negative charge covalently located on an attached
atom (5). This characteristic of a
mesoionic structure having distinct separation of positive and
negative charges configured on a heterocyclic ring structure,
suggests the capability of mesoionic compounds to interact with
biological polymers such as carbohydrates, lipids, nucleic acids,
and proteins (6). While mesoionic
compounds have internal positive and negative charges, their
overall neutral charge allows them to cross the cell membranes.
The potential use of these compounds as cancer
chemotherapeutic agents has been investigated primarily in the
context of melanoma (7,8). Treatment of melanoma cells with a
1,3,4-thiadiazolium mesoionic compound (MI-D) resulted in
significant decreases in viability and proliferation. MI-D acts as
an uncoupler of oxidative phosphorylation in the mitochondria by
inhibiting the electron transport chain between complex II and
complex III, resulting in a collapse of the transmembrane proton
gradient and stimulating ATPase activity in the mitochondria
(9). This disruption in
mitochondrial energy appears to be associated with increased
membrane permeability and fluidity (10).
Sydnones are mesoionic compounds containing a
1,2,3-oxadiazole core with a keto group in the 5 position. A series
of sydnone derivatives were synthesized by Dunkley and Thoman
(11). In screening a panel of
cancer cell lines (MCF-7, NCI-H460, and SF-268 cells), an
N-(4′-F-3′-nitrophenyl) sydnone was shown to exhibit significant
cytotoxic activity (11). The
anticancer activity of another sydnone compound, Sydnone 1 (SYD-1),
was tested in vitro (12) and
in vivo in using a rat Walker-256 carcinosarcoma model
(13). Treatment of rats with SYD-1
decreased tumor volumes and tumor weight compared with the
untreated animals (13).
Investigation of the underlying mechanism of action of SYD-1
suggested that the anticancer activity of mesoionic compounds may
be associated with changes in mitochondrial metabolism and
activation of apoptotic pathways resulting in tumor cell death
(12). The results of these studies
confirm the potential role of mesoionic compounds as anticancer
agents in the treatment of several types of cancer.
In our previous study, a 1,3-thiazolium-5-thiolate
derivative of a mesoionic compound, MIH 2.4Bl was synthesized
(14), and its potential selective
anti-cancer properties were assessed using a panel of breast cancer
cell lines and cells derived from normal human breast lineage.
Treatment with MIH 2.4Bl inhibited growth of most of the breast
cancer cell lines tested compared with normal human mammary
epithelial cells. A focus was placed on the MCF-7 cell line as it
is one of the most widely used cell lines as a model for
hormone-receptor-positive breast cancer both in vitro and
in vivo (15). The MCF-7 cell
line is associated with differentiated features such as hormone
receptor expression patterns typical of mammary epithelial cells
(16), and its characterization also
includes detailed transcriptome analysis (17,18).
Additionally, the extensive body of published literature on MCF-7
cells provides contextual relevance in studying breast cancer
biology and drug development (15,16).
Treatment of MCF-7 cells with MIH 2.4Bl resulted in alterations in
cell cycle distribution with an increased proportion of cells in
the G2/M phase compared with untreated cells. MCF-7 cells treated
with MIH 2.4Bl also showed morphological changes consistent with
apoptotic cell death, a finding confirmed by the results of a TUNEL
assay.
The present study was designed to investigate
further how the mesoionic compound MI 2.4Bl induced cell death in
MCF-7 cells and explore possible underlying mechanisms of action.
One such mechanism associated with mesoionic compounds is its
effects on mitochondrial function (19), an organelle that has been
functionally targeted in cancer treatment (20). Importantly, cells have a mechanism to
protect against dysfunctional mitochondria, which can be
deleterious to normal function. This mechanism, known as autophagy,
involves the selective segregation and subsequent degradation of
defective mitochondria before the onset of programmed cell death
(21). In cancer, the autophagic
process can be both pro- and anti-apoptotic, dependent on external
stimuli and alterations in the microenvironment. Based on these
considerations, the aim of the present study was to assess the
cytotoxic effects of MIH 2.4 Bl on the mitochondria in MCF-7 cells,
to understand the effects of the compound on autophagic cell death
and to identify the genes involved in the process of cell death and
proliferation following treatment.
Materials and methods
Synthesis of MIH 2.4Bl
All reagents and solvents for the synthesis of MIH
2.4Bl were purchased from Sigma-Aldrich; Merck KGaA, and used
without further purification. The method for synthesis of the free
base mesoionic compound MIH 2.4Bl [chemical formula,
2-(4-chlorophenyl)-3-methyl
4-(4-methylphenyl)-1,3-thiazolium-5-thiolate], has been previously
described (22). The chemical
identity of the product was confirmed as previously described
(22) by Fourier-transform infrared
spectroscopy using an IFS 66 instrument (Bruker Corp.) and nuclear
magnetic resonance spectroscopy using an Avance Ultrashield NMR
spectrophotometer (Bruker Corporation) at 300 MHz for 1H
and 75 MHz for 13C.
Cell line and culture of cells
MCF-7 cells were obtained from American Type Culture
Collection (ATCC) and maintained in DMEM (Genesee Scientific)
supplemented with 10% FBS (Gemini Bio-Products), 1% non-essential
amino acids (Gemini Bio-Products), and 1% antibiotic/antimycotic
solution (Gemini Bio-Products) containing 100 IU/ml penicillin, 100
µg/ml streptomycin and 25 µg/ml amphotericin B. Primary human
mammary epithelial cells (HMECs) were obtained from ATCC, and
cultured in Mammary Epithelial Cell Basal Medium (ATCC)
supplemented with a Mammary Epithelial Cell Growth kit (ATCC)
containing 5 µg/ml hH-insulin, 6 mM L-glutamine, 0.5 µM
epinephrine, 5 µg/ml apo-transferrin, 5 ng/ml rH-TGF-α, 0.4%
ExtractP and 100 ng/ml hydrocortisone hemisuccinate. MCF-10A cells
were obtained from ATCC and cultured in DMEM/Ham's F12 media
supplemented with 5% Equine Serum (Gemini Bio-Products), 20 ng/ml
epidermal growth factor (Sigma-Aldrich; Merck KGaA), 10 µg/ml
insulin (Sigma-Aldrich; Merck KGaA), 0.5 mg/ml hydrocortisone
(Sigma-Aldrich; Merck KGaA), 100 ng/ml cholera toxin
(Sigma-Aldrich; Merck KGaA) and 1% antibiotic/antimycotic solution.
All cells were maintained in 5% CO2 at 37°C in a
humidified incubator.
Cytotoxicity assay
For the evaluation of cytotoxicity, a crystal violet
assay was used as described previously (23). The crystal violet assay uses a
non-specific dye whose staining pattern is directly proportional to
the number of viable adherent cells. A total of 5×103
cells were seeded and cultured for 24 h in 96-well tissue culture
plates. Subsequently, the cells were treated with eight different
concentrations of MIH 2.4Bl: 150 µM and seven serial dilutions
(resulting in 75, 37.5, 18.8, 9.4, 4.7, 2.3, and 1.2 µM,
respectively). As a positive control, cells were treated with 1.34
µM doxorubicin, a concentration representing ~x10 the reported
IC50 in MCF-7 cells (24–26).
Cells were treated with vehicle (DMSO) alone at 0.1% (v/v) in media
as a negative control. The cells were treated at each concentration
using eight replicate wells for 72 h. At each time point, the media
was removed, and the wells were washed twice with Dulbecco's PBS
(Gemini Bio-Products). Subsequently, the plates were gently
inverted on filter paper to remove any remaining liquid and dried
at room temperature. The dried cells were fixed with 20% methanol
for 30 min at room temperature and stained with 0.5% w/v crystal
violet solution for 5 min at room temperature. The plates were
washed of excess crystal violet with deionized water, and the
washed plates were air-dried. Subsequently, the crystal violet
stained cells were solubilized with 200 µl of 95% ethanol/40 mM HCl
for 30 min at room temperature. The optical density (OD) of each
well was measured at 595 nm using a SpectraMax 190 absorbance
microplate reader (Molecular Devices, LLC).
Oxygen consumption rate (OCR)
measurements
OCR measurements were performed using a Seahorse
XF96 Extracellular Flux Analyzer (Seahorse Bioscience). MCF-7 cells
were initially seeded at a density of 5×103 cells per
well into Seahorse 96-well microplates for 24 h. Subsequently, the
cells were treated for 24 h with MIH 2.4Bl using the eight
aforementioned concentrations, and the same positive and negative
controls were used as described above. After treatment, the cell
media was changed to unbuffered DMEM containing 20 mM carnosine and
2.0 mM sodium pyruvate. The MCF-7 cells were maintained at 37°C
under normal atmospheric conditions. After baseline measurements,
OCR was measured following the addition of 1 µM oligomycin, then 1
µM FCCP, and finally 1 µM rotenone/5 µM antimycin A to each well.
After baseline measurements, the ATP-linked OCR was determined as
the measurements after inhibition of ATP synthase by oligomycin.
FCCP (an uncoupler of mitochondrial oxidative phosphorylation) was
added to determine maximal respiration. The maximal respiration was
normalized by subtracting the measurement of the non-mitochondrial
respiration, which was identified by addition of rotenone/antimycin
A. The basal respiration was subtracted from the maximal
respiration to obtain the reserve respiratory capacity. OCR data
was reported in units of picomoles per minute and was generated
using the Wave Desktop and Controller Software (version 2.6.1;
Seahorse Bioscience).
Western blot analysis
MCF-7 cells were seeded in 60 mm tissue culture
dishes at a density of 3×106 cells/dish for 24 h.
Subsequently, the cells were treated for 48 or 72 h with 75 µM MIH
2.4Bl. The MCF-7 cells were also treated with vehicle (DMSO) alone
at 0.1% (v/v) in media as a negative control. After each treatment
time point, the cells were harvested by scraping into ice-cold RIPA
buffer (composed of 50 mM Tris, pH 8.0, 150 mM NaCl, 1%NP-40, 0.5%
deoxycholate, and 0.1% sodium dodecyl sulfate,) containing a
cocktail of protease inhibitors (Pierce Protease Inhibitor Tablets;
Thermo Fisher Scientific, Inc.). Total protein concentrations of
the cell lysates were determined using a Bradford protein assay
(Bio-Rad Laboratories, Inc.). Normalized protein samples (20 µg
each) were denatured at 95°C using Laemmli sample buffer (Bio-Rad
Laboratories, Inc.), separated by electrophoresis on 4–20% gradient
polyacrylamide mini gels (Bio-Rad Laboratories, Inc.) and
transferred using a Trans-Blot Turbo transfer apparatus (Bio-Rad
Laboratories, Inc.) to PVDF membranes. The membranes were
subsequently incubated at room temperature for 1 h in blocking
buffer of TBS (composed of 50 mM Tris-Cl, pH 7.5 and 150 mM NaCl),
containing 0.01% Tween-20 (TBS-T) and 5% non-fat milk (w/v).
Subsequently, the membranes were washed three times using TBS-T for
5 min each. The membranes were incubated overnight with gentle
agitation at 4°C in 5 ml blocking buffer containing mouse
monoclonal anti-Beclin-1 (cat. no. 2A4 MA5-15825; 1:500;
Invitrogen; Thermo Fisher Scientific, Inc.), rabbit polyclonal
anti-ATG5 (cat. no. PA5-35201; 1:1,000; Invitrogen; Thermo Fisher
Scientific, Inc.), rabbit polyclonal anti-LC3 II/I (cat. no.
PA5-46286; 1:1,000; Invitrogen; Thermo Fisher Scientific, Inc.) or
mouse monoclonal anti-GAPDH antibody (cat. no. GA1R, MA5-15738;
1:1,000; Invitrogen; Thermo Fisher Scientific, Inc.). The membranes
were washed five times with TBS-T for 5 min each, and subsequently
incubated with a goat polyclonal horseradish peroxidase-conjugated
secondary anti-rabbit antibody (IgG; cat. no. A28177, 1:10,000;
Invitrogen; Thermo Fisher Scientific, Inc.) or goat polyclonal
anti-mouse antibody (IgG; cat. no. A28177; 1:10,000; Invitrogen;
Thermo Fisher Scientific, Inc.) for 30 min at room temperature.
Bands associated with specific proteins were detected using the
SuperSignal West PICO plus chemiluminescent substrate (Thermo
Fisher Scientific, Inc.) and analyzed using a ChemiDoc imaging
system (Bio-Rad Laboratories, Inc.). Densitometry analysis was
performed using Quantity One software (version 4.6.6; Bio-Rad
Laboratories, Inc.).
Total RNA isolation
MCF-7 cells were cultured in 6-well tissue culture
plates at a density of 1×106 cells per well and
incubated for 24 h. Subsequently, the cells were treated for 24 h
with 75 µM MIH 2.4Bl. As a control, cells were treated with vehicle
(DMSO) alone at 0.1% (v/v) in media. A Zymo Research Direct-Zol RNA
Purification kit (Zymo Research Corp.) was used to extract and
purify total RNA from each sample well for subsequent analysis. The
quantity and integrity of the RNA samples were assessed using an
Agilent Tape Station RNA Assay (Agilent Technologies, Inc.).
Microarray analysis
Total RNA samples were reverse transcribed using an
Affymetrix 3′IVT Express kit (Affymetrix; Thermo Fisher Scientific,
Inc.) with T7-Oligo(dT) primer, which contains a T7 RNA polymerase
promoter. Then, second-strand cDNA synthesis was performed and was
followed by antisense RNA (aRNA) amplification of the cDNA
templates prepared using biotinylated nucleotide analogs according
to the manufacturer's protocol. Biotin-labeled aRNA was fragmented
by incubating the samples for 35 min at 94°C in reaction buffer (40
mM Tris-acetate, pH 8.1, 100 mM potassium acetate and 30 mM
magnesium acetate). Subsequently, 15 µg of the fragmented aRNA was
hybridized at 45°C for 16 h to a Human Genome U133 Plus 2.0 Array
(Affymetrix; Thermo Fisher Scientific, Inc.) in a GeneChip
Hybridization Oven 645 (Thermo Fisher Scientific, Inc.). After the
hybridization was complete, the gene chips were automatically
washed and stained for expression detection with
streptavidin-phycoerythrin in a GeneChip Fluidics Station 450
(Thermo Fisher Scientific, Inc.) using the FS450_0004 protocol.
Finally, the probe arrays were scanned and processed using a
GeneChip Scanner 3000 7G (Thermo Fisher Scientific, Inc.).
Transcriptome Analysis Console version 4.0.2 (Thermo Fisher
Scientific, Inc.) was used to analyze the fold-change of each gene
based on the relative differences between signal intensities. Only
genes with a fold-change ≥2.0 were considered differentially
expressed genes (DEGs).
The Database for Annotation, Visualization and
Integrated Discovery (DAVID; david.abcc.ncifcrf.gov/) and analytics tools (27,28) was
used to map the DEGs to relevant pathway networks in the Gene
Ontology (GO; geneontology.org/) and Kyoto Encyclopedia of Genes and
Genomes (KEGG; genome.jp/kegg/pathway.html/) databases (29–31). The
P-values were calculated by DAVID using the Benjamini-Hochberg
method (32) to control the False
Discovery Rate, P<0.05 was considered to indicate a
statistically significant difference. The top ontologies in the GO
subsets (molecular functions, biological processes, and cellular
components), as well as the top KEGG pathways, were selected based
on a hierarchy of the P-values calculated.
Reverse transcription-quantitative
(RT-q)PCR
Expression levels of select genes identified as up
or downregulated following MIH 2.4Bl treatment compared with
vehicle (control) cells based on the microarray analysis were
quantified by RT-qPCR. A panel of four upregulated genes
[activating transcription factor 3 (ATF3), acidic repeat-containing
protein (ACRC), heparin-binding EGF-like growth factor (HBEGF),
regulator of G-protein signaling 2 (RGS2)] and two downregulated
genes [Dickkopf WNT signaling pathway inhibitor 1 (DKK1) and
adhesion molecule with Ig like domain 2 (AMIGO2)] was used and
compared to the expression of GAPDH as a control. A Verso 1-step
RT-qPCR kit (Thermo Fisher Scientific) was used to synthesize the
first-strand cDNA and perform quantitative PCR in a single step
assay according to the manufacturer's protocol. An Applied
Biosystems 7300 Real-Time PCR system (Thermo Fisher Scientific,
Inc.) was used to detect the transcripts using PrimePCR primers and
FAM-labeled probe mixtures (Bio-Rad Laboratories, Inc.). The
sequences of the primers and probes are not disclosed by the
manufacturer; however, target sequence information is presented in
Table I. The thermocycling
conditions for the qPCR reactions were: 15 min at 50°C for the cDNA
synthesis; initial denaturation at 95°C for 5 min; followed by 40
cycles of 95°C for 15 sec and 60°C for 60 sec. The comparative
quantification cycle (Cq) method was used to quantify the relative
changes in gene expression (33).
 | Table I.Target sequence identity of selected
genes used in reverse transcription-quantitative PCR analysis. |
Table I.
Target sequence identity of selected
genes used in reverse transcription-quantitative PCR analysis.
| Gene name | Gene symbol | Amplicon
length | Unique assay
ID |
|---|
| Activating
transcription factor 3 | ATF3 | 76 | qHsaCEP0053273 |
| Acidic
repeat-containing protein | ACRC | 149 | qHsaCIP0029745 |
| Adhesion molecule
with Ig like domain 2 | AMIGO2 | 96 | qHsaCEP0052159 |
| Dickkopf WNT
signaling pathway inhibitor 1 | DKK1 | 96 | qHsaCEP0050470 |
|
Gyceraldehyde-3-phosphate
dehydrogenase | GAPDH | 117 | qHsaCEP0041396 |
| Heparin-binding
EGF-like growth factor | HBEGF | 117 | qHsaCEP0050730 |
| Regulator of
G-protein signaling 2, 24 kDa | RGS2 | 112 | qHsaCEP0024158 |
Statistical analysis
The cell viability data were calculated from the
mean OD readings obtained for each concentration of the MIH 2.4Bl
mesoionic compound assayed normalized to the mean OD readings of
the negative control and mean blank values, and is expressed as the
mean percentage ± the standard error of the mean. The dose-response
curves were plotted and analyzed to calculate the IC50
values using a four parameters logistic (4PL) model for curve
fitting, in which a variable slope was employed that allowed
non-linear fitting of the equation. The IC50 values
(concentrations that reduced cell viability to 50% compared with
the negative control) were determined using nonlinear regression
analysis in the GraphPad Prism version 7.0 (GraphPad Software,
Inc.) and standard error of the mean was calculated from the cell
viability data. The mean percent inhibition values were plotted
against the MIH 2.4Bl concentrations allowing for the generation of
a sigmoidal dose-response curve using the following equation:
Y=100/[1+10^((LogIC50-X) × HillSlope))]. The
IC50 values were obtained by the interpolation of
curve-fit data from each curve using the GraphPad Prism.
Replicates of eight OCR measurements are expressed
as the mean ± the standard error of the mean. Data from RT-qPCR
analysis were calculated from three replicates. For the comparison
between data, a Student's t-test was used. P<0.05 was considered
to indicate a statistically significant difference.
Results
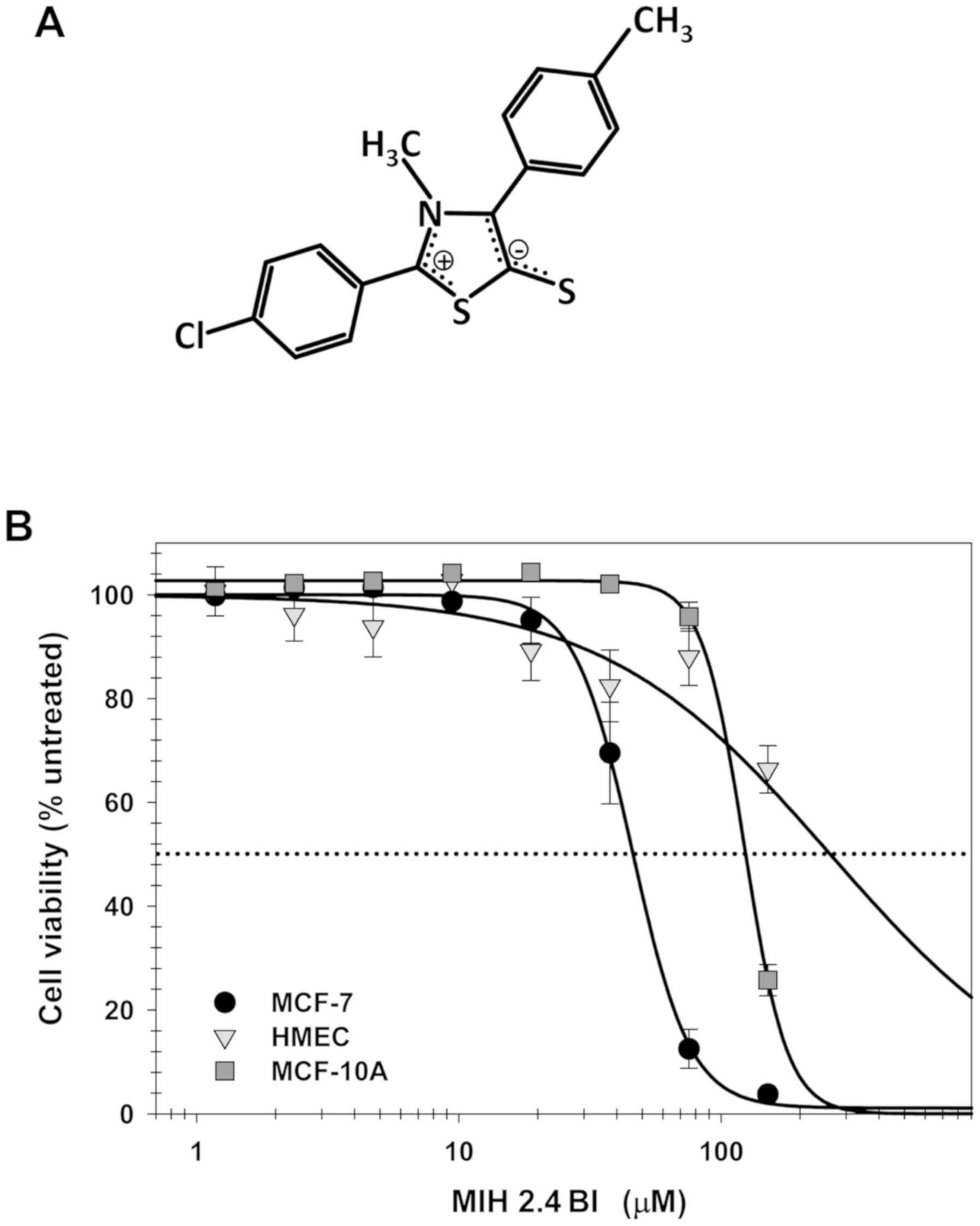
Effect of MIH 2.4Bl on cellular
cytotoxicity
To determine the cytotoxic effects of MIH 2.4Bl on
the MCF-7 cells, a crystal violet assay was used, based on the
simplicity of the assay and its suitability for examining the
effects of chemotherapeutics on cell survival and growth inhibition
(23). The dose-response curve
presented in Fig. 1 indicates that
MIH 2.4Bl decreased MCF-7 cell viability after 72 h treatment.
Based on the curve fitting, the IC50 for MIH 2.4Bl was
45.9±1.0 µM. MCF-10A (a spontaneously immortalized non-tumorigenic
cell line derived from benign proliferative breast tissue) required
a significantly higher concentration of MIH 2.4 Bl required for
inhibition of cell viability, with an IC50 of 123.8±1.0
µM. Primary HMEC only exhibited slight inhibitory activity
following MIH 2.4Bl treatment, with an IC50 of 917±19 µM
(Fig. 1). These results demonstrate
the potential cancer specificity of cytotoxic inhibition on MCF-7
cells mediated by MIH 2.4Bl.
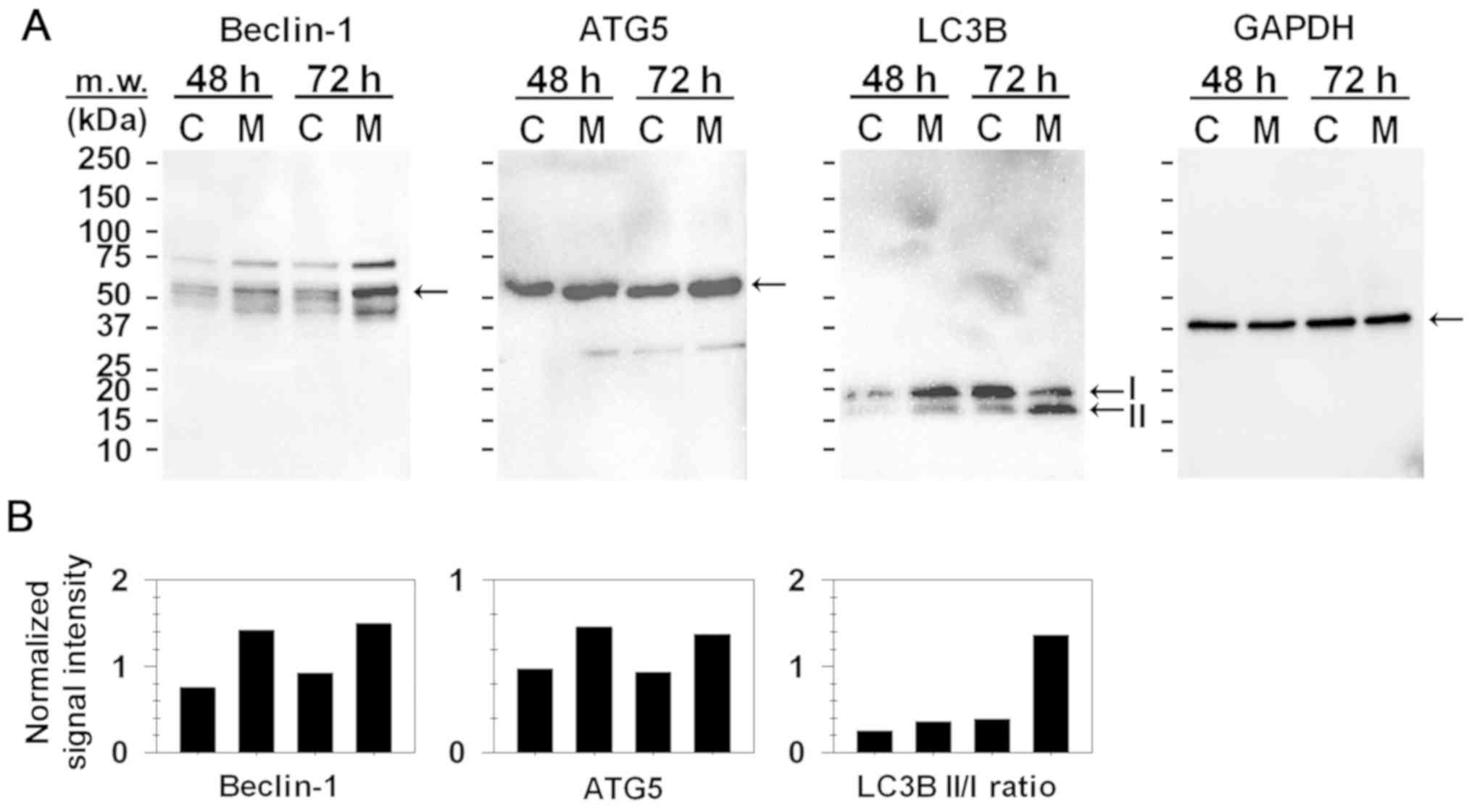
Effect of MIH 2.4Bl on the induction
of autophagy
To determine the significance of autophagy in the
treatment of MCF-7 cells with MIH 2.4Bl, the expression of Beclin-1
and ATG5 were assessed due to their role in autophagy. As shown in
Fig. 2A, western blotting analysis
showed that Beclin-1 and ATG5 expression were increased following
treatment with MIH 2.4Bl. Microtubule-associated protein 1A/1B
light chain 3B (LC3B) is a specific autophagy marker widely
utilized for measuring the autophagy level in cells and tissues. As
shown in Fig. 2A, both LC3B-I and
LC3B-II were present as 18 and 16 kDa bands, respectively. LC3B-I
is the cytosolic precursor form of LC3B-II, whereas LC3B-II is an
active form conjugated with phosphatidylethanolamine and is
membrane-bound. A widely accepted indicator of the number of
autophagosomes is an increase in the LC3B-II to LC3B-I ratio
(34). As shown in Fig. 2B, there was a significant increase in
the LC3B-II to LC3B-I ratio in MCF-7 cells treated for 72 h with
MIH 2.4Bl. Although the LC3B levels increased in control cells
between 48 and 72 h, there was no increase in the ratio of LC3B-II
to LC3B-I. These results suggest that induction of autophagy is a
potential mechanism of cell death mediated by MIH 2.4Bl.
Effect of MIH 2.4Bl on mitochondrial
OCR
Tumor cells are highly dependent on glycolytic
metabolism since this bioenergetic pathway produces energy and
metabolites capable of supporting the tumor's high growth and
proliferation rates relatively quickly (35). Thus, measuring changes in
mitochondrial OCR is a reliable approach for evaluating cellular
energy metabolism (36). Therefore,
the mitochondrial OCR was measured in MCF-7 cells following
treatment with increasing concentrations of MIH 2.4 from 4.7–75.0
µM for 24 h. Untreated MCF-7 cells were used as the negative
control, and doxorubicin treated MCF-7 cells (at a concentration of
1.5 µM) were used as the positive control. The initial
mitochondrial OCR was measured using a Seahorse XF96 analyzer and
after sequential addition of oligomycin, FCCP, and a
rotenone/antimycin A combination (Fig.
3A). These additions allowed for the determination of the
following mitochondrial parameters: Basal respiration, maximal
respiration, ATP production, proton leak, reserve respiratory
capacity, and non-mitochondrial respiration. As shown in Fig. 3B, treatment of MCF-7 cells with
increasing concentrations of MIH 2.4Bl resulted in a significant
reduction in all the mitochondrial respiratory parameters compared
with the control cells, indicative of an overall decrease in
mitochondrial membrane potential. At 75.0 µM MIH 2.4Bl treatment,
almost all mitochondrial respiration was abolished, leaving only
residual oxygen consumption (non-mitochondrial respiration). These
results highlight the potential of therapeutic targeting of
mitochondrial metabolism using MIH 2.4Bl for cancer therapy.
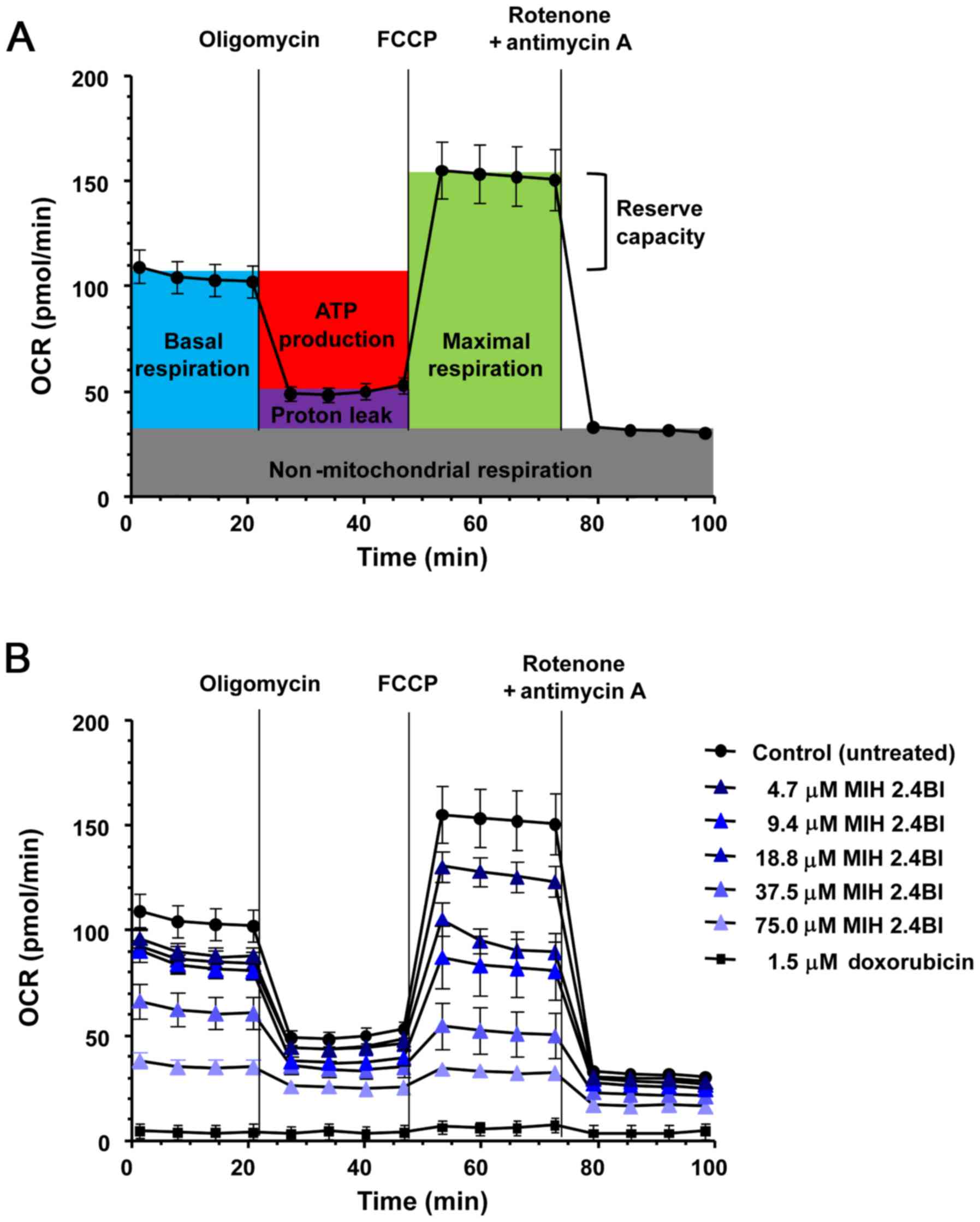 | Figure 3.Dose-dependent effects of MIH 2.4Bl
on the OCR of MCF-7 cells. (A) Representative schematic of OCR
using specific inhibitors to assess mitochondrial function. After
four initial measurements to acquire the baseline OCR, sequential
injections of oligomycin, FCCP, and a rotenone/antimycin A mixture
were performed with four OCR measurements obtained after each
injection. Shown in the OCR profile are calculations of basal
respiration, maximal respiration, ATP production, proton leak,
reserve respiratory capacity and non-mitochondrial respiration. (B)
OCR profiles were determined in MCF-7 cells after treatment for 24
h with 4.7, 9.4, 18.8, 37.7 or 75.4 µM MIH 2.4Bl. Treatment with
1.5 µM doxorubicin for 24 h was used as a positive control for
maximum cytotoxic effect. Each data point represents the mean ± the
standard error of the mean of eight replicates. OCR, oxygen
consumption rate. |
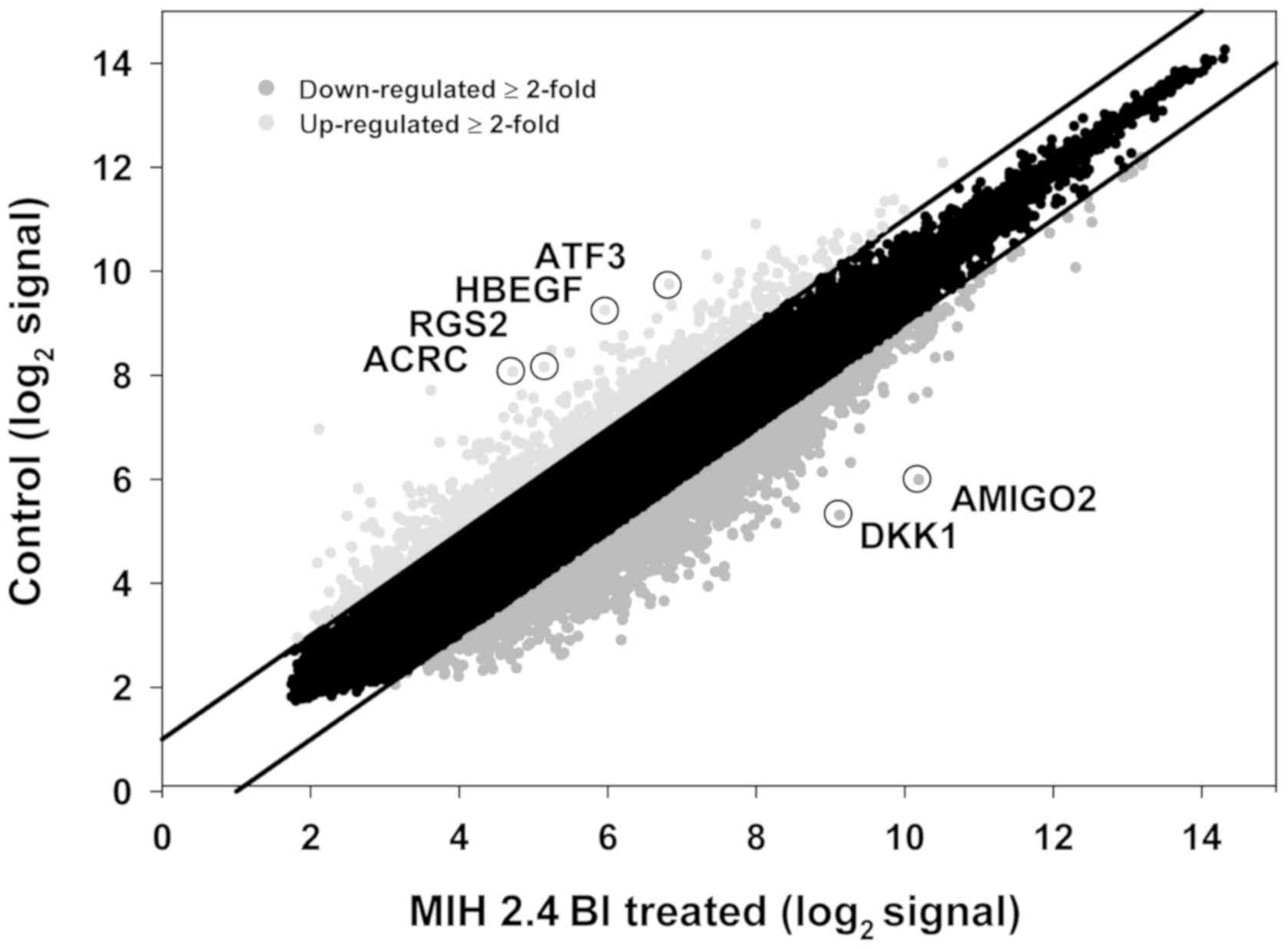
Analysis of DEGs in MCF-7 cells
treated with MIH 2.4Bl
A DNA microarray analysis of mRNA levels in MCF-7
cells was performed following treatment with MIH 2.4Bl to determine
changes in global genome-wide gene expression patterns. Microarray
analysis of mRNA isolated from control and MIH 2.4Bl treated cells
was performed using human Affymetrix Human Genome U133 Plus 2.0
Arrays. A scatter plot of the mean of the normalized gene
expression of RNA from MIH 2.4Bl treated cells vs. RNA from control
cells shows the DEGs (Fig. 4). A
total of 3,659 DEGs were detected between the two groups with a
fold-change ≥2.0. Among these genes, 779 were upregulated and 2,880
were downregulated in cells treated with MIH 2.4Bl compared with
the control cells (Tables SI and
SII). There were 144 genes detected
with a fold-change ≥5.0, including 37 upregulated genes and 107
downregulated genes. Based on the identity of the transcripts and
fold-change of expression, 6 genes were selected for verification
by RT-qPCR, as shown in Table
II.
 | Table II.Identity of selected genes
differentially regulated by MIH 2.4Bl treatment. |
Table II.
Identity of selected genes
differentially regulated by MIH 2.4Bl treatment.
| Probe Set ID | Fold-change | Gene symbol | Description | Transcript ID |
|---|
| 238825_at | 10.2 | ACRC | Acidic repeat
containing | Hs.135167.0 |
| 202672_s_at | 7.5 | ATF3 | Activating
transcription factor 3 | g4502262 |
| 203821_at | 9.8 | HBEGF | Heparin-binding
EGF-like growth factor | g4503412 |
| 202388_at | 8.1 | RGS2 | Regulator of
G-protein signaling 2 | g4506516 |
| 222108_at | −18.3 | AMIGO2 | Adhesion molecule
with Ig-like domain 2 | Hs.121520.0 |
| 204602_at | −14.0 | DKK1 | Dickkopf WNT
signaling pathway inhibitor 1 | g7110718 |
RT-qPCR analysis
The differences in the expression of these 6 genes
between treated and untreated cells is shown in Fig. 5. RT-qPCR analysis showed consistent
results with the results of the microarray analysis. ACRC, ATF3,
HBEGF and RGS2 genes were significantly upregulated in cells
treated with MIH 2.4Bl cells compared with the control cells. In
contrast, expression levels of AMIGO2 and DKK1 genes were
significantly downregulated in MIH 2.4Bl treated cells compared
with the control cells. The RT-qPCR results confirmed the trend in
changes of gene expression identified using microarray analysis,
although the fold-differences detected were different between the
two analyses.
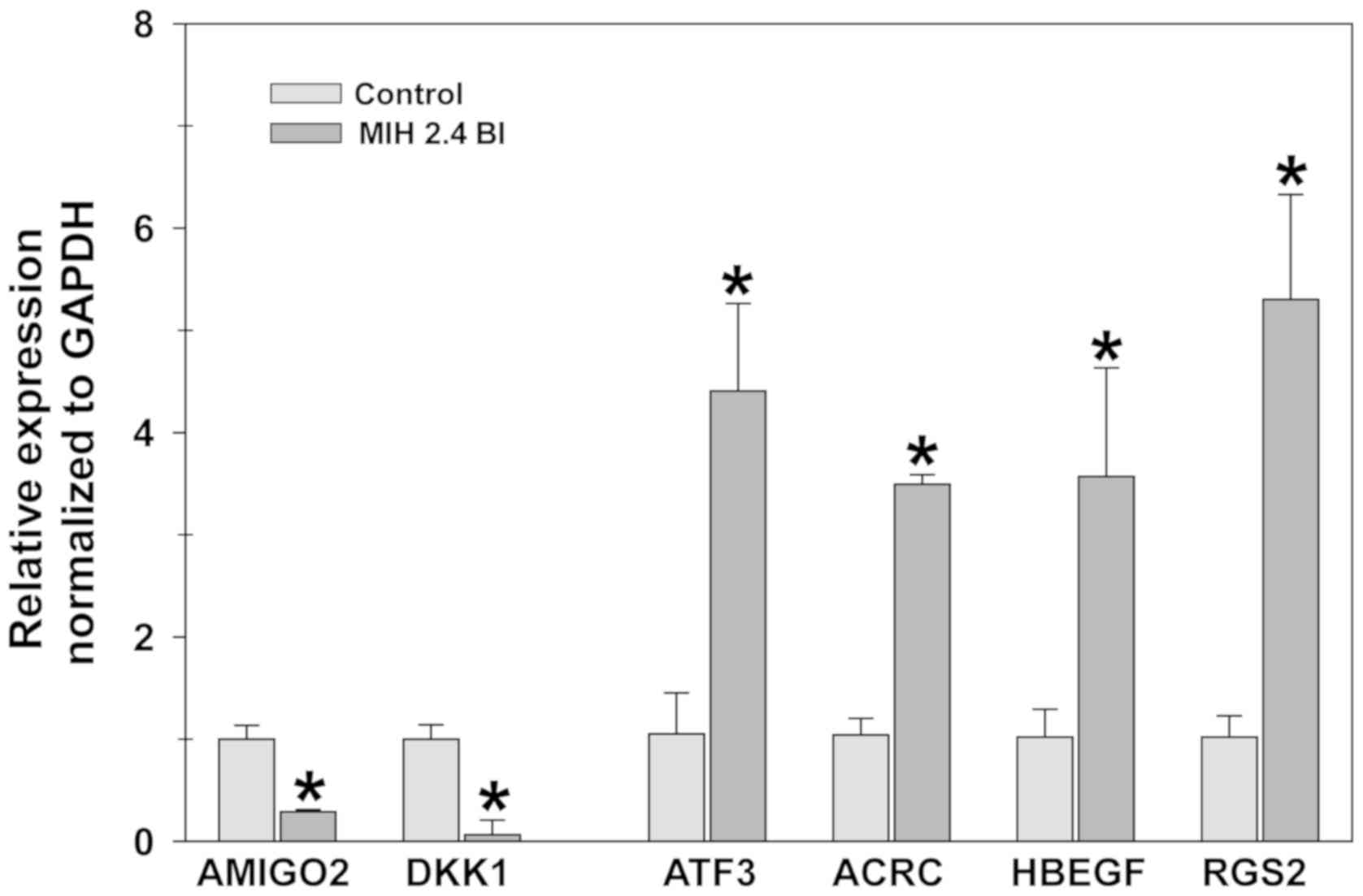 | Figure 5.Effect of MIH 2.4Bl on the mRNA
expression levels of six differentially expressed genes in MCF-7
cells. Relative expression levels of ACRC, HBEGF, ATF3, RGS2, DKK1
and AMIGO2 were determined in control and MIH 2.4Bl treated cells.
Data were calculated as ΔΔCq values based on Cq
expression levels in MIH 2.4Bl treated cells compared with control
cells, and normalized to GAPDH expression. Each data point
represents the mean ± the standard error of the mean of three
replicates. *P<0.05. ACRC, acidic repeat-containing protein;
HBEGF, heparin-binding EGF-like growth factor; ATF3, activating
transcription factor 3; RGS2, Regulator of G-protein signaling 2;
DKK1, Dickkopf WNT signaling pathway inhibitor 1; AMIGO2, adhesion
molecule with Ig like domain 2. |
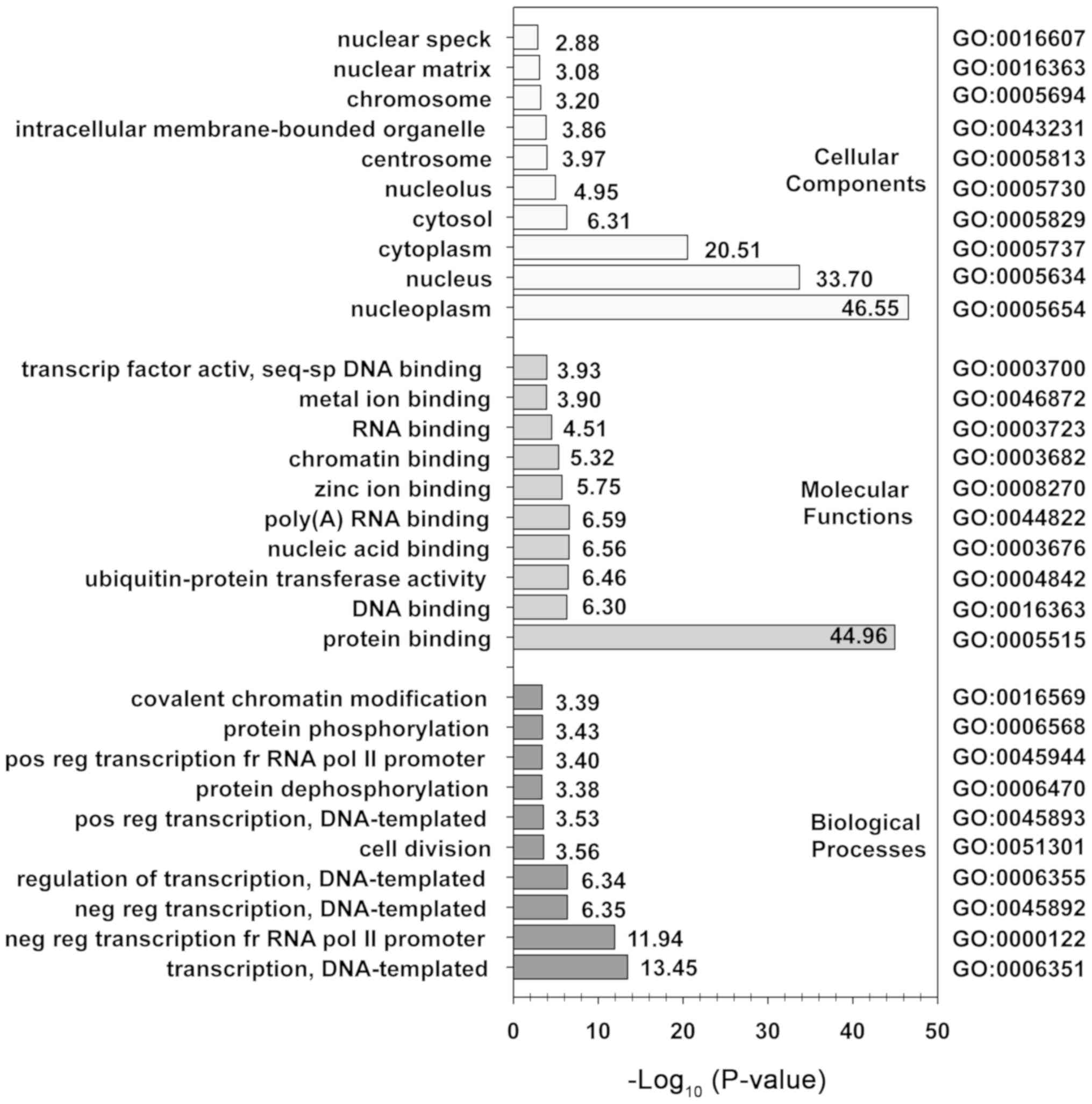
To gain further insight into the pattern of gene
expression changes following MIH 2.4Bl treatment, GO ontology
analysis and KEGG pathway analysis were performed using DAVID. Of
the 3,659 DEGs identified from the microarray analysis, 2,643 genes
were analyzed further using DAVID; 290 were unable to be mapped
with HUGO Gene Nomenclature Committee gene symbols, and 726 were
duplicate genes that were ignored in favor of lower adjusted
P-values. As shown in Fig. 6, the
biological processes of the DEGs were primarily enriched
DNA-template transcription as well as positive and negative
regulation of transcription. The cellular components of the DEGs
were primarily enriched in cytoplasm, nucleoplasm, and nucleus
components. The molecular functions of the DEGs were identified to
be primarily enriched in protein binding. The results of KEGG
analysis showed 12 pathways were significantly enriched by the DEGs
(Table III). Of these pathways,
three involved mechanisms regulating cell signaling (hsa04068,
hsa04550, and hsa04350), and two involved pathways regulating
cancer (hsa05202 and hsa05211). The most significant pathway
mediating FoxO signaling (hsa04068) serves a role in regulating a
wide variety of cellular processes, including cell cycle,
apoptosis, regulation of autophagy, and oxidative stress
resistance.
 | Table III.KEGG pathways with significant
enrichment of differentially expressed genes in MCF-7 cells treated
with MIH 2.4Bl. |
Table III.
KEGG pathways with significant
enrichment of differentially expressed genes in MCF-7 cells treated
with MIH 2.4Bl.
| Pathway name | KEGG | P-value | Hit | Total genes | Percent |
|---|
| FoxO signaling
pathway | hsa04068 | 0.0037 | 35 | 134 | 26.1 |
| Signaling pathways
regulating pluripotency of stem cells | hsa04550 | 0.0121 | 34 | 140 | 24.3 |
| Adherens
junction | hsa04520 | 0.0161 | 21 | 71 | 29.6 |
| RNA
degradation | hsa03018 | 0.0138 | 22 | 77 | 28.6 |
| TGF-β signaling
pathway | hsa04350 | 0.0149 | 23 | 84 | 27.4 |
| Ubiquitin mediated
proteolysis | hsa04120 | 0.0143 | 32 | 137 | 23.4 |
| Protein processing
in endoplasmic reticulum | hsa04141 | 0.0147 | 37 | 169 | 21.9 |
| Insulin
resistance | hsa04931 | 0.0279 | 26 | 108 | 24.1 |
| Lysine
degradation | hsa00310 | 0.0257 | 16 | 52 | 30.8 |
| Transcriptional
misregulation in cancer | hsa05202 | 0.0341 | 35 | 167 | 21.0 |
| Endocytosis | hsa04144 | 0.0376 | 46 | 241 | 19.1 |
| Renal cell
carcinoma | hsa05211 | 0.0361 | 18 | 66 | 27.3 |
Discussion
Despite the existence of a considerable number of
drugs for the treatment of cancer, in several cases, therapeutic
success is not achieved due to treatment failures, resulting in
high relapse rates, poor patient survival, or adverse effects.
These outcomes necessitate a continuous search for novel more
efficacious treatments with reduced side effects (37). Investigators have focused on studying
the potential uses of mesoionic compounds due to of their unique
structure and reaction properties, which make their biological
activities particularly amenable to pharmaceutical use (38,39). The
synthesis of a new mesoionic compound, MIH 2.4Bl has been described
in our previous study (4), and the
selective cytotoxicity against the MCF-7 breast cancer cells was
demonstrated in the present study.
A critical step in pharmaceutical development is to
identify the direct targets of potential drug candidates (40). Mitochondria function as energy
producers of energy in the form of ATP and as a result, have
received attention as a potential target for novel cancer therapies
(20). During physiological cell
function, mitochondria produce ATP via oxidative phosphorylation,
which constitutes the primary mechanism underlying the accumulation
of (ROS. Due to the rapid proliferation rate of cancer cells,
mitochondrial function in ROS production and oxidative stress
serves a definitive role in carcinogenesis (41). Importantly, the dramatic reduction of
mitochondrial respiration we observed in MCF-7 cells treated with
MIH 2.4Bl in the present study may be associated with mitochondrial
dysfunction resulting in ROS accumulation (42).
Autophagy is a fundamental mechanism of cellular
homeostasis, initiated to protect cells by promoting turnover of
organelles and proteins (43).
Autophagy is also associated with the induction of programmed cell
death (44), and also contributes to
functions as a tumor suppressor by degrading damaged mitochondria
that would otherwise contribute to ROS accumulation (45). In the present study, it was
demonstrated that the mesoionic compound MIH2.4 Bl induced the
expression of two autophagy-related proteins (Beclin-1 and ATG5),
which serve an important role in the autophagic process (46) in response to stress caused by
mitochondrial dysfunction. During the formation of autophagosomes,
ATG5 is required for the conversion of LC3B-I to LC3B-II, a
participant in autophagosome membrane expansion and fusion
(47). Beclin 1 has been identified
as a mammalian protein associated with autophagy and involved in
cancer processes such as tumor suppression and cell death (48). Cheng et al (49) identified the role of autophagy in
MCF-7 cells treated with Icariin (a flavonoid isolated from plants)
by measuring the expression levels of ATG5, Beclin 1 and LC3-I to
LC3-II proteins. Liang et al (50) demonstrated that exogenous Beclin 1
expression in MCF-7 cells resulted in decreased cell division and
inhibition of tumorigenesis in a mouse model. Given the results of
the present study, MIH2.4Bl may have also induced autophagy in
MCF-7 cells.
The results of mitochondrial respiration suggest a
possible induction of cellular cytotoxicity by mitochondrial
dysfunction induced by MIH 2.4Bl treatment. In support of these
results, studies using the mesoionic compound SYD-1 in isolated rat
liver mitochondria showed a functional decrease in electron
transport and oxidative phosphorylation efficiency (19). Another mesoionic compound, MI-D, was
also shown to act as an uncoupler of the respiratory chain between
complexes II and III (9).
Mechanistic studies demonstrated that alteration of membrane
fluidity and elasticity were associated with the disruption of
mitochondrial function by mesoionic compounds (10). Interestingly, altered mitochondrial
metabolism has been suggested as an approach to overcome the
resistance to apoptotic cell death observed in cancer cells
(51).
Four genes that were shown to be highly upregulated
and two genes that were highly downregulated based on the
microarray data were selected for verification by RT-qPCR. Of the
genes selected, ACRC (also known as germ cell nuclear acidic
peptidase), is localized in the nucleus and serves a role in
chromatin structure (52). ATF3 is a
member of the mammalian ATF/CREB family of transcription factors
that modulate TGFβ signaling in breast cancer (53). HBEGF is synthesized as a
membrane-anchored glycoprotein that serves a role as a growth
factor in breast cancer metastasis (54). RGS2 is a member of a family of genes
that act as GTPase activating proteins for G proteins and may have
an inhibitory effect in breast cancer (55). AMIGO2 is a member of a family of cell
surface transmembrane proteins that function in cell adhesion and
serves a survival role in cancer growth (56). DKK1 is a member of the Dickkopf
family of proteins that are characterized by two cysteine-rich
domains functioning to interact with the LRP6 co-receptor resulting
in disruption of Wnt signaling (57). DKK1 has been suggested to have
differential effects on lung and bone metastasis in breast cancer
(58).
In conclusion, it was demonstrated that the
mesoionic compound MIH 2.4Bl exhibits cytotoxic activity against
MCF-7 breast cancer cells and reduced cell growth. Analysis of the
functional effects of MIH 2.4Bl support a mechanism of action
mediated by the induction of mitochondrial dysfunction and
autophagy. Additional studies are required for elucidation of the
specific mechanistic pathways involved in regulating autophagy. In
particular, further studies are necessary to understand the
specific role of different genes and pathways regulated by MIH
2,4Bl. Nonetheless, these studies confirm the potential therapeutic
use of MIH 2.4Bl for treatment of breast cancer.
Supplementary Material
Supporting Data
Acknowledgments
The authors would like to thank Ms. Paula Polk in
the Research Core Facility at LSU Health Shreveport for her
technical assistance.
Funding
The present study was supported by Coordenação de
Aperfeiçoamento de Pessoal de Nível Superior, through an
international scholarship awarded through the Science without
Borders program (grant no. 88887.122971/2016-00), and through
research bridge funding provided by the Department of Cellular
Biology and Anatomy at LSU School of Veterinary Medicine.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LAMC performed the experiments and analyzed and
interpreted the data. DD assisted in designing and performing the
cytotoxicity and OCR experiments and in interpreting the results.
ACH assisted in designing and performing the autophagy experiments
and in interpreting the results. SSA assisted in the statistical
analysis of the data and in interpretation of the results. HDSS
synthesized the mesoionic compound used in the experiments. PFAF
directed the synthesis of the mesoionic compound used in the
experiments and interpreted the analysis of the results. AW and
MAGF assisted in designing the study and oversaw the analysis and
interpretation of the data and wrote the manuscript. JMM directed
the project, designed the study, oversaw the analysis and
interpretation of the data, and assisted in writing the manuscript.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
World Health Organization: WHO report on
cancer: Setting priorities, investing wisely and providing care for
all World Health Organization, 2020.
|
|
2
|
Harbeck N, Penault-Llorca F, Cortes J,
Gnant M, Houssami N, Poortmans P, Ruddy K, Tsang J and Cardoso F:
Breast cancer. Nat Rev Dis Primers. 5:662019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Santini D, Vincenzi B, Galluzzo S,
Battistoni F, Rocci L, Venditti O, Schiavon G, Angeletti S, Uzzalli
F, Caraglia M and Dicuonzo G: Repeated intermittent low-dose
therapy with zoledronic acid induces an early, sustained, and
long-lasting decrease of peripheral vascular endothelial growth
factor levels in cancer patients. Clin Cancer Res. 13:4482–4486.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bhosale SK, Deshpande SR, Wagh RD and
Dhake AS: Biological activities of 1, 2, 3-oxadiazolium-5-olate
derivatives. Der Chem Sin. 6:79–95. 2015.
|
|
5
|
Badami BV: Mesoionic compounds. Resonance.
11:40–48. 2006. View Article : Google Scholar
|
|
6
|
Kier LB and Roche EB: Medicinal chemistry
of the mesoionic compounds. J Pharm Sci. 56:149–168. 1967.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Senff-Ribeiro A, Echevarria A, Silva EF,
Sanches Veiga S and Oliveira MB: Effect of a new
1,3,4-thiadiazolium mesoionic compound (MI-D) on B16-F10 murine
melanoma. Melanoma Res. 13:465–471. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Senff-Ribeiro A, Echevarria A, Silva EF,
Franco CR, Veiga SS and Oliveira MB: Cytotoxic effect of a new
1,3,4-thiadiazolium mesoionic compound (MI-D) on cell lines of
human melanoma. Br J Cancer. 91:297–304. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cadena SMSC, Carnieri EGS, Echevarria A
and de Oliveira MBM: Effect of MI-D, a new mesoionic compound, on
energy-linked functions of rat liver mitochondria. FEBS Lett.
440:46–50. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cadena SMSC, Carnieri EGS, Echevarria A
and de Oliveira MBM: Interference of MI-D, a new mesoionic
compound, on artificial and native membranes. Cell Biochem Funct.
20:31–37. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dunkley CS and Thoman CJ: Synthesis and
biological evaluation of a novel phenyl substituted sydnone series
as potential antitumor agents. Bioorg Med Chem Lett. 13:2899–2901.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gozzi GJ, Pires Ado R, Martinez GR, Rocha
ME, Noleto GR, Echevarria A, Canuto AV and Cadena SM: The
antioxidant effect of the mesoionic compound SYD-1 in mitochondria.
Chem Biol Interact. 205:181–187. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Galuppo LF, dos Reis Lívero FA, Martins
GG, Cardoso CC, Beltrame OC, Klassen LMB, Canuto AV, Echevarria A,
Telles JE, Klassen G and Acco A: Sydnone 1: A mesoionic compound
with antitumoral and haematological effects in vivo. Basic Clin
Pharmacol Toxicol. 119:41–50. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Amaral de Mascena Costa L, Cássio Silva de
Lima F, da Silva Viana R, de Sousa Araújo S, Wischral A, Diógenes
da Silva Souzad H, Filgueiras de Athayde Filhod P, Araújo de
Azevedo L, Alves Junior S, Adrião Gomes Filho M and Mathis JM:
Abstract 5877: Antitumor activity of the mesoionic compound MI H
2.4 on breast cancer cell lines. Cancer Res. 78 (Suppl
13):2018.
|
|
15
|
Lee AV, Oesterreich S and Davidson NE:
MCF-7 cells-changing the course of breast cancer research and care
for 45 years. J Natl Cancer Inst. 107:djv0732015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Comşa Ş, Cîmpean AM and Raica M: The story
of MCF-7 breast cancer cell line: 40 years of experience in
research. Anticancer Res. 35:3147–3154. 2015.PubMed/NCBI
|
|
17
|
Anvar SY, Allard G, Tseng E, Sheynkman GM,
de Klerk E, Vermaat M, Yin RH, Johansson HE, Ariyurek Y, den Dunnen
JT and Turner SW: Full-length mRNA sequencing uncovers a widespread
coupling between transcription initiation and mRNA processing.
Genome Biol. 19:462018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chiang YS, Huang YF, Midha MK, Chen TH,
Shiau HC and Chiu KP: Single cell transcriptome analysis of MCF-7
reveals consistently and inconsistently expressed gene groups each
associated with distinct cellular localization and functions. PLoS
One. 13:e01994712018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Halila GC, de Oliveira MB, Echevarria A,
Belém AC, Rocha ME, Carnieri EG, Martinez GR, Noleto GR and Cadena
SM: Effect of sydnone SYD-1, a mesoionic compound, on energy-linked
functions of rat liver mitochondria. Chem Biol Interact.
169:160–170. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Weinberg SE and Chandel NS: Targeting
mitochondria metabolism for cancer therapy. Nat Chem Biol. 11:9–15.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kubli DA and Gustafsson ÅB: Mitochondria
and mitophagy: The yin and yang of cell death control. Circ Res.
111:1208–1221. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lira BF, de Athayde Filho PF, Miller J,
Simas AM, de Farias Dias A and Vieira MJ: Synthesis and
characterization of some new mesoionic 1, 3-thiazolium-5-thiolates
via cyclodehydration and in situ 1, 3-dipolar
Cycloaddition/cycloreversion. Molecules. 7:791–800. 2002.
View Article : Google Scholar
|
|
23
|
Feoktistova M, Geserick P and Leverkus M:
Crystal violet assay for determining viability of cultured cells.
Cold Spring Harbor Protoc. 2016:pdb–rot087379. 2016. View Article : Google Scholar
|
|
24
|
Tsou SH, Chen TM, Hsiao HT and Chen YH: A
critical dose of doxorubicin is required to alter the gene
expression profiles in MCF-7 cells acquiring multidrug resistance.
PLoS One. 10:e01167472015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jokar F, Mahabadi JA, Salimian M, Taherian
A, Hayat SMG, Sahebkar A and Atlasi MA: Differential expression of
HSP90β in MDA-MB-231 and MCF-7 cell lines after treatment with
doxorubicin. J Pharmacopuncture. 22:28–34. 2019.PubMed/NCBI
|
|
26
|
Fornari FA, Randolph JK, Yalowich JC,
Ritke MK and Gewirtz DA: Interference by doxorubicin with DNA
unwinding in MCF-7 breast tumor cells. Mol Pharmacol. 45:649–656.
1994.PubMed/NCBI
|
|
27
|
Huang DW, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang DW, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT
and Harris MA: Gene ontology: Tool for the unification of biology.
Nat Genet. 25:25–29. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
The Gene Ontology Consortium, . The gene
ontology resource: 20 years and still GOing strong. Nucleic Acids
Res. 47:D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kanehisa M: Post-genome informatics.
Oxford University Press. (Oxford). 2000.
|
|
32
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc Series B (Methodological).
57:289–300. 1995. View Article : Google Scholar
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kabeya Y, Mizushima N, Ueno T, Yamamoto A,
Kirisako T, Noda T, Kominami E, Ohsumi Y and Yoshimori T: LC3, a
mammalian homologue of yeast Apg8p, is localized in autophagosome
membranes after processing. EMBO J. 19:5720–5728. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sica V, Bravo-San Pedro JM, Stoll G and
Kroemer G: Oxidative phosphorylation as a potential therapeutic
target for cancer therapy. Int J Cancer. 146:10–17. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Brand MD and Nicholls DG: Assessing
mitochondrial dysfunction in cells. Biochem J. 435:297–312. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tang Y, Wang Y, Kiani MF and Wang B:
Classification, treatment strategy, and associated drug resistance
in breast cancer. Clin Br Cancer. 16:335–343. 2016. View Article : Google Scholar
|
|
38
|
Kaur G and Singh R: Thiadiazole analogs as
potential pharmacological agents: A brief review. Int J Pharm Sci.
6:35–46. 2014.
|
|
39
|
Abdualkader AM, Taher MU and Yusoff NI:
Mesoionic sydnones. A review in their chemical and biological
properties. Int J Pharm Pharm Sci. 9:1–9. 2017. View Article : Google Scholar
|
|
40
|
Rogers GW, Burroughs SE and Dranka BP:
Direct measurements of cellular metabolism for identification of
mitochondrial drug targets. Agilent Technologies. 14–Nov;2018.
|
|
41
|
Czupiela PP, Delplace V and Shoicheta MS:
Cationic block amphiphiles show anti-mitochondrial activity in
multi-drug resistant breast cancer cells. J Control Realeas.
305:210–219. 2019. View Article : Google Scholar
|
|
42
|
Storz P: Reactive oxygen species in tumor
progression. Front Biosci. 10:1881–1896. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mizushima N, Levine B, Cuervo AM and
Klionsky DJ: Autophagy fights disease through cellular
self-digestion. Nature. 451:1069–1075. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pampliega O, Orhon I, Patel B, Sridhar S,
Dıaz-Carretero A, Beau I, Codogno P, Satir BH, Satir P and Cuervo
AM: Functional interaction between autophagy and ciliogenesis.
Nature. 502:194–200. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Matsuda N, Sato S, Shiba K, Okatsu K,
Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F, et al:
PINK1 stabilized by mitochondrial depolarization recruits Parkin to
damaged mitochondria and activates latent Parkin for mitophagy. J
Cell Biol. 189:211–221. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Akar U, Chaves-Reyez A, Barria M, Tari A,
Sanguino A, Kondo Y, Kondo S, Arun B, Lopez-Berestein G and Ozpolat
B: Silencing of Bcl-2 expression by small interfering RNA induces
autophagic cell death in MCF-7 breast cancer cells. Autophagy.
4:669–679. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Nakagawa I, Amano A, Mizushima N, Yamamoto
A, Yamaguchi H and Kamimoto T: Autophagy defends cells against
invading group A Streptococcus. Science. 306:1037–1040. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
He C and Levine B: The Beclin 1
interactome. Curr Opin Cell Biol. 22:140–149. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Cheng X, Tan S, Duan F, Yuan Q, Li Q and
Deng G: Icariin induces apoptosis by suppressing autophagy in
Tamoxifen-resistant breast cancer cell line MCF-7/TAM Breast
Cancer. 26:766–775. 2019.PubMed/NCBI
|
|
50
|
Liang XH, Jackson S, Seaman M, Brown K,
Kempkes B, Hibshoosh H and Levine B: Induction of autophagy and
inhibition of tumorigenesis by beclin 1. Nature. 402:672–676. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Vyas S, Zaganjor E and Haigis MC:
Mitochondria and cancer. Cell. 166:555–566. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bhargava V, Goldstein CD, Russell L, Xu L,
Ahmed M, Li W, Casey A, Servage K, Kollipara R, Picciarelli Z and
Kittler R: GCNA preserves genome integrity and fertility across
species. Dev Cell. 52:38–52. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yin X, Wolford CC, Chang YS, McConoughey
SJ, Ramsey SA, Aderem A and Hai T: ATF3, an adaptive-response gene,
enhances TGFβ signaling and cancer-initiating cell features in
breast cancer cells. J Cell Sci. 123:3558–3565. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhou ZN, Sharma VP, Beaty BT, Roh-Johnson
M, Peterson EA, Van Rooijen N, Kenny PA, Wiley HS, Condeelis JS and
Segall JE: Autocrine HBEGF expression promotes breast cancer
intravasation, metastasis and macrophage-independent invasion in
vivo. Oncogene. 33:3784–3793. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lyu JH, Park DW, Huang B, Kang SH, Lee SJ,
Lee C, Bae YS, Lee JG and Baek SH: RGS2 suppresses breast cancer
cell growth via a MCPIP1-dependent pathway. J Cell Biochem.
116:260–267. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Fontanals-Cirera B, Hasson D, Vardabasso
C, Di Micco R, Agrawal P, Chowdhury A, Gantz M, de
Pablos-Aragoneses A, Morgenstern A, Wu P, et al: Harnessing BET
inhibitor sensitivity reveals AMIGO2 as a melanoma survival gene.
Mol Cell. 68:731–744.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Niida A, Hiroko T, Kasai M, Furukawa Y,
Nakamura Y, Suzuki Y, Sugano S and Akiyama T: DKK1, a negative
regulator of Wnt signaling, is a target of the beta-catenin/TCF
pathway. Oncogene. 23:8520–8526. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhuang X, Zhang H, Li X, Li X, Cong M,
Peng F, Yu J, Zhang X, Yang Q and Hu G: Differential effects on
lung and bone metastasis of breast cancer by Wnt signalling
inhibitor DKK1. Nat Cell Biol. 19:1274–1285. 2017. View Article : Google Scholar : PubMed/NCBI
|